3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(7):846-853. doi:10.7150/jca.13437 This issue Cite
Research Paper
Combining Telomerase Reverse Transcriptase Genetic Variant rs2736100 with Epidemiologic Factors in the Prediction of Lung Cancer Susceptibility
1. Cancer and Stem Cell Center, First Affiliated Hospital, Jilin University, Changchun, Jilin 130061, P.R. China.
2. Stanford University Medical School Stanford, Palo Alto Veterans Institute for Research, Palo Alto, CA94305, USA.
3. Second Department of Neurology, China-Japan Union Hospital of Jilin University, Changchun , Jilin 130033, P.R. China.
4. School of Public Health, Jilin University, Changchun 130021, Jilin, P. R. China.
*Contributed equally.
Received 2015-8-3; Accepted 2016-3-15; Published 2016-4-28
Abstract
Genetic variants from a considerable number of susceptibility loci have been identified in association with cancer risk, but their interaction with epidemiologic factors in lung cancer remains to be defined. We sought to establish a forecasting model for identifying individuals with high-risk of lung cancer by combing gene single-nucleotide polymorphisms with epidemiologic factors. Genotyping and clinical data from 500 lung cancer cases and 500 controls were used for developing the logistic regression model. We found that lung cancer was associated with telomerase reverse transcriptase (TERT) rs2736100 single-nucleotide polymorphism. The TERT rs2736100 model was still significantly associated with lung cancer risk when combined with environmental and lifestyle factors, including lower education, lower BMI, COPD history, heavy cigarettes smoking, heavy cooking emission, and dietary factors (over-consumption of meat and deficiency in fish/shrimp, vegetables, dairy products, and soybean products). These data suggest that combining TERT SNP and epidemiologic factors may be a useful approach to discriminate high and low-risk individuals for lung cancer.
Keywords: Lung cancer, forecasting model, telomerase, TERT, WWOX, single nucleotide polymorphism, epidemiologic factors, Chinese population.
Introduction
Lung cancer is the most common cancer by incidence (1.82 million in 2012) and a leading cause of cancer-related deaths (1.6 million deaths in 2012) worldwide 1. Most lung cancer patients are diagnosed at an advanced stage, and hence are not able to undergo surgical removal of tumors2. Systemic chemotherapy and radiotherapy are currently the main treatment options for lung cancer, but most patients eventually develop resistance to these treatments. As a result, the overall 5-year survival rate for lung cancer patients is still low 3. Since early stage detection is essential for effective therapy, it would be important to develop an accurate lung cancer risk forecasting model that could identify individuals at high-risk.
Several models4-7 have been developed to predict individual risk for lung cancer within a specified period by using a patient's characteristics, epidemiologic, social, and clinical risk factors, including age, body mass index (BMI), socioeconomic status, cigarette smoking history, second-hand smoke exposure in never-smokers, asbestos exposure, chronic obstructive pulmonary disease (COPD) history, pneumonia history, and a family history of lung cancer. Other factors associated with lung cancer in epidemiological studies might also be useful for forecasting, including dietary factors8, exposure to cooking emissions9, and occupational exposure to diesel or gasoline10, pesticides11, and ink12.
Recent advances in genetic epidemiology have led to identification of genetic and molecular variants affecting the risk of diseases, suggesting that genetic markers, such as single-nucleotide polymorphisms (SNP), can be added to risk models to improve forecasting of future risk of disease7,13. Genome-wide association studies (GWAS) and large case-control studies have identified a considerable number of genetic susceptibility loci associated with lung cancer risk14-24, including telomerase reverse transcriptase (TERT).
TERT is a reverse transcriptase that is critical for telomere replication and stabilization by controlling telomere length. The telomerase enzyme protein is highly expressed in many tumor tissues, including lung cancer, whereas it is tightly repressed in most normal human cells 25-26. High TERT expression predicts poor prognosis in patients with lung cancer27. When TERT is inhibited, cancer cells undergo telomere shortening, senescence or apoptosis, and eventually lose their oncogenic potential. However, the exact role of rs2736100 in the pathogenesis of lung cancer remains to be defined.
Nearly all of the published models have focused on either genetic variables (e.g. SNPs) or epidemiological variables. Lung cancer is a multi-etiological disease initiated by both genetic and lifestyle factors. Thus, in this study we attempted to establish an ethnicity-specific lung cancer risk forecasting model by combining gene polymorphisms, like TERT rs2736100, with epidemiologic factors, aiming to provide a more precise prediction in a Chinese population.
