3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(4):446-452. doi:10.7150/jca.13065 This issue Cite
Research Paper
PCDH8 is Frequently Inactivated by Promoter Hypermethylation in Liver Cancer: Diagnostic and Clinical Significance
Institute of Pathology and Pathophysiology, Shandong University School of Medicine, Jinan 250012, P. R. China.
#These two authors contributed equally to this work.
Received 2015-6-27; Accepted 2015-12-8; Published 2016-1-29
Abstract
AIM: Protocadherin-8 (PCDH8) plays an important role in signaling pathways of cell adhesin, proliferation, and migration. It has been reported that PCDH8 is mutated or methylated in several human cancers. However, little is known about PCDH8 in liver cancer. The aim of this study was to investigate the protein expression and promoter methylation status of PCDH8 in liver cancer and evaluate the association between PCDH8 methylation and the clinicopathological features.
METHODS: The methylation status of PCDH8 in 42 hepatocellular carcinoma (HCC), 8 Cholangiocarcinoma (CC) and 50 normal liver tissues were examined using methylation-specific PCR (MSP) and the protein expression of PCDH8 was detected by immunohistochemistry. The relationships between PCDH8 methylation and clinicopathological features as well as overall survival of patients were evaluated.
RESULTS: The PCDH8 methylation was more frequent in liver cancer tissues than that in the normal liver tissues (88% vs. 32%, P < 0.001), and is significantly associated with loss of its protein expression (P = 0.004). Moreover, there is a significant correlation between PCDH8 methylation and the alpha-fetoprotein (AFP) level (P = 0.008). Kaplan-Meier survival analysis revealed that patients with PCDH8 methylation have shorter OS and PFS than those without PCDH8 methylation (P = 0.041 and P = 0.028, respectively).
CONCLUSION: PCDH8 is often inactivated by promoter methylation in liver cancer. PCDH8 methylation can serve as a valuable diagnostic biomarker for early detection of liver cancer and might be useful to predict an unfavorable clinical feature.
Keywords: PCDH8, Protocadherin, Promoter methylation, Liver cancer, Tumor suppressor.
Introduction
Liver cancer is one of the most common human cancers worldwide, especially in Asia and Africa. Hepatocellular carcinoma (HCC) accounts for about 75% of liver cancers. Cholangiocarcinoma (CC) is the second most common primary liver malignant tumour arising from cholangiocytes and CC accounts for about 10% of primary liver cancers. There are often no symptoms of liver cancer in the early stage and the poor survival rate is largely due to the delayed diagnosis. Only 10% to 20% of liver cancers are detected at an early enough stage for potentially curative therapy. Therefore, new early diagnosis strategies are urgently needed.
Inactivation of tumor suppressor genes (TSGs) is a key molecular event in the multistep process of carcinogenesis. It has been shown that either genetic or epigenetic mechanisms contribute to inactivate TSGs [1-3]. Emerging evidence suggests that hypermethylation of TSGs is one of the hallmarks in cancer initiation and progression [4-6], some of the tumor-specific methylated TSGs detected in early or late stage of cancer could function as diagnostic or prognostic biomarkers [7-9]. In liver cancer, epigenetic changes occur more frequently than genetic mutations [10]. Many studies have suggested that methylation of tumor suppressor genes in liver cancer may contribute to the pathogenesis of this disease [11-13]. Such epigenetic defects also have been observed in noncancerous liver tissues, supporting the concept that methylation-induced silencing may play a role in the early stages of liver cancer [14,15].
PCDH8 is a member of the cadherin family which plays multiple roles in cell adhesion, proliferation, differentiation and migration [16]. Several members of the protocadherin family (PCDH10, 17, and 20) are frequently silenced by promoter methylation in nasopharynx carcinoma, gastric and colorectal cancers, or non-small-cell lung cancers [17-19]. It has reported that PCDH8 can function as a candidate tumor suppressor and is inactivated in many cancers through mutation or promoter methylation [20-23]. However, the PCDH8 methylation status in liver cancer and its role in liver tumorigenesis remain unknown.
