3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2010; 1:70-79. doi:10.7150/jca.1.70 This volume Cite
Research Paper
Protein Profiling of Isolated Leukocytes, Myofibroblasts, Epithelial, Basal, and Endothelial Cells from Normal, Hyperplastic, Cancerous, and Inflammatory Human Prostate Tissues
1. Department of Chemistry and Biochemistry and Institute of Molecular Biophysics, Florida State University, Tallahassee, FL 32306, USA;
2. Department of Pathology, University of Colorado Health Science Center, Aurora, CO 80045, USA
Published 2010-6-15
Abstract
In situ neoplastic prostate cells are not lethal unless they become invasive and metastatic. For cells to become invasive, the prostate gland must undergo degradation of the basement membrane and disruption of the basal cell layer underneath the luminal epithelia. Although the roles of proteinases in breaking down the basement membrane have been well-studied, little is known about the factors that induce basal cell layer disruption, degeneration, and its eventual disappearance in invasive cancer. It is hypothesized that microenvironmental factors may affect the degradation of the basal cell layer, which if protected may prevent tumor progression and invasion. In this study, we have revealed differential protein expression patterns between epithelial and stromal cells isolated from different prostate pathologies and identified several important epithelial and stromal proteins that may contribute to inflammation and malignant transformation of human benign prostate tissues to cancerous tissues using matrix-assisted laser desorption ionization time-of-flight mass spectrometry and proteomics methods. Cellular retinoic acid-binding protein 2 was downregulated in basal cells of benign prostate. Caspase-1 and interleukin-18 receptor 1 were highly expressed in leukocytes of prostate cancer. Proto-oncogene Wnt-3 was downregulated in endothelial cells of prostatitis tissue and tyrosine phosphatase non receptor type 1 was only found in normal and benign endothelial cells. Poly ADP-ribose polymerase 14 was downregulated in myofibroblasts of prostatitis tissue. Interestingly, integrin alpha-6 was upregulated in epithelial cells but not detected in myofibroblasts of prostate cancer. Further validation of these proteins may generate new strategies for the prevention of basal cell layer disruption and subsequent cancer invasion.
Keywords: Prostate cancer, tumor microenvironment, inflammation, tumorigenesis, protein biomarkers, magnetic bead cell separation
Introduction
Prostate cancer is the most common non-skin malignancy in men and the second leading cause of cancer-death in the United States. It develops in a series of morphologic and genetic steps that begins with the transformation of normal tissue to hyperplastic lesions, and continues to high-grade prostatic intraepithelial neoplasia (HGPIN), invasion and metastasis.
In addition to neoplastic epithelial cells, tumors also contain a highly complex microenvironment composed of a mixture of fibroblasts, myofibroblasts, endothelial cells, inflammatory cells and extracellular matrix. As tumor progresses, carcinoma cells proliferate and invade the surrounding stroma through the basement membrane. As a response to the disruption of normal tissue homeostasis, the stroma will undergo several modifications creating new stromal microenvironment termed 'reactive' stroma which differs from the stroma in normal tissue and can promote tumorigenesis 1. These modifications include proliferation of fibroblasts 2, lymphocyte infiltration 3, extracellular matrix remodeling and increased angiogenesis 1.
Transformed carcinoma cells which result from accumulation of genetic mutations have been the focus of cancer research for many years and they were claimed to be the trigger of malignant phenotype. However, this epithelial cell-driven tumorigenesis viewed the surrounding stroma as a passive support structure and neglected its contribution in the malignant transformation. Historically, Paget recognized the importance of tumor microenvironment in cancer progression and proposed the “seed and soil” hypothesis in 1889 4. In this hypothesis, Paget pointed out the importance of the interaction between tumor cells and microenvironment in oncogenesis and cancer progression. Recent studies provided insight into this crosstalk and identified key role of stromal cells in many steps of tumor development and invasion 5,6. It is now understood that microenvironmental interactions in carcinomas are crucial in cancer progression. Carcinoma associated fibroblasts (CAFs) enhanced tumor progression and transformation of nontumorigenic, immortalized human prostatic cell line 2. Another principal component of reactive stroma is tumor associated macrophages. A localized increase of leukocyte infiltration was observed in malignant prostate tissue as compared to adjacent benign areas 3,7,8. A global expression analysis of reactive stroma in prostate cancer shows differential gene expression pattern in tumor associated stroma versus normal stroma 9,10. Taken together, these studies underscore the stromal-epithelial interface as a critical mediator of oncogenic potential. While stromal cells are influenced by the adjacent epithelium via paracrine mechanisms, they are not just responding to these stimuli but also actively affecting the fate of adjacent epithelium. These data demonstrate that targeting the tumor microenvironment can be a feasible therapeutic strategy.
Almost all current cancer therapies target the tumor epithelial cells from which prostate carcinomas are thought to arise. However, the genetic instability of epithelial cells caused them to acquire resistance to most therapies. Now, extensive efforts are dedicated to explore therapies that target more genetically stable stromal cells that are less likely to develop resistance. Therefore, targeting tumor microenvironment in an attempt to identify novel stromal biomarkers may provide a promising opportunity for prostate cancer prevention and treatment. Toward that end, the present study focuses on proteomic analysis of stromal cells and epithelial cells in normal, benign, cancerous and inflammatory prostate tissues. Different cell types including leukocytes, myofibroblasts, endothelial cells, basal cells and epithelial cells are isolated and profiled. The identification of some molecular markers may help to understand the molecular mechanisms that govern the initiation of invasion.
Materials and Methods
Tissue preparation
Fresh frozen prostate tumor specimens were collected from our collaborator Dr. Kenneth Iczkowski. The pathologies of the prostate tissues used are normal, benign prostatic hyperplasia (BPH), prostatitis, and prostate cancer. Collectively, the tissues were minced and cell suspension was prepared as described in Allinen et al. 11. Briefly, tissues were digested into 1-2 mm pieces, incubated in 5 x volume of DMEM that contains 2 mg/mL collagenase I and 2 mg/mL hyaluronidase at 37 ºC for 2 hours. Tubes were then centrifuged at 3000 x g for 10 minutes. The supernatants were discarded. Pellets were washed with 1 x HBSS. Tubes were then centrifuged at 3000 x g for 10 minutes. Supernatant were discarded and pellets were collected. Pellets were then incubated in 10 x volume of trypsin/EDTA at 37 ºC for 5 minutes. Ice cold medium including 10% FBS was then added to the mixture as needed to filter the pellets. The mixture was then filtered through a 100 μm than 40 μm and then a 20 μm filter mesh strainer. Flow through was then centrifuged at 3000 x g for 5 minutes. Cell mixture was then collected, washed with HBSS, centrifuged at 3000 x g for 5 minutes. Cells were collected and resuspended in 200 μL/ 1 million cells with PBE.