Materials and Methods
Study population
This study was a hospital-based case-control study involving a total of 1000 subjects from northeastern China (Changchun, Jilin Province). All subjects were local residents of Han descent, comprising 500 lung cancer patients and 500 cancer-free controls. Eligible patients had histologically confirmed primary lung cancer without previous cancer history and with no radiotherapy or chemotherapy for other conditions. Control participants were randomly selected from individuals receiving routine physical examinations in our hospital. The selection of controls was frequency-matched to cases by age, gender, and residential area (urban or rural). The study was approved by the Ethics Committee of the First Hospital of Jilin Medical University, and conducted according to Declaration of Helsinki principles. All subjects signed a written informed consent at the beginning of the study.
Data collection
Standard interviews were conducted by trained physicians at either the hospital or the participants' homes. A standardized lifestyle questionnaire was used to collect socioeconomic status, medical history, family history, lifestyle history, and cancer diagnosis. Risk factor information and peripheral blood lymphocytes were collected prior to and up to the time of diagnosis for cancer patients and at the interview date for control subjects.
According to the Chinese food pagoda 28, the recommended daily intake levels are 300-500 g for vegetables, 200-400 g for fruits, 50-75 g for meats, 50-100 g for fish and shrimp, 30-50 g for soybean products and nuts, and 300 g for dairy. Based on these recommended levels, we divided the amounts of meat, soybean products and nuts, and dairy products into three (inadequate, normal, and excess) or two (inadequate and adequate) levels, respectively, which were used as inputs for model development. For vegetable consumption, fruits, fish and shrimp, amounts of daily intakes were entered into the model directly. Since there were limited data on the exposure to asbestos and recent chest x-rays in either cases or controls, these items were excluded from analysis.
Genotyping and quality control
DNA was extracted from the peripheral blood lymphocytes using a Wizard® Genomic DNA Purification Kit A1125 (Promega, WI, USA). MassArray (Sequenom, CA, USA) was employed for genotyping all markers by using allele specific matrix-assisted laser desorption/ionization time-of-flight mass spectrometry29 according to the manufacturer's instructions. Primers and multiplex reactions were designed by using the RealSNP.com Website.
All lung cancer patients as well as healthy controls were genotyped for TP6 rs4488809, TERT rs2736100, MIPEP-TNFRSF19 rs753955, MTMR3-HORMAD2-LIF rs17728461, CHRNA3 rs6495309, and WWOX rs3764340 polymorphisms. These SNPs have been reported previously in association with lung cancer risk by GWAS analyses in case-control studies20-24. Concordance among the three genomic control DNA samples presented in duplicate was 100%. Of SNPs with genotyping data, call rates were >95%.
Statistical analysis
Hardy-Weinberg equilibrium was tested by a goodness-of-fit chi-square (χ2) test. The χ2 test was used to determine genotype frequencies with observed genotype frequencies in cancer-free controls vs lung cancer cases and association of SNP with clinicopathological data. Multiple logistic regression model was performed to identify independent risk factors for lung cancer. A forward stepwise likelihood ratio method was employed to screen variables in the model selection, where the cut-off for variables in the model was 0.05 and the cut-off for variables out of model was 0.10. We selected the optimal model with the maximum Cox & Snell R square and Nagelkerke R square. All categorical variables were set as dummy variables, and the first category of each variable was selected as baseline. All analyses were conducted by SPSS v19.0 software (SPSS, Inc., Chicago, IL, USA). All p-values were two-sided, and those <0.05 were considered statistically significant. Multiplicative models were used for rs4488809, rs2736100, rs753955, rs3764340, and rs17728461 in the multiple logistic regression model. The classification ability of the model was evaluated using the area under the receiver operating characteristic (ROC) curve (AUC).
Results
Characteristics, genotypes and risk factors of participants
A total of 500 lung cancer patients and 500 controls were recruited between 2010 and 2012. Tables 1-2 shows the distribution of study-specific risk factors and the genotype distributions of SNPs between cases and controls. We focused on six SNPs that were reported previously in association with lung cancer 21-24,28, including TP63 rs4488809, TERT rs2736100, MIPEP-TNFRSF19 rs753955, MTMR3-HORMAD2-LIF rs17728461, CHRNA3 rs6495309, and WWOX rs3764340. All SNPs followed Hardy-Weinberg equilibrium in the control groups (P>0.05).