In this study, we investigated protein expression and promoter methylation status of the PCDH8 gene in primary HCC and CC tissues as well as normal liver tissues, and the correlation between the methylation status and clinicopathological features was evaluated. To determine the potential prognostic significance of the PCDH8 gene in patients with liver cancer, the association between the overall survival and the PCDH8 methylation was also analyzed.
Materials and Methods
Patients and tissue samples
A total of 100 liver tissues, including 42 HCCs, 8 CCs, and 50 normal liver tissues were analyzed. Samples were obtained from the Department of Pathology, Shandong University Qilu Hospital, China between January 2013 and March 2014. All the liver cancer tissue specimens were confirmed histologically. Thirteen cases were small HCC (< 3cm) and 29 cases were advanced (> 3cm) HCC. Two CC were poor differentiation and six were well&moderate differentiation. Fifty normal liver tissues were from 34 cases of matched noncancerous specimens and 16 of hepatolithus. The mean ages of the patients with liver cancers and those with normal liver were 55.7 years (range from 36-78) and 51.5 years (range from 25-67) respectively. The follow-up data was available for all 50 liver cancer patients with a median time of 19.4 months (range from 1 to 27 months). This study was approved by the ethics committee of Shandong University.
Immunohistochemistry
Immunohistochemisty was performed on 5-μm-thick formalin-fixed, paraffin-embedded liver cancer and normal liver tissue sections. The sections were deparaffinized in xylene, rehydrated in a series of alcohol, and washed in phosphate-buffered saline (PBS). Antigen retrieval was achieved by heat treatment in a pressure cooker for 5 min in EDTA (pH 8.9). After blocking of endogenous peroxidase with 3% hydrogen peroxide, slides were blocked to avoid nonspecific binding by addition of a 10% normal goat serum at 37°C for 30 min and then incubated for 4°C overnight with the monoclonal antibody against PCDH8 (Abcam, Cambridge, UK) at a 1:150 dilution. In the next day, the sections were washed three times with 0.01 mol/L PBS (pH 7.4) for 15 min, and then the primary antibody was detected using an secondary antibody (Universal PV9000 Kit, Zhongshan Biotechnology, Beijing, China) for 30 min at 37°C and visualized using DAB. PCDH8 staining was expressed diffusely in the cytoplasm.
Methylation-specific PCR (MSP)
To determine the methylation status within the promoter region of the PCDH8 gene, genomic DNAs were extracted using a Dneasy tissue kit (Qiagen, Valencia, CA) following the manufacturer's instructions. 1 μg of purified genomic DNA from each specimen was used. DNA modification was performed using EZ DNA Methylation Kit (Zymo Research, USA), and all procedures were done according to manufactures protocol. The bisufite modifited DNA was then used for MSP. The primers specific for PCDH8 unmethylated and methylated sequences were as follows: unmethylated: forward 5'-GGTTATAAAGGTAAAGGTGGT-3' and reverse 5'-AAAATCACACTCTTTACAAAC-3'; methylated: forward 5'-GTTATAAAGGTAAAGGCGGC-3' and reverse 5'-AAATCACGCTCTTTACGAAC-3'. The PCR amplification of the modified DNA consisted of one cycle of 95°C for 12 min, 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s, and 1 cycle of 72°C for 10 min. The PCR products were electrophoresed on 3% agarose gel and visualized under UV illumination.
Statistical analysis
The SPSS statistical package (16.0, Chicago, IL, USA) was used for data analysis. Frequencies of methylation of two groups were compared using the χ2 test. The associations between the PCDH8 methylation and clinicopathological features were determined by Fisher's exact test. Survival analysis (including overall survival (OS) and progression-free survival (PFS)) was performed according to Kaplan-Meier survival analysis and log-rank test. Multivariate Cox proportional hazard model analysis was used to assess the independent prognostic effect of liver cancer. All P-values were 2-sided, and P < 0.05 was considered statistically significant.