Cell separation
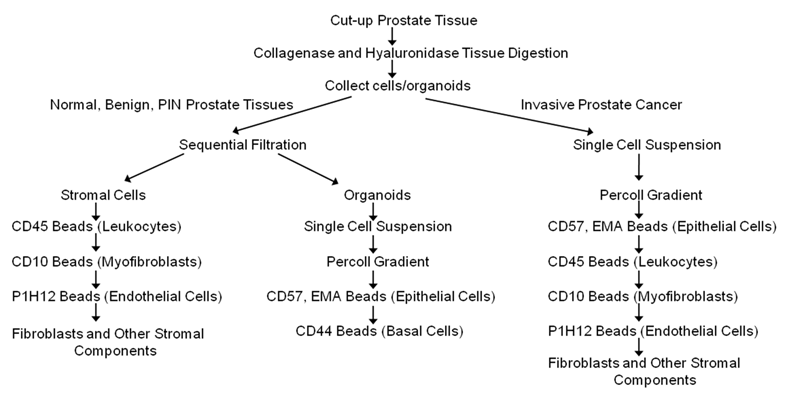
Cells were separated according to the diagram in Figure 1 using dynal beads. The beads are attached to antibodies specific to surface marker of the cells to be separated. Following the manufacturer's instructions, 100 μl of the dynal beads are added to the cell suspension, incubated for 20 minutes at 4 ºC with gentle tilting and rotation and placed on the magnet for 2 minutes. The bead bound cells are washed 3 times by resuspending in PBE buffer to remove contaminating cells, and then the captured cells are frozen immediately on dry ice. Unbound cells are collected by centrifugation and reconstituted in PBE buffer. Epithelial enrich dynabeads (Cat # 161.02, Invitrogen, Carlsbad, CA), CD31 beads (Cat # 111.55D Invitrogen, Carlsbad, CA), and equal mixture of CD45 (Cat # 111.53D, Invitrogen, Carlsbad, CA) and CD15 (Cat # 111.37D, Invitrogen, Carlsbad, CA) beads were used to isolate epithelial cells, endothelial cells and leukocytes respectively. CELLection Biotin Binder dynabeads (Cat # 115.33D, Invitrogen, Carlsbad, CA) were incubated for 2 hours at RT with CD44 (Cat # CL89153B, Cedarlane, Burlington, NC) or CD10 biotinylated antibodies (Cat # 13-0108-82, eBioscience, San Diego, CA) to isolate basal cells or myofibroblasts respectively.
A schematic diagram/flow chart for the isolation of epithelial cells, basal cells, endothelial cells, leukocytes, and other types of cells from normal, benign, cancerous and chronic inflammatory human prostate tissues using magnetic beads conjugated to antibodies targeting specific membrane cell markers (Adapted from Allinen et al. 23).
Protein extraction and quantification
Cells were lysed by adding cell lysis buffer (30 mM Tris, 7 M Urea, 4% CHAPS, protease inhibitor), followed by vortexing for 1 hour then centrifugation for 15 min at 15000g. The supernatant was used for protein quantification. Microplate bicinchoninic acid (BCA) Protein assay kit (Pierce, Rockford, IL) was used to determine the protein concentration using bovine serum albumin as a standard according to manufacturer's instructions. A triplicate of each sample was utilized and the mean absorbance was used to calculate the concentration of the protein using the standard curve.
Two-dimensional gel electrophoresis
Two-dimensional gel electrophoresis was performed as described previously 12. Briefly, a total of 50 µg of proteins were vacuum-dried and reconstituted in 200 µL of rehydration buffer (30 mM Tris, 7 M Urea, 4% CHAPS, 50 mM DTT). Isoelectric focusing was run as first dimension using immobilized pH gradient (IPG) strips, pH 4 to 7 (Bio-Rad). This was followed by rehydration of the strips in two equilibration buffers using 2 mL for 15 min each. The first buffer constituted of 375 mM Tris-HCL pH 8.8, 6 M urea, 2% SDS and 2% DTT. The second buffer was composed of 375 mM Tris-HCL pH 8.8, 6 M urea, 2% SDS and 2% iodoacetamide. Proteins were then separated using Polyacrylamide Gel Electrophoresis (PAGE) where the strips were placed on 10% polyacrylamide gel and allowed to electrophorese at 50 V for 30 min then at 100 V till the end of separation. The cathode buffer used consisted of 0.1 M Tricine, 0.1 M Tris-HCl pH 8.2, and 0.1% SDS; whereas the anode buffer was 0.2 M Tris-HCl pH 8.9. Gels were fixed in 40% ethanol and 10% acetic acid and stained with SYPRO Ruby (Bio-Rad) for 3 hours at room temperature. Gels were destained in 10% methanol and 7% acetic acid to decrease background staining followed by washing 3 times in ddH2O. Typhoon 9410 Scanning Systems (GE Healthcare) was used to scan the gels. The excitation wavelength was 457 nm and the signals were detected at 550 V.
Trypsin Digestion and Matrix-assisted Laser Desorption Ionization Time-of-flight Mass Spectrometry (MALDI-TOF MS)
Protein digestion was performed as described previously 13. Briefly, protein spots from 2DE gels were excised under UV light, combined, and incubated with trypsin overnight (Cat # T7575, Sigma Aldrich). Peptides generated from the trypsin digestion were then mixed with a matrix (α-cyano-hydroxycinnamic acid) 14,15 at a 100-1000 fold molar excess over the analyte 16. This mixture was then spotted onto a target plate and allowed to evaporate until crystals were formed. Spectra were collected in reflectron positive ion mode over the mass range of 500 to 3000 Da. MASCOT software was used to identify the proteins. The search was performed against the SwissProt database for the Homo sapiens taxonomy. The following parameters were used: one missed cleavage, mass tolerance of 1.3 Da, oxidation of methionine (variable modification), and carbamidomethylation (fixed modification).