Association between SNPs and lung cancer risk by univariate analysis
No significant differences of SNPs between the lung cancer group and the healthy control group were found in five SNPs, including rs4488809, rs753955, rs6495309, rs3764340, and rs17728461 (Table 2). However, the C allele of TERT rs2736100 was significantly associated with the increased risk of lung cancer (p=0.000).
Characteristic in case and healthy control groups and the associated distribution of risk factors.
| Characteristic | Case group (n=500) | Control group (n=500) | P-value | |
|---|---|---|---|---|
| Gender | Male | 305 (61.0%) | 302 (60.4%) | |
| Female | 195 (39.0%) | 198 (39.6%) | 0.85 | |
| Age (years) | <30 | 2 (0.4%) | 5 (1.0%) | |
| 30-39 | 14 (2.8%) | 16 (3.2%) | ||
| 40-49 | 64 (12.8%) | 70 (14.0%) | ||
| 50-59 | 176 (35.2%) | 196 (39.2%) | ||
| 60-69 | 174 (34.8%) | 148 (19.7%) | ||
| ≥70 | 70 (14.0%) | 65 (13.0%) | 0.42 | |
| Education | Junior high school or lower | 318 (63.6%) | 130 (26.0%) | |
| High school | 97 (19.4%) | 144 (28.8%) | ||
| Greater than high school | 85 (17.0%) | 226 (45.2%) | 0.00 | |
| Smoking (pack-years) | 14.25 (0.0-36.0) | 0.00 (0.0-6.9) | 0.00 | |
| Fish and shrimps (g/day) | 4.00 (2.5-17.1) | 14.29 (3.3-28.6) | 0.00 | |
| Vegetable (g/day) | 177.10 (148.3-199.2) | 205.60 (174.8-407.9) | 0.00 | |
| Fruit (g/day) | 60.70 (31.40-101.90) | 99.50 (49.80-201.30) | 0.00 | |
| Meat | Deficient | 296 (59.2%) | 304 (60.8%) | |
| Normal | 108 (21.6%) | 128 (25.6%) | ||
| Over-sufficient | 96 (19.2%) | 68 (13.6%) | 0.04 | |
| Dairy products | Deficient | 499 (99.8%) | 457 (91.4%) | |
| Sufficient | 1 (0.2%) | 43 (8.6%) | 0.00 | |
| Soybean products and nuts | deficient | 327 (65.4%) | 228 (45.6%) | |
| Normal | 93 (18.6%) | 80 (16.0%) | ||
| Over-sufficient | 80 (16.0%) | 192 (38.4%) | 0.00 | |
| Alcohol (times/week) | 0 | 276 (55.2%) | 297 (59.4%) | |
| 1-2 | 104 (20.8%) | 130 (26.0%) | ||
| 3-6 | 31 (6.2%) | 43 (8.6%) | ||
| ≥7 | 89 (17.8%) | 30 (6.0%) | 0.00 | |
| Exposure to pesticide | Absent | 398 (79.6%) | 473 (94.6%) | |
| Present | 102 (20.4%) | 27 (5.4%) | 0.00 | |
| Exposure to gasoline/diesel | Absent | 487 (97.4%) | 496 (99.2%) | |
| Present | 13 (2.6%) | 4 (0.8%) | 0.04 | |
| Cooking emissions (total dish-years) | Absent | 244 (48.8%) | 250 (50.0%) | |
| ≤50 | 149 (29.8%) | 152 (30.4%) | ||
| 51-100 | 61 (12.2%) | 80 (16.0%) | ||
| 101-150 | 46 (9.2%) | 18 (3.6%) | 0.00 | |
| Pneumonia history | Absent | 477 (95.4%) | 490 (98.0%) | |
| Present | 23 (4.6%) | 10 (2.0%) | ||
| COPD history | Absent | 449 (89.8%) | 489 (97.8%) | |
| Present | 51 (10.2%) | 11 (2.2%) | 0.00 | |
| Pulmonary tuberculosis history | Absent | 470 (94.0%) | 486 (97.2%) | |
| Present | 30 (6.0%) | 14 (2.8%) | 0.02 | |
| Bronchial asthma history | Absent | 488 (97.6%) | 495 (99.0%) | |
| Present | 12 (2.4%) | 5 (1.0%) | 0.10 | |
| Cancer family history | Absent | 330 (66.0%) | 397 (79.4%) | |
| Present | 170 (34.0%) | 103 (20.6%) | 0.00 | |
| BMI (kg/m2) | <18.5 | 49 (9.8%) | 15 (3.0%) | |
| 18.5-24 | 302 (60.4%) | 230 (46.0%) | ||
| ≥24 | 149 (29.8%) | 255 (51.0%) | 0.00 | |
| Histology types | Squamous cell | 141(28.2%) | ||
| Adenocarcinomas | 176(35.2%) | |||
| Small cell | 126(25.2%) | |||
| Other carcinomas* | 57(11.4%) | |||
Genotype of SNPs in case and healthy control groups and the association of SNPs with lung cancer risk in univariate analysis.