Result
Reduced PCDH8 protein expression in liver cancer
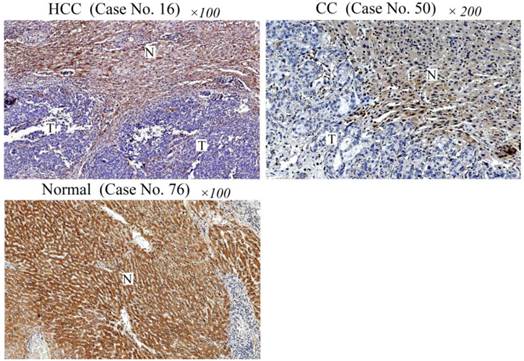
To study whether PCDH8 may play a role in liver cancer, we first examined the protein expression of PCDH8 in liver cancer and normal liver tissues. As shown in Fig. 1, PCDH8 was strongly positive in the normal liver tissues (only 7/50 cases were negative) but loss its expression in 35/42 HCC cases and 7/8 CC cases.
Frequency of PCDH8 methylation in liver cancer and normal liver specimens
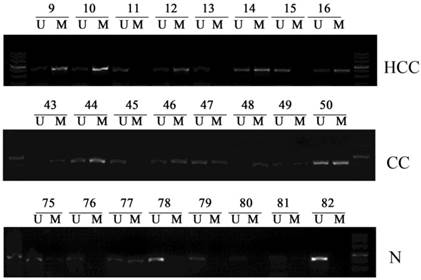
Subsequently, we examined the promoter methylation status of PCDH8 in all the liver cancer tissues and normal liver tissues. PCDH8 promoter methylation was present in 37/42 HCC (88.1%) and 7/8 (87.5%) CC, whereas 16/50 (32%) of the normal liver tissues. There was a significant difference in PCDH8 promoter methylation between the liver cancer and normal liver tissues (P < 0.001). Representative results of MSP assays for PCDH8 methylation are shown in Fig. 2.
Immunohistochemical analysis of PCDH8 protein expression in liver cancer and normal liver tissues. Case No. 16 is a HCC case showing negative immunoreactivity of the HCC cells (indicated as T) for PCDH8 protein. Note the strong immunostaining of non-cancerous cells (indicated with N) in this section. Case No. 50 is a CC case showing negative immunoreactivity of the CC cells for PCDH8 protein. Normal liver tissue section (Case No. 76) shows a strong immunoreactivity for PCDH8 protein. HCC: hepatocellular carcinomas; CC: cholangiocarcinoma.
Representative samples of the methylation status of PCDH8 in 42 hepatocellular carcinomas (HCCs) and 8 cholangiocarcinoma (CCs) as well as 50 normal liver tissues. The methylation-specific PCR (MSP) products in the M lanes indicate the presence of methylated alleles, and those in the U lanes indicate the presence of unmethylated allele; N, normal liver tissues.
DNA methylation of the PCDH8 promoter and regulation of expression
To determine whether transcriptional silencing of the PCDH8 gene is caused by promoter hypermethylation, we compared the methylation status of the PCDH8 gene with its expression in liver cancer tissues. In the normal liver tissue sections, PCDH8 protein was detected in 43/50 cases. However, there was 42 cases lost their protein expression in all the 50 cases of liver cancer tissues, and among the 42 PCDH8 negative cases, 40 cases occurred PCDH8 promoter methylation. As expected, PCDH8 protein expression was lost in most of the cases with promoter methylation, which indicated a significant correlation between promoter hypermethylation and transcriptional silencing (P = 0.004).
Correlation between PCDH8 methylation and clinicopathological features in liver cancer
To clarify the potential clinical significance of PCDH8 methylation, the associations of PCDH8 methylation status with the clinicopathological characteristics were also analyzed. The result was summarized in Table 1. The aberrant methylation status of PCDH8 in liver cancer tissues was significantly associated with AFP level (P = 0.008), while there were no statistically significant associations between PCDH8 promoter methylation and the other clinicopathological parameters such as age, gender, tumor size, or tumor differentiation (P > 0.05).