Densitometric analysis
The density of the spots was measured using ImageJ Java-based software developed at National Institutes of Health (NIH) and available on the internet (http://rsb.info.nih.gov/nih-image/). It was based on the area under the curve (AUC) from rectangular image captures. Fold change was calculated as the ratio of the area of malignant state to that of normal state. Fold regulation was then measured as the following: if fold change >1, it remained unchanged; if fold change <1, the negative inverse of it was calculated to show downregulation.
Results
Differential protein expression in different prostate tissue pathologies
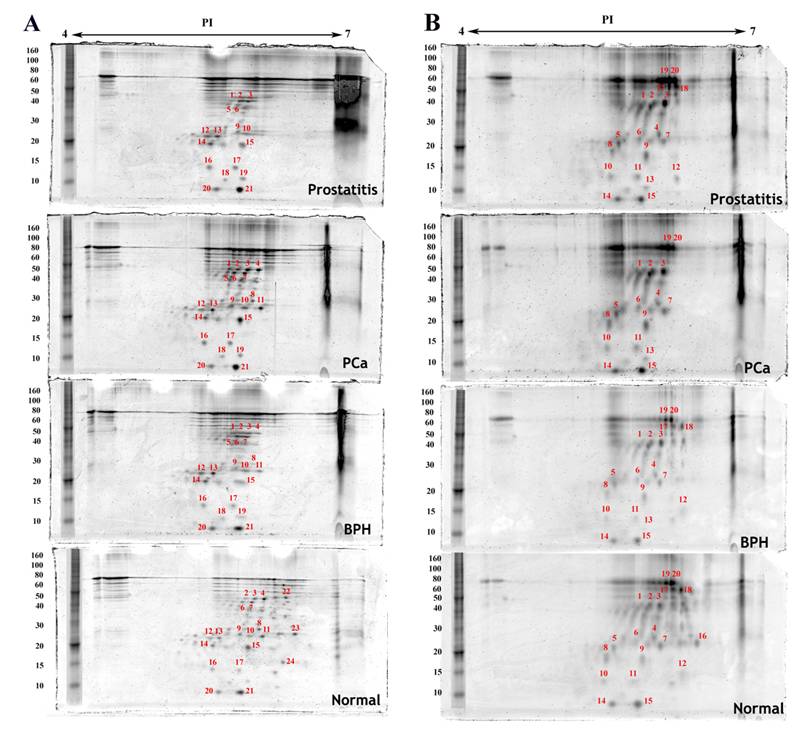
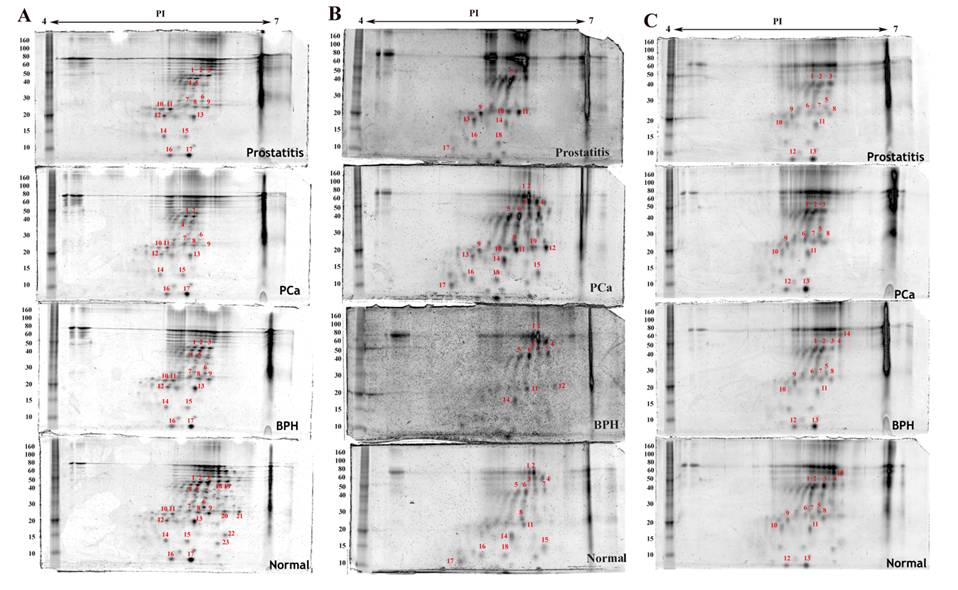
Densitometric analysis of proteins expressed in the same cell type in normal, benign, cancerous and prostatitis tissues shows different levels of expression when compared to normal tissue expression levels (Table 1). Some proteins are upregulated, others are downregulated. Moreover, few proteins are found to be exclusively expressed either in the normal tissue or in the malignant one (Figure 2 and Figure 3). For example, four proteins are upregulated and ten are downregulated in endothelial cells of the hyperplastic tissue compared to the normal one. In prostate cancer, seven spots are overexpressed and four are downexpressed, while two spots were not detected. In prostatitis, five spots are upregulated, seven spots are downregulated, and two proteins were not detected (Figure 3C).
Identification of some putative protein biomarkers
MALDI-TOF analysis followed by Mascot search resulted in the identification of a subset of proteins in different cell types (Table 2). In leukocytes, we were able to identify GTP-binding protein (GTPB1), junctophilin, leucine-rich repeat serine/threonine-protein kinase (LRRK2), caspase1, interleukin 18 receptor (IL18R), and Rab5 GDP/GTP exchange factor (Rabex5 or RAP1). Proto-oncogene Wnt-3, uncharacterized serine/threonine-protein kinase, tyrosine-protein phosphatase non-receptor type 1(PTPN1 or PTP1B) and Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 1 were found in endothelial cells. Integrin alpha-6 and poly ADP-ribose polymerase 14 were discovered in epithelial cells and myofibroblasts. Cellular retinoic acid-binding protein 2 (CRABP2) was identified in basal cells. Actin-related protein complex was found in all cell types.