| SNP(Reference) | Genotype | Case | Control | Crude OR (95% CI) | P-value | ||
|---|---|---|---|---|---|---|---|
| N | % | N | % | ||||
| rs4488809(21,22) | CC | 133 | 26.6 | 140 | 28.0 | ||
| 3q28,TP63 | CT | 252 | 50.4 | 258 | 51.6 | ||
| TT | 115 | 23.0 | 102 | 20.4 | |||
| Multiplicative model | C vs T | 0.92(0.77-1.10) | 0.37 | ||||
| rs2736100(22,23) | CC | 112 | 22.4 | 80 | 16.0 | ||
| 5p15.33,TERT | CA | 257 | 51.4 | 242 | 48.4 | ||
| AA | 131 | 26.2 | 178 | 35.6 | |||
| Multiplicative model | C vs A | 1.39 (1.16-1.66) | 0.00 | ||||
| rs753955(22) | GG | 65 | 13.0 | 65 | 13.0 | ||
| 13q12.12,MIPEP-TNFRSF19 | GA | 214 | 42.8 | 223 | 44.6 | ||
| AA | 221 | 44.2 | 212 | 42.4 | |||
| Multiplicative model | G vs A | 0.96 (0.80-1.15) | 0.68 | ||||
| rs6495309(25) | CC | 160 | 32.0 | 155 | 31.0 | ||
| 15q25,CHRNA3 | CT | 253 | 50.6 | 241 | 48.2 | ||
| TT | 87 | 17.4 | 104 | 20.8 | |||
| Multiplicative model | C vs T | 1.09 (0.92-1.31) | 0.32 | ||||
| rs3764340(24) | GG | 1 | 0.2 | 3 | 0.6 | ||
| 16q23,WWOX | GC | 74 | 14.8 | 91 | 18.2 | ||
| CC | 425 | 85.0 | 406 | 81.2 | |||
| Multiplicative model | G vs C | 0.77 (0.55-1.04) | 0.09 | ||||
| rs17728461(22) | GG | 27 | 5.4 | 21 | 4.2 | ||
| 22q12.2,MTMR3-HORMAD2-LIF | GC | 164 | 32.8 | 194 | 38.8 | ||
| CC | 309 | 61.8 | 285 | 57.0 | |||
| Multiplicative model | G vs C | 0.90 (0.73-1.11) | 0.33 | ||||
Multivariate logistic regression model
Multivariate logistic repression analysis was then performed to select risk factors and SNPs for the prediction model. A forward stepwise likelihood ratio method was employed to screen variables in the regression model. The final optimal model was selected by including a total of 12 risk factors, with the maximum Cox & Snell R square (0.43) and Nagelkerke R square (0.57) (Table 3). In addition to TERT rs2736100, the WWOX rs3764340 genotype was also significantly included in the model in combination with other epidemiological factors.
The fitted final multivariate logistic regression model is presented in Table 4. Significantly increased lung cancer risk in the multivariate analysis was found to be associated with the following factors: lower education, lower BMI, COPD history, heavy cigarettes smoking, heavy cooking emission, and dietary factors, including deficient in fish/shrimp, vegetables, dairy products, soybean products and nuts, and over-sufficient meat. TERT rs2736100 was still significantly associated with a high-risk of lung cancer even after adjusting for confounding factors. For the gene-gene and gene-environment interaction analyses, only the WWOX rs3764340 genotype was correlated with the TERT rs2736100 genotype (p<0.05) and cooking emissions (p<0.01).