Survival analysis
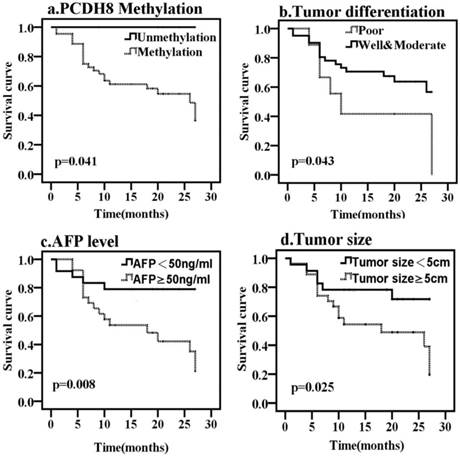
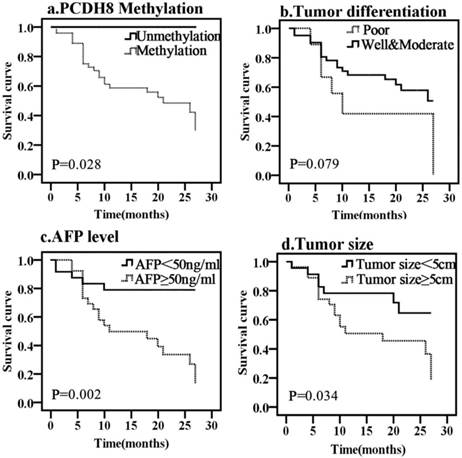
To determine whether any of the clinicopathological features and the PCDH8 methylation is of prognostic significance, the survival of all patients with liver cancer was analyzed. Kaplan-Meier survival analysis revealed that the overall median survival of patients with methylated PCDH8 in HCC was 15 months (Fig. 3). On the other hand, those without methylated PCDH8 tended to have a longer survival (all are alive at the end of follow up, P = 0.041). Moreover, tumor size, tumor differentiation and AFP level were also found to be associated with OS of liver cancer. In addition, the median progression-free survival of patients with methylated PCDH8 in liver cancer was 13.5 months (Fig. 4), and none of patients with unmethylated PCDH8 have progression at the end of follow up (P = 0.028). Similarly, tumor size and AFP level were found to be associated with PFS of liver cancer. Next, Univariate and Multivariate Cox proportional analysis were performed to determine whether these markers can act as an independent prognostic factor for liver cancer. The result showed that although the HR (26.231) was very high, the PCDH8 methylation could not act as an independent predictor (P > 0.05). Similar results were obtained from tumor size and tumor differentiation: Although Univariate Cox proportional analysis revealed that tumor size and tumor differentiation are risk factors for predicting survival of liver cancer, Multivariate Cox proportional analysis couldn't confirm this result. Multivariate Cox proportional analysis revealed that only AFP level could serve as an independent prognostic factor.
Discussion
Epigenetic events of hypermethylation are emerging as one of the most promising molecular strategies for cancer detection, prognosis prediction or gene target therapy [24-26]. Cadherins is known to be critical for creating and maintaining tissue architecture in cancer and development. The cadherin family is classified into classical cadherins, desmosomal cadherins and protocadherins (PCDHs). E-cadherin is a classical tumor suppressor that plays a key role in carcinogenesis such as breast cancer and gastric cancer [27,28]. PCDHs are the largest subgroup within the cadherin superfamily. Recently, aberrant promoter methylation of cadherin family genes has been implicated in various cancers. Previous studies have suggested that epigenetic modifications regulate the expression of PCDHs, several members of the PCDHs (PCDH10, 17 and 20) had been reported to be often inactivated by promoter methylation in human cancers, and the aberrant promoter methylation can be used as potential biomarker for tumor diagnosis, surveillance, or prognosis [19,29,30].