Densitometric analysis of spots in Epithelial, Basal cells, Myofibroblasts, Leukocytes and Endothelial cells. The fold change is shown as compared to the normal. † NA: reflects no expression of the spot in the normal tissue sample. BPH, benign prostate hyperplasia. PCa, cancer of prostate. Titis, prostatitis.
| Spot# | Epithelial | Basal | Myofibroblast | Leukocytes | Endothelial | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BPH | PCA | Titis | BPH | PCA | Titis | BPH | PCA | Titis | BPH | PCA | Titis | BPH | PCA | Titis | |
| 1 | NA | NA | NA | 2.2 | 6.85 | 7.1 | 1.92 | 1.79 | -1.64 | 2.16 | 1.54 | 1.76 | -1.18 | 1.30 | 1.01 |
| 2 | 2.01 | 3.26 | 4.07 | -1.2 | 2.63 | 1.76 | 1.37 | 1.09 | -2.21 | 1.83 | 1.06 | 1.36 | 1.02 | 1.17 | -1.04 |
| 3 | 1.03 | 2.39 | 1.60 | -1.24 | 2.04 | 1.79 | -1.10 | 0 | -1.40 | 1.44 | 1.47 | 1.94 | -3.41 | -1.26 | 1.03 |
| 4 | -5.65 | 1.16 | 0 | -4.21 | 1.38 | 1.7 | 1.04 | -1.06 | 1.85 | 1.85 | 2.21 | 0 | -2.13 | 0 | 0 |
| 5 | NA | NA | NA | -1.25 | 1.44 | 1.33 | 1.08 | 0 | 1.29 | 1.28 | 1.75 | 1.73 | -1.52 | -1.85 | -2.46 |
| 6 | 1.91 | 1.83 | 1.67 | -1.13 | 1.09 | -1.12 | -1.63 | -2.09 | -1.57 | 1.41 | 1.64 | 0 | 2.37 | 2.60 | 1.58 |
| 7 | -1.29 | 1.46 | 0 | -2.26 | 1.07 | -1.24 | 1.01 | -1.19 | -1.40 | 1.70 | 2.79 | 0 | 1.62 | 1.29 | -1.55 |
| 8 | -2.51 | 1.08 | 0 | -2 | -1.3 | -1.34 | 1.32 | -1.16 | 1.01 | 0 | 2.35 | 0 | -1.25 | -1.50 | -2.74 |
| 9 | 1.62 | 1.65 | -1.25 | -1.15 | 1.69 | 2.24 | 1.03 | -1.64 | -1.31 | 0 | NA | NA | 1.62 | -1.04 | 1.37 |
| 10 | 1.50 | 2.27 | 1.26 | -5.26 | 1.22 | 1.13 | 2.03 | 1.71 | 1.76 | 0 | NA | NA | -1.32 | -1.15 | -1.38 |
| 11 | 1.15 | 1.29 | 0 | -1.78 | 1.94 | 1.4 | 1.72 | 1.29 | 1.48 | 1.08 | 2.24 | 1.56 | -1.50 | 1.87 | -1.27 |
| 12 | 1.68 | 1.77 | 2.16 | -1.52 | 0 | 2.15 | -1.07 | -1.35 | -1.19 | NA | NA | 0 | -1.44 | 1.12 | -1.3 |
| 13 | 1.51 | 1.88 | 1.88 | -1.56 | 1.02 | 1 | 1.02 | -2.25 | -1.12 | 0 | NA | NA | -1.13 | 1.12 | 1.05 |
| 14 | 1.50 | 1.04 | 2.58 | 1.33 | 1.28 | 1.66 | 1.13 | 1.24 | 1.10 | -2.65 | 2.05 | -1.71 | -1.40 | 0 | 0 |
| 15 | -1.24 | 1.17 | -1.06 | -1.16 | -1.07 | 1.15 | -1.07 | 1.38 | -1.25 | 0 | 2.47 | 0 | |||
| 16 | 1.18 | 1.63 | 1.68 | 0 | 0 | 0 | 1.70 | 1.02 | 1.05 | 0 | 1.15 | -1.29 | |||
| 17 | 1.46 | 1.96 | 2.08 | -3.21 | 1 | -1.44 | -1.04 | -1.01 | 1.04 | 0 | -1.22 | -2.17 | |||
| 18 | NA | NA | NA | -2.13 | 1 | 1.01 | 0 | 1.15 | -1.29 | ||||||
| 19 | NA | NA | NA | -2.64 | -1.97 | -1.2 | 0 | NA | 0 | ||||||
| 20 | 1.55 | 1.06 | 1.16 | -1.43 | -1.05 | 1.04 | |||||||||
| 21 | 1.12 | -1.04 | -1.02 | ||||||||||||
Proteins detected by two dimensional gel electrophoresis with SYPRO Ruby staining of epithelial cell fractions isolated from normal, benign (BPH), cancerous (PCa) and inflammatory (prostatitis) prostate tissues. A) Epithelial cells, B) Basal cells. The first dimension separation was performed on an 11cm linear IPG strip and the second dimension separation was accomplished on a 10% polyacrylamide gel. Annotated spots were quantified using imageJ, excised, trypsin digested, and analyzed by MALDI-TOF MS.
Proteins detected by two dimensional gel electrophoresis with SYPRO Ruby staining of stromal cell fractions isolated from normal, benign (BPH), cancerous (PCa) and inflammatory (prostatitis) prostate tissues under the same experimental conditions as described in figure 2. A) Myofibroblasts, B) Leukocytes, C) Endothelial cells.
Proteins identified using MALDI-TOF MS and their respective densities in benign (BPH), cancerous (PCa) and inflammatory (Titis) prostate tissues.