The optimal regression model with the maximum Cox & Snell R square and Nagelkerke R square.
| Procedure | Risk factors | Cox & Snell R square | Nagelkerke R square | Percentage Correct | P-value |
|---|---|---|---|---|---|
| 1 | Vegetable | 0.25 | 0.33 | 0.73 | 0.00 |
| 2 | Education | 0.31 | 0.41 | 0.73 | 0.00 |
| 3 | Smoking | 0.35 | 0.46 | 0.77 | 0.00 |
| 4 | Fish and shrimps | 0.36 | 0.48 | 0.77 | 0.00 |
| 5 | Meat | 0.38 | 0.50 | 0.78 | 0.00 |
| 6 | BMI | 0.39 | 0.52 | 0.78 | 0.00 |
| 7 | COPD | 0.40 | 0.53 | 0.78 | 0.00 |
| 8 | rs2736100 | 0.41 | 0.54 | 0.78 | 0.00 |
| 9 | Soybean products and nuts | 0.41 | 0.55 | 0.78 | 0.01 |
| 10 | Dairy products | 0.42 | 0.56 | 0.79 | 0.00 |
| 11 | rs3764340 | 0.42 | 0.56 | 0.79 | 0.01 |
| 12 | Cooking emissions | 0.43 | 0.57 | 0.80 | 0.01 |
The final multivariate logistic regression model with adjusted ORs and 95% CI.
| Risk factors | Exp (B) | 95% CI* | P-value |
|---|---|---|---|
| Vegetable (g/day) | 0.99 | 0.99-0.99 | 0.00 |
| Education | 0.00 | ||
| Junior high school and lower | 1.00 | Reference | - |
| High school | 0.36 | 0.24-0.55 | 0.00 |
| Greater than high school | 0.29 | 0.19-0.44 | 0.00 |
| Smoking (pack-years) | 1.03 | 1.02-1.04 | 0.00 |
| Fish and shrimps (g/day) | 0.97 | 0.96-0.98 | 0.00 |
| Meat | 0.00 | ||
| Deficient | 1.00 | Reference | - |
| Normal | 1.51 | 0.98-2.32 | 0.06 |
| Over-sufficient | 4.91 | 2.76-8.75 | 0.00 |
| BMI (kg/m2) | 0.00 | ||
| <18.5 | 1.00 | Reference | - |
| 18.5-24 | 0.54 | 0.25-1.17 | 0.12 |
| ≥24 | 0.27 | 0.12-0.59 | 0.00 |
| COPD history | |||
| Absent | 1.00 | Reference | - |
| Present | 3.67 | 1.59-8.44 | 0.00 |
| rs2736100 | |||
| C allele | 1.51 | 1.18-1.93 | 0.00 |
| Soybean products and nuts | 0.00 | ||
| Deficient | 1.00 | Reference | - |
| Normal | 0.80 | 0.50-1.28 | 0.36 |
| Over-sufficient | 0.48 | 0.31-0.74 | 0.00 |
| Dairy products | |||
| Deficient | 1.00 | Reference | - |
| Sufficient | 0.10 | 0.01-0.77 | 0.03 |
| rs3764340 | |||
| G allele | 1.70 | 1.11-2.59 | 0.02 |
| Cooking emissions (total dish-years) | 0.05 | ||
| ≤50 | 1.00 | Reference | - |
| 51-100 | 1.74 | 1.16-2.62 | 0.01 |
| 101-150 | 1.07 | 0.64-1.80 | 0.79 |
| >150 | 2.76 | 1.26-6.06 | 0.01 |
ROC analysis
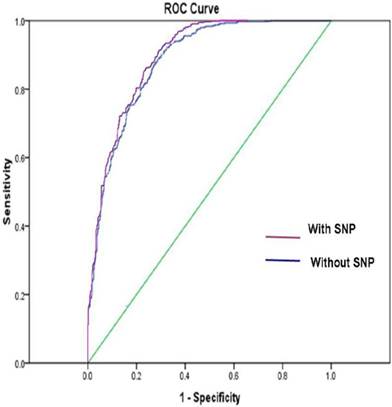
Figure 1 shows the ROC curve derived from our model with or without SNP variables, respectively. The ROC AUC of the model was 0.89 with SNPs and was 0.88 without SNPs, respectively.
The ROC AUC