The correlation between PCDH8 methylation and clinical features in liver cancers.
| Clinical feature | PCDH8 (M) Case number | PCDH8 (U) Case number | P |
|---|---|---|---|
| Age(years) | 3 | 21 | 1.000 |
| <55 | 3 | 23 | |
| ≥55 | |||
| Gender | 0.317 | ||
| Female | 0 | 11 | |
| Male | 6 | 33 | |
| Tumor size (cm) | 0.674 | ||
| <5 | 2 | 21 | |
| ≥5 | 4 | 23 | |
| AFP level (ng/ml) | 0.008** | ||
| <50 | 6 | 18 | |
| ≥50 | 0 | 26 | |
| Tumor differentiation | 1.000 | ||
| Well&Moderate | 5 | 36 | |
| Poor | 1 | 8 |
M: methylation; U: unmethylation. **P < 0.01
Kaplan-Meier curves predicting the OS of liver cancer, by (a) PCDH8 methylation status; (b) tumor size; (c) tumor differentiation; (d) AFP level.
Kaplan-Meier curves predicting the PFS of liver cancer, by (a) PCDH8 methylation status; (b) tumor size; (c) tumor differentiation; (d) AFP level.
In recent years, it has been revealed that PCDH8 acts as a tumor suppressor and is often mutated or methylated in many cancers including bladder cancer, renal cell carcinoma and breast cancer [21,23,31]. JS Yu et al reported that loss of PCDH8 occurred early in tumor development in breast ductal carcinoma in situ (DCIS), indicating that inactivated PCDH8 appears to be an early step in breast tumor progression, and this inactivation is caused through either mutation or hypermethylation [23]. In our current study, the methylation of PCDH8 was more frequently detected in liver cancer but much low in normal liver tissues, suggesting that PCDH8 promoter methylation can serve as a valuable diagnostic biomarker for liver cancer. However, this methylation occurred in both early and late stage of liver cancer, indicating that it might be essential in the early development of liver cancer and persist throughout the course of this development. While in other studies, Lin et al. reported that methylation of PCDH8 was significantly correlated with advanced tumor stage, high grade, larger tumor size, or tumor recurrence, suggesting that the PCDH8 methylation may be used as a potential biomarker to predict the prognosis of bladder cancer and renal cell carcinoma [31,32], consistent with the findings of Zhang et al who found that methylation of PCDH8 was significantly correlated with lymph node metastasis and tumor differentiation in gastric cancer [33]. We do not find any statistically significant associations between PCDH8 promoter methylation and tumor size or tumor differentiation. But interestingly, our data shows that the methylation of PCDH8 is significantly correlated with the level of AFP. AFP has been the serum marker that is most widely used for diagnosis as well as surveillance of liver cancer. However, AFP level may be normal in up to 40% of patients with liver cancer, particularly during the early stages (low sensitivity). Furthermore, elevated AFP levels may be seen in patients with cirrhosis or exacerbations of chronic hepatitis (low specificity) [34,35]. In general, AFP level > 400 ng/ml will be considered to have diagnostic significance for liver cancer [36]. Our study shows that PCDH8 methylation seems to more easily occur in patients with AFP level > 50 ng/ml, supplying a possibility for early diagnosis of liver cancer by combing AFP level and PCDH8 promoter methylation status.
In addition, the present study also assessed the association between aberrant PCDH8 methylation and protein expression. Immunohistochemical results revealed that in all the 44 liver cancer cases with PCDH8 promoter methylation, forty cases had complete loss of PCDH8 immunoreactivity within the tumor tissue, indicating that PCDH8 promoter methylation is the main mechanism of loss of expression in liver cancer.