| Spot# | Cell type | Protein | Score | Mw | PI | BPH | PCa | Titis |
|---|---|---|---|---|---|---|---|---|
| 14 | Leukocytes | Caspase-1 | 69 | 45814 | 5.63 | -2.65 | 2.05 | -1.71 |
| 8 | Leukocytes | GTP-binding protein 1 (GTPB1) | 62 | 73035 | 8.6 | 0 | 2.35 | 0 |
| 19 | Leukocytes | Junctophilin-1 | 76 | 72098 | 9.37 | 0 | NA | 0 |
| 12 | Leukocytes | leucine-rich repeat protein kinase (LRRK2) | 61 | 289568 | 6.35 | NA | NA | 0 |
| 4 | Leukocytes | Interleukin-18 receptor 1(IL18R1) | 63 | 63119 | 8.06 | 1.85 | 2.21 | 0 |
| 3 | Leukocytes | Rab5 GDP/GTP exchange factor | 59 | 80575 | 6.42 | 1.44 | 1.47 | 1.94 |
| 2 | Endothelial | Proto-oncogene Wnt-3 | 66 | 40988 | 7.46 | 1.02 | 1.17 | -1.04 |
| 3 | Endothelial | Uncharacterized serine/threonine-protein kinase | 61 | 31480 | 9.12 | -3.41 | -1.26 | 1.03 |
| 14 | Endothelial | Tyrosine-protein phosphatase non-receptor type 1 (PTPN1) | 59 | 50505 | 5.88 | -1.04 | 0 | 0 |
| 1 | Endothelial | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 1(ARAP1) | 59 | 163743 | 5.86 | -1.18 | 1.3 | 1.01 |
| 15 | Basal | Actin-related protein 2/3 complex subunit 3 | 62 | 20761 | 8.78 | -1.16 | -1.07 | 1.15 |
| 18 | Basal | Cellular retinoic acid-binding protein 2 (CRABP2) | 59 | 15854 | 5.42 | -2.13 | 1 | 1.01 |
| 13 | Epithelial | Uncharacterized protein | 58 | 108689 | 6.68 | 1.51 | 1.88 | 1.88 |
| 3 | Epithelial | Integrin alpha-6 | 60 | 127751 | 6.39 | 1.03 | 2.39 | 1.6 |
| 2 | Epithelial | Poly ADP-ribose polymerase 14 (PARP14) | 59 | 195627 | 8.21 | 2.01 | 3.26 | 4.07 |
| 13 | Myofibroblasts | Uncharacterized protein | 58 | 108689 | 6.68 | 1.02 | -2.25 | -1.12 |
| 3 | Myofibroblasts | Integrin alpha-6 | 60 | 127751 | 6.39 | -1.1 | 0 | -1.4 |
| 2 | Myofibroblasts | Poly ADP-ribose polymerase 14 (PARP14) | 59 | 195627 | 8.21 | 1.37 | 1.09 | -2.21 |
Discussion
The epithelium of normal and preneoplastic prostate gland is underpinned by a basal cell layer and a basement membrane which separate it from the stroma and act as messengers mediating its communication with the surrounding microenvironment 3. To become invasive, carcinoma cells must first pass through the basal cell layer and the underlying basement membrane. A common diagnostic feature of prostate cancer progression from in situ to invasive tumor is the gradual disappearance of fully differentiated basal cell layer 8. This implies that abrogation of the basal cell layer is an absolute prerequisite for tumor invasion. Whilst most of the concepts regarding disruption of the basal cell layer and the underlying basement membrane defined a prominent role of proteolytic enzymes 17,18, clinical trials using proteinase inhibitors have been disappointing 3,19 and only 10-30% of PIN lesions expressing high levels of proteinases become invasive 20,21. This suggests an alternative pathway of prostate cancer progression and invasion. Recently, numerous studies support the hypothesis that tumor microenvironment plays important roles in cancer initiation, growth, progression, invasion, and metastasis. The stroma is no longer perceived as a reactive responder to the neoplastic process, but rather as an interactive component that actively affects the fate of the adjacent epithelium.
So far, it is unknown what mechanisms lead to focal basal cell layer disruptions and its contribution to tumor progression. To identify the intrinsic mechanism of tumor initiation and progression, Man et al. suggested a potential role of autoimmune reactions in malignant transformation. Focally disrupted basal cell layers were correlated with higher leukocytes influx. They exhibited lower proliferation rate, lower tumor suppressor genes expression and higher apoptosis. The tumor epithelial cells overlying disrupted basal cells also showed substantial differences from other epithelial cells in the ducts. They displayed a higher proliferation rate and a higher expression of invasion and cell cycle related genes 3,8. Whether this hypothesis reflects the sole trigger of tumor initiation is vague and requires further investigation.
Recently, Polyak et al. proposed two alternative models of in situ to invasive carcinoma transition in mammary gland 22. The “Escape” model suggests that a genetically distinct tumor epithelial cell clone will disrupt the myoepithelial cell layer and the underlying basement membrane, escape from the duct and migrate into the stroma. In the “Release” model, myoepithelial cells will cooperate with leukocytes and myofibroblasts to break down the ducts and release tumor epithelial cells. Polyak purported that the two models may not be mutually exclusive and myoepithalial cells together with infiltrating leukocytes and myofibroblasts may affect the clonal evolution of tumor epithelial cells. Moreover, recent genomic characterization of tumor microenvironment was done by the purification of all major cell types from normal breast tissue, ductal carcinoma in situ (DCIS), and invasive carcinomas 23. They found genetic changes in all cell types of the mammary gland suggesting that tumor progression results from reciprocal interactions between epithelial and stromal cells challenging the old paradigm of epithelial cell-driven tumorigenesis. Due to similarities between mammary and prostate glands, the potential involvement of all stromal cell types in initiation of invasion in prostate cancer needs to be addressed. To elucidate this mechanism, epithelial and stromal cells were isolated from different prostate tissue pathologies. This study aims to recognize specific protein signatures that may govern the breakdown of basal cell layer, thereby leading to the initiation of tumor invasion.
2-DE of proteins extracted from epithelial cells (Figure 2) and stromal cells (Figure 3) isolated from normal, benign, cancerous, and inflammatory prostate tissues shows differential protein signatures within the same cell type in different prostate tissue pathologies that support the differential gene expression pattern found in epithelial cells and stromal cells in normal versus cancerous prostate 9,10. Densitometric analysis of the spots reveals distinct expression pattern among the proteins in benign, cancerous and prostatitis tissues as compared to normal tissue. Some proteins are only expressed in normal tissues, others start to appear in BPH and increase in prostate cancer suggesting their involvement in tumor initiation and progression (Table 1).
We were able to identify some proteins in leukocytes, endothelial cells, basal cells, epithelial cells, and myofibroblasts (Table 2). In leukocytes, caspase-1 is found in spot # 14 and appears to be highly expressed in prostate cancer. It is a proteolytic enzyme that cleaves some components of the inflammatory response such as interleukin-1β and interleukin-18 into active peptides 24. Caspase-1, also known as IL-1β-converting enzyme, is synthesized as a zymogen which is activated by cleavage and dimerization of its 10 KDa and 20 KDa subunits. It has been shown to be involved in the formation of inflammasome and the activation of inflammatory response and apoptosis 25. Caspase-1 expression level was investigated in epithelial cells of hyperplastic 26 and malignant prostate 27. The expression of caspase-1 was dramatically diminished in the hyperplastic tissue as compared to the normal control 26. Moreover, its expression was completely lost in prostate cancer specimens 27. However, caspase-1 expression was not reported in stromal cells of prostate cancer tissues.
Interleukin-18 receptor 1 (IL18R1), identified in leukocytes (spot # 4), is a receptor for the proinflammatory cytokine, IL18, that determines the development and function of T and NK cells 28,29 and is activated by caspase-1 30. Its expression increases from normal to benign to cancerous prostate tissues. It has been reported that IL18R is expressed in spleen, thymus, leukocytes, liver, lung, heart, prostate, and placenta 31. Recently, a study showed that IL18 is produced by prostate cancer cell lines and its receptor IL-18R1 was found to be expressed in the poorly differentiated prostate cancer cell lines, PC-3 and DU-145, but is absent in the well differentiated LNCaP cells suggesting a correlation with tumor progression 32.
Rab5 GDP/GTP exchange factor or Rabex-5, also identified in leukocytes, is involved in membrane trafficking 33. It displays GDP/GTP exchange activity on Rab5, a member of Ras superfamily of G proteins, by complexing with Rabaptin-5 which is a direct effector of Rab5 34. Rabex-5-Rabaptin-5 complex helps to translocate the Rab5 GTPase to the membrane enhancing membrane docking and fusion 33.
The proto-oncogene Wnt-3 is identified in endothelial cells. It is a member of Wnt family of proteins which are involved in oncogenesis and various developmental processes including cell proliferation, differentiation, and cell fate 35. Aberrant Wnt signaling through β-catenin is known to affect prostate cancer progression and bone metastasis 36. PTPN1 (protein tyrosine phosphatase non-receptor type 1, or PTP1B) is expressed in endothelial cells of normal and benign prostate. Contradictory data suggests that PTP1B has both inhibitory and stimulatory effects on tumors based on the associated proteins and the cellular context 37. As a tumor suppressor, PTP1B was found to decrease tumorigenecity of v-src-expressing cells 38 and neu TM-transformed cells 39. On the contrary, PTP1B was reported to be an oncogene associated with breast tumorigenesis 40 and capable of activating src oncogene 41. Whether PTP1B is a tumor suppressor or tumor promoter or both is still debatable and requires further investigation. In the prostate, PTP1B has been linked to neuroendocrine differentiation which is associated with androgen independent tumors suggesting an implication in prostate cancer progression 42. ARAP1 contains Arf-GAP, RHO-GAP, Ankyrin repeat, Ras-associating and five PH domains. Associated with the Golgi, ARAP1 can mediate cell rounding, loss of stress fibers, changes in golgi structure and filopodia 43. It regulates endocytic trafficking of the epidermal growth factor receptor (EGFR) 44 and DR4, a receptor protein involved in apoptosis 45.
Cellular retinoic acid-binding protein 2 (CRABP2), identified in normal and benign prostate tissues of basal cells, is an intracellular lipid binding protein that associates with retinoic acid and mediates its actions in cell growth, differentiation, apoptosis, and anti-tumorigenesis 46. CRABP2 is found to be down regulated in prostate cancer and thus, can be a novel therapeutic marker for prostate cancer 47.
Integrin α-6 is a member of the integrins which are cell surface receptors for extracellular matrix proteins involved in cell migration, differentiation, cell survival, and tissue organization. The laminin binding integrin α-6 expression increases in the epithelial cells as normal tissue progresses into benign and cancerous. However, its expression remained almost the same in the myofibroblasts of hyperplastic prostate but diminished in the myofibroblasts of tumorigenic prostate. Several studies have reported higher expression of α-6 integrin in invasive and migrating human prostate cancers as opposed to other integrins 48. Recently, a novel variant of α-6 integrin called α-6p generated by urokinase plasminogen activator (uPA) mediated cleavage of laminin binding domain was reported to increase cellular migration and invasion on laminin substrates 49,50, and to enhance tumor cells tropism to the laminin rich bone microenvironment 51,52. Moreover, inhibition of uPA-dependent α-6 integrin cleavage delayed human prostate cancer metastasis 52. PARP14 is a poly ADP-ribose polymerase which catalyzes the transfer of ADP-ribose from NAD to target proteins involved in DNA damage and repair, apoptosis, and transcription and it was reported to signal B-cell survival 53.
Conclusion
The concept of microenvironmental effects on tumor invasion constitutes a paradigm-shift from the “protease-centered” hypothesis that focuses on basement membrane degradation and the development of protease inhibitors to prevent tumor invasion and metastasis. The role of the stroma in malignant transformation is growing steadily which focuses on reciprocal interactions between the epithelium and the surrounding microenvironment. To elucidate this role in prostate cancer, we have isolated epithelial cells, basal cells, endothelial cells, myofibroblasts, and leukocytes, from human normal, prostatitis, BPH, and prostate cancer tissues to study their protein expression. The protein profiles of the different cells show differential protein signatures for each pathology. Furthermore, we were able to identify some novel molecules that may be responsible for basal cell layer disruption and disappearance during the progression of in situ tumors to invasive cancers. Further validation of these proteins' identity may lead to verification of some early invasion-promoting protein biomarkers that may be targeted to prevent invasion.
Acknowledgements
This work was supported in part by grants W81XWH-07-1-0225 and DAMD17-02-1-0238 from DOD, US Congressionally Directed Medical Research Programs, a grant from the Elsa U. Pardee Foundation, and grants from Florida State University (to Q.-X.S.) and a DOD grant W81XWH-07-1-0300 (to K.A.I.).