Kaplan-Meier survival analysis reveals that hypermethylation of PCDH8 statistically correlates with shorter OS and PFS compared with unmethylation of PCDH8 in liver cancer. Moreover, it also shows that tumor size, tumor differentiation and AFP level also correlate with survival of liver cancer. Because there are very few cases (only 6 cases) without PCDH8 methylation in liver cancer and all the 6 patients are still alive at the end of the follow up, so it is difficult to confirm PCDH8 methylation as an independent predictor of poor prognosis for liver cancer by further Cox proportional analysis. Neither does tumor size or tumor differentiation due to the limited sample size. Multivariate Cox proportional analysis only confirms AFP level as an independent predictor for liver cancer due to its larger sample size.
In summary, our data demonstrate for the first time that PCDH8 plays an important role in the carcinogenesis of liver cancer and is often inactivated by promoter methylation. PCDH8 methylation is a frequent event in liver cancer and could serve as a valuable diagnostic biomarker for early detection of this disease and might be useful to predict an unfavorable clinical feature.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 81272277) and the Shandong Province Postdoctoral Innovation Project (No. 127682).
Conflict of Interest
There are no conflicts of interest.
References
1. Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, Kasper LH, Lerach S, Tang H, Ma J, Rossi D, Chadburn A, Murty VV, Mullighan CG, Gaidano G, Rabadan R, Brindle PK, Dalla-Favera R. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471(7337):189-95
2. Nephew KP, Huang TH. Epigenetic gene silencing in cancer initiation and progression. Cancer Lett. 2003;190(2):125-33
3. Wang Q, Peng D, Zhu S, Chen Z, Hu T, Soutto M, Saad R, Zhang S, Ei-Rifai W. Regulation of Desmocollin3 Expression by Promoter Hypermethylation is Associated with Advanced Esophageal Adenocarcinomas. J Cancer. 2014;5(6):457-64
4. Li W, Li X, Wang W, Li X, Tan Y, Yi M, Yang J, McCarthy JB, Xiong W, Wu M, Ma J, Su B, Zhang Z, Liao Q, Xiang B, Li G. NOR1 is an HSF1- and NRF1-regulated putative tumor suppressor inactivated by promoter hypermethylation in nasopharyngeal carcinoma. Carcinogenesis. 2011;32(9):1305-14
5. Gokul G, Gautami B, Malathi S, Sowjanya AP, Poli UR, Jain M, Ramakrishna G, Khosla S. DNA methylation profile at the DNMT3L promoter: a potential biomarker for cervical cancer. Epigenetics. 2007;2(2):80-5
6. Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet. 2007;16:R50-9
7. Zhu X, Shan L, Wang F, Wang J, Wang F, Shen G, Liu X, Wang B, Yuan Y, Ying J, Yang H. Hypermethylation of BRCA1 gene: implication for prognostic biomarker and therapeutic target in sporadic primary triple-negative breast cancer. Breast Cancer Res Treat. 2015;150(3):479-86
8. Otani K, Dong Y, Li X, Lu J, Zhang N, Xu L, Go MY, Ng EK, Arakawa T, Chan FK, Sung JJ, Yu J. Odd-skipped related 1 is a novel tumour suppressor gene and a potential prognostic biomarker in gastric cancer. J Pathol. 2014;234(3):302-15
9. Summers T, Langan RC, Nissan A, Brucher BL, Bilchik AJ, Protic M, Daumer M, Avital I, Stojadinovic A. Serum-based DNA methylation biomarkers in colorectal cancer: potential for screening and early detection. J Cancer. 2013;4(3):210-6