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Tuxhorn JA, Ayala GE, Rowley DR. Reactive stroma in prostate cancer progression. Journal of Urology. 2001;166(6):2472-2483
2. Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tisty TD, Cunha GR. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Research. 1999;59(19):5002-5011
3. Man YG, Shen T, Zhao YG, Sang QXA. Focal prostate basal cell layer disruptions and leukocyte infiltration are correlated events: A potential mechanism for basal cell layer disruptions and tumor invasion. Cancer Detection and Prevention. 2005;29(2):161-169
4. Fidler IJ. Timeline - The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nature Reviews Cancer. 2003;3(6):453-458
5. Mueller MM, Fusenig NE. Friends or foes - Bipolar effects of the tumour stroma in cancer. Nature Reviews Cancer. 2004;4(11):839-849
6. Tlsty TD, Hein PW. Know thy neighbor: stromal cells can contribute oncogenic signals. Current Opinion in Genetics & Development. 2001;11(1):54-59
7. Shimura S, Yang G, Ebara S, Wheeler TM, Frolov A, Thompson TC. Reduced infiltration of tumor-associated macrophages in human prostate cancer: Association with cancer progression. Cancer Research. 2000;60(20):5857-5861
8. Man YG, Gardner WA. Focal degeneration of basal cells and the resultant auto-immunoreactions: A novel mechanism for prostate tumor progression and invasion. Medical Hypotheses. 2008;70(2):387-408
9. Dakhova O, Ozen M, Creighton CJ, Li RL, Ayala G, Rowley D, Ittmann M. Global Gene Expression Analysis of Reactive Stroma in Prostate Cancer. Clinical Cancer Research. 2009;15(12):3979-3989
10. Richardson AM, Woodson K, Wang Y, Rodriguez-Canales J, Erickson HS, Tangrea MA, Novakovic K, Gonzalez S, Velasco A, Kawasaki ES, Emmert-Buck MR, Chuaqui RF, Player A. Global expression analysis of prostate cancer-associated stroma and epithelia. Diagnostic Molecular Pathology. 2007;16(4):189-197
11. Min H, Yao J, Allinen M, Cai L, Polyak K. The role of the tumor microenvironment in breast cancer progression. Breast Cancer Research. 2005;7:S7-S7
12. Sahab ZJ, Suh Y, Sang QXA. Isoelectric point-based prefractionation of proteins from crude biological samples prior to two-dimensional gel electrophoresis. Journal of Proteome Research. 2005;4(6):2266-2272
13. Sahab ZJ, Iczkowski KA, Sang QXA. Anion exchange fractionation of serum proteins versus albumin elimination. Analytical Biochemistry. 2007;368(1):24-32
14. Karas M, Hillenkamp F. Laser Desorption Ionization of Proteins with Molecular Masses Exceeding 10000 Daltons. Analytical Chemistry. 1988;60(20):2299-2301
15. Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y, Yohida T. Protein and polymer analyses up to m/z 100,000 by laser ionization time-of-flight mass spectrometry. Rapid Communications in Mass Spectrometry. 1988;2(8):151-153
16. Schiller J, Arnhold J, Benard S, Muller M, Reichl S, Arnold K. Lipid analysis by matrix-assisted laser desorption and ionization mass spectrometry: A methodological approach. Analytical Biochemistry. 1999;267(1):46-56
17. Goldfarb RH, Liotta LA. Proteolytic-Enzymes in Cancer Invasion and Metastasis. Seminars in Thrombosis and Hemostasis. 1986;12(4):294-307
18. Dawson LA, Maitland NJ, Turner AJ, Usmani BA. Stromal-epithelial interactions influence prostate cancer cell invasion by altering the balance of metallopeptidase expression. British Journal of Cancer. 2004;90(8):1577-1582
19. Coussens LM, Fingleton B, Matrisian LM. Cancer therapy - Matrix metalloproteinase inhibitors and cancer: Trials and tribulations. Science. 2002;295(5564):2387-2392
20. Ashida S, Nakagawa H, Katagiri T, Furihata M, Iiizumi M, Anazawa Y, Tsunoda T, Takata R, Kasahara K, Miki T, Fujioka T, Shuin T, Nakamura Y. Molecular features of the transition from prostatic intraepithelial neoplasia (PIN) to prostate cancer: Genome-wide gene-expression profiles of prostate cancers and PINs. Cancer Research. 2004;64(17):5963-5972
21. Liu AJ, Wei LX, Gardner WA, Deng CX, Man YG. Correlated alterations in prostate basal cell layer and basement membrane. International Journal of Biological Sciences. 2009;5(3):276-285
22. Polyak K, Hu M. Do myoepithelial cells hold the key for breast tumor progression? Journal of Mammary Gland Biology and Neoplasia. 2005;10(3):231-247
23. Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang HY, Porter D, Hu M, Chin L, Richardson A, Schnitt S, Sellers WR, Polyak K. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6(1):17-32
24. Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, Miller DK, Molineaux SM, Weidner JR, Aunins J, Elliston KO, Ayala JM, Casano FJ, Chin J, Ding GJF, Egger LA, Gaffney EP, Limjuco G, Palyha OC, Raju SM, Rolando AM, Salley JP, Yamin TT, Lee TD, Shively JE, Maccross M, Mumford RA, Schmidt JA, Tocci MJ. A Novel Heterodimeric Cysteine Protease Is Required for Interleukin-1-Beta Processing in Monocytes. Nature. 1992;356(6372):768-774
25. Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, Roose-Girma M, Erickson S, Dixit VM. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430(6996):213-218
26. Deng CH, Zang ZJ, Sun XZ, Huang XH, Wang D, Wang H, Su S. The expression and significance of caspase-1 in tissue of benign prostatic hyperplasia. International Journal of Andrology. 2005;28:120-120
27. Winter RN, Kramer A, Borkowski A, Kyprianou N. Loss of caspase-1 and caspase-3 protein expression in human prostate cancer. Cancer Research. 2001;61(3):1227-1232
28. Akira S. The role of IL-18 in innate immunity. Current Opinion in Immunology. 2000;12(1):59-63
29. Bazan JF, Timans JC, Kastelein RA. A newly defined interleukin-1? Nature. 1996;379(6566):591-591
30. Gu Y, Kuida K, Tsutsui H, Ku G, Hsiao K, Fleming MA, Hayashi N, Higashino K, Okamura H, Nakanishi K, Kurimoto M, Tanimoto T, Flavell RA, Sato V, Harding MW, Livingston DJ, Su MSS. Activation of interferon-gamma inducing factor mediated by interleukin-1 beta converting enzyme. Science. 1997;275(5297):206-209
31. Parnet P, Garka KE, Bonnert TP, Dower SK, Sims JE. IL-1Rrp is a novel receptor-like molecule similar to the type I interleukin-1 receptor and its homologues T1/ST2 and IL-1R AcP. Journal of Biological Chemistry. 1996;271(8):3967-3970
32. Lebel-Binay S, Thiounn N, De Pinieux G, Vieillefond A, Debre B, Bonnefoy JY, Fridman WH, Pages F. IL-18 is produced by prostate cancer cells and secreted in response to interferons. International Journal of Cancer. 2003;106(6):827-835
33. Horiuchi H, Lippe R, McBride HM, Rubino M, Woodman P, Stenmark H, Rybin V, Wilm M, Ashman K, Mann M, Zerial M. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 1997;90(6):1149-1159
34. Stenmark H, Vitale G, Ullrich O, Zerial M. Rabaptin-5 Is a Direct Effector of the Small Gtpase Rab5 in Endocytic Membrane-Fusion. Cell. 1995;83(3):423-432
35. Emami KH, Corey E. When prostate cancer meets bone: Control by wnts. Cancer Letters. 2007;253(2):170-179
36. Hall CL, Kang SN, MacDougald OA, Keller ET. Role of Wnts in prostate cancer bone metastases. Journal of Cellular Biochemistry. 2006;97(4):661-672
37. Lessard L, Stuible M, Tremblay ML. The two faces of PTP1B in cancer. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2010;1804(3):613-619
38. Woodfordthomas TA, Rhodes JD, Dixon JE. Expression of a Protein Tyrosine Phosphatase in Normal and Nu-Src-Transformed Mouse 3T3 Fibroblasts. Journal of Cell Biology. 1992;117(2):401-414
39. Brownshimer S, Johnson KA, Hill DE, Bruskin AM. Effect of Protein Tyrosine Phosphatase-1B Expression on Transformation by the Human Neu Oncogene. Cancer Research. 1992;52(2):478-482
40. Wiener JR, Kerns BJM, Harvey EL, Conaway MR, Iglehart JD, Berchuck A, Bast RC. Overexpression of the Protein-Tyrosine-Phosphatase Ptp1B in Human Breast-Cancer - Association with P185(C-Erbb-2) Protein Expression. Journal of the National Cancer Institute. 1994;86(5):372-378
41. Bjorge JD, Pang A, Fujita DJ. Identification of protein-tyrosine phosphatase 1B as the major tyrosine phosphatase activity capable of dephosphorylating and activating c-Src in several human breast cancer cell lines. Journal of Biological Chemistry. 2000;275(52):41439-41446
42. Wu CY, Zhang L, Bourne PA, Reeder JE, di Sant'Agnese PA, Yao JL, Na YQ, Huang JT. Protein tyrosine phosphatase PTPIB is involved in neuroendocrine differentiation of prostate cancer. Prostate. 2006;66(11):1125-1135
43. Miura K, Jacques KM, Stauffer S, Kubosaki A, Zhu KJ, Hirsch DS, Resau J, Zheng Y, Randazzo PA. ARAP1: A point of convergence for Arf and Rho signaling. Molecular Cell. 2002;9(1):109-119
44. Daniele T, Di Tullio G, Santoro M, Turacchio G, De Matteis MA. ARAP1 Regulates EGF Receptor Trafficking and Signalling. Traffic. 2008;9(12):2221-2235
45. Simova S, Klima M, Cermak L, Sourkova V, Andera L. Arf and Rho GAP adapter protein ARAP1 participates in the mobilization of TRAIL-R1/DR4 to the plasma membrane. Apoptosis. 2008;13(3):423-436
46. Budhu AS, Noy N. Direct channeling of retinoic acid between cellular retinoic acid-binding protein II and retinoic acid receptor sensitizes mammary carcinoma cells to retinoic acid-induced growth arrest. Molecular and Cellular Biology. 2002;22(8):2632-2641
47. Okuducu AF, Janzen V, Ko Y, Hahne JC, Lu H, Ma ZL, Albers P, Sahin A, Wellmann A, Scheinert P, Wernert N. Cellular retinoic acid-binding protein 2 is down-regulated in prostate cancer. International Journal of Oncology. 2005;27(5):1273-1282
48. Rabinovitz I, Nagle RB, Cress AE. Integrin Alpha-6 Expression in Human Prostate Carcinoma-Cells Is Associated with a Migratory and Invasive Phenotype in-Vitro and in-Vivo. Clinical & Experimental Metastasis. 1995;13(6):481-491
49. Demetriou MC, Pennington ME, Nagle RB, Cress AE. Extracellular alpha 6 integrin cleavage by urokinase-type plasminogen activator in human prostate cancer. Experimental Cell Research. 2004;294(2):550-558
50. Pawar SC, Demetriou MC, Nagle RB, Bowden GT, Cress AE. Integrin alpha 6 cleavage: A novel modification to modulate cell migration. Experimental Cell Research. 2007;313(6):1080-1089
51. King TE, Pawar SC, Majuta L, Sroka IC, Wynn D, Demetriou MC, Nagle RB, Porreca F, Cress AE. The Role of Alpha 6 Integrin in Prostate Cancer Migration and Bone Pain in a Novel Xenograft Model. Plos One. 2008;3(10):e3535
52. Ports MO, Nagle RB, Pond GD, Cress AE. Extracellular engagement of alpha 6 integrin blocks uPA mediated cleavage and prevents human prostate bone metastasis. Clinical & Experimental Metastasis. 2009;26(7):871-871
53. Cho SH, Goenka S, Henttinen T, Gudapati P, Reinikainen A, Eischen CM, Lahesmaa R, Boothby M. PARP-14, a member of the B aggressive lymphoma family, transduces survival signals in primary B cells. Blood. 2009;113(11):2416-2425
Author contact
![]() Corresponding author: Prof. Qing-Xiang Amy SANG, Ph.D., Department of Chemistry and Biochemistry and Institute of Molecular Biophysics, Florida State University, 102 Varsity Way, Tallahassee, FL 32306-4390, USA; Tel: +1-850-644-8683; Fax: +1-850-644-8281; E-mail: sangfsu.edu
Corresponding author: Prof. Qing-Xiang Amy SANG, Ph.D., Department of Chemistry and Biochemistry and Institute of Molecular Biophysics, Florida State University, 102 Varsity Way, Tallahassee, FL 32306-4390, USA; Tel: +1-850-644-8683; Fax: +1-850-644-8281; E-mail: sangfsu.edu

 Global reach, higher impact
Global reach, higher impact