10. Nishida N, Goel A. Genetic and epigenetic signatures in human hepatocellular carcinoma: a systematic review. Curr Genomics. 2011;12(2):130-7
11. Matsumura S, Imoto I, Kozaki K, Matsui T, Muramatsu T, Furuta M, Tanaka S, Sakamoto M, Arii S, Inazawa J. Integrative array-based approach identifies MZB1 as a frequently methylated putative tumor suppressor in hepatocellular carcinoma. Clin Cancer Res. 2012;18(13):3541-51
12. Nishida N, Kudo M, Nagasaka T, Ikai I, Goel A. Characteristic patterns of altered DNA methylation predict emergence of human hepatocellular carcinoma. Hepatology. 2012;56(3):994-1003
13. Zhang C, Li H, Wang Y, Liu W, Zhang Q, Zhang T, Zhang X, Han B, Zhou G. Epigenetic inactivation of the tumor suppressor gene RIZ1 in hepatocellular carcinoma involves both DNA methylation and histone modifications. J Hepatol. 2010;53(5):889-95
14. Zhang X, Li HM, Liu Z, Zhou G, Zhang Q, Zhang T, Zhang J, Zhang C. Loss of heterozygosity and methylation of multiple tumor suppressor genes on chromosome 3 in hepatocellular carcinoma. J Gastroenterol. 2013;48(1):132-43
15. Nishida N, Nagasaka T, Nishimura T, Ikai I, Boland CR, Goel A. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology. 2008;47(3):908-18
16. Etzrodt J, Krishna KK, Redies C. Expression of classic cadherins and delta-protocadherins in the developing ferret retina. BMC Neurosci. 2009;10:153
17. Chen T, Long B, Ren G, Xiang T, Li L, Wang Z, He Y, Zeng Q, Hong S, Hu G. Protocadherin20 acts as a tumor suppressor gene: epigenetic inactivation in nasopharyngeal carcinoma. J Cell Biochem. 2015
18. Hu X, Sui X, Li L, Huang X, Rong R, Su X, Shi Q, Mo L, Shu X, Kuang Y, Tao Q, He C. Protocadherin 17 acts as a tumour suppressor inducing tumour cell apoptosis and autophagy, and is frequently methylated in gastric and colorectal cancers. J Pathol. 2013;229(1):62-73
19. Imoto I, Izumi H, Yokoi S, Hosoda H, Shibata T, Hosoda F, Ohki M, Hirohashi S, Inazawa J. Frequent silencing of the candidate tumor suppressor PCDH20 by epigenetic mechanism in non-small-cell lung cancers. Cancer Res. 2006;66(9):4617-26
20. He D, Zeng Q, Ren G, Xiang T, Qian Y, Hu Q, Zhu J, Hong S, Hu G. Protocadherin8 is a functional tumor suppressor frequently inactivated by promoter methylation in nasopharyngeal carcinoma. Eur J Cancer Prev. 2012;21(6):569-75
21. Morris MR, Ricketts CJ, Gentle D, McRonald F, Carli N, Khalili H, Brown M, Kishida T, Yao M, Banks RE, Clarke N, Latif F, Maher ER. Genome-wide methylation analysis identifies epigenetically inactivated candidate tumour suppressor genes in renal cell carcinoma. Oncogene. 2011;30(12):1390-401
22. Leshchenko VV, Kuo PY, Shaknovich R, Yang DT, Gellen T, Petrich A, Yu Y, Remache Y, Weniger MA, Rafiq S, Suh KS, Goy A, Wilson W, Verma A, Braunschweig I, Muthusamy N, Kahl BS, Byrd JC, Wiestner A, Melnick A, Parekh S. Genomewide DNA methylation analysis reveals novel targets for drug development in mantle cell lymphoma. Blood. 2010;116(7):1025-34
23. Yu JS, Koujak S, Nagase S, Li CM, Su T, Wang X, Keniry M, Memeo L, Rojtman A, Mansukhani M, Hibshoosh H, Tycko B, Parsons R. PCDH8, the human homolog of PAPC, is a candidate tumor suppressor of breast cancer. Oncogene. 2008;27(34):4657-65
24. Jeschke J, Van Neste L, Glockner SC, Dhir M, Calmon MF, Deregowski V, Van Criekinge W, Vlassenbroeck I, Koch A, Chan TA, Cope L, Hooker CM, Schuebel KE, Gabrielson E, Winterpacht A, Baylin SB, Herman JG, Ahuja N. Biomarkers for detection and prognosis of breast cancer identified by a functional hypermethylome screen. Epigenetics. 2012;7(7):701-9
25. Morris MR, Ricketts C, Gentle D, Abdulrahman M, Clarke N, Brown M, Kishida T, Yao M, Latif F, Maher ER. Identification of candidate tumour suppressor genes frequently methylated in renal cell carcinoma. Oncogene. 2010;29(14):2104-17
26. Tang YA, Lin RK, Tsai YT, Hsu HS, Yang YC, Chen CY, Wang YC. MDM2 overexpression deregulates the transcriptional control of RB/E2F leading to DNA methyltransferase 3A overexpression in lung cancer. Clin Cancer Res. 2012;18(16):4325-33
27. Su YJ, Chang YW, Lin WH, Liang CL, Lee JL. An aberrant nuclear localization of E-cadherin is a potent inhibitor of Wnt/beta-catenin-elicited promotion of the cancer stem cell phenotype. Oncogenesis. 2015;4:e157
28. Ribeiro AS, Sousa B, Carreto L, Mendes N, Nobre AR, Ricardo S, Albergaria A, Cameselle-Teijeiro JF, Gerhard R, Soderberg O, Seruca R, Santos MA, Schmitt F, Paredes J. P-cadherin functional role is dependent on E-cadherin cellular context: a proof of concept using the breast cancer model. J Pathol. 2013;229(5):705-18
29. Haruki S, Imoto I, Kozaki K, Matsui T, Kawachi H, Komatsu S, Muramatsu T, Shimada Y, Kawano T, Inazawa J. Frequent silencing of protocadherin 17, a candidate tumour suppressor for esophageal squamous cell carcinoma. Carcinogenesis. 2010;31(6):1027-36
30. Yu B, Yang H, Zhang C, Wu Q, Shao Y, Zhang J, Guan M, Wan J, Zhang W. High-resolution melting analysis of PCDH10 methylation levels in gastric, colorectal and pancreatic cancers. Neoplasma. 2010;57(3):247-52
31. Lin YL, Wang YL, Ma JG, Li WP. Clinical significance of protocadherin 8 (PCDH8) promoter methylation in non-muscle invasive bladder cancer. J Exp Clin Cancer Res. 2014;33:68
32. Lin YL, Ma JH, Luo XL, Guan TY, Li ZG. Clinical significance of protocadherin-8 (PCDH8) promoter methylation in bladder cancer. J Int Med Res. 2013;41(1):48-54
33. Zhang D, Zhao W, Liao X, Bi T, Li H, Che X. Frequent silencing of protocadherin 8 by promoter methylation, a candidate tumor suppressor for human gastric cancer. Oncol Rep. 2012;28(5):1785-91
34. Roncalli M, Bianchi P, Bruni B, Laghi L, Destro A, Di Gioia S, Gennari L, Tommasini M, Malesci A, Coggi G. Methylation framework of cell cycle gene inhibitors in cirrhosis and associated hepatocellular carcinoma. Hepatology. 2002;36(2):427-32
35. Kanai Y. Genome-wide DNA methylation profiles in precancerous conditions and cancers. Cancer Sci. 2010;101(1):36-45
36. Talwalkar JA, Gores GJ. Diagnosis and staging of hepatocellular carcinoma. Gastroenterology. 2004;127(5 Suppl 1):S126-32
Author contact
![]() Corresponding author: Cuijuan Zhang, Institute of Pathology and Pathophysiology, Shandong University School of Medicine, Wenhuaxi Road 44#, Jinan 250012, P. R. China. E-mail: cuijuanzhangedu.cn Tel.: 86-531-88382574 Fax: 86-531-88383168.
Corresponding author: Cuijuan Zhang, Institute of Pathology and Pathophysiology, Shandong University School of Medicine, Wenhuaxi Road 44#, Jinan 250012, P. R. China. E-mail: cuijuanzhangedu.cn Tel.: 86-531-88382574 Fax: 86-531-88383168.

 Global reach, higher impact
Global reach, higher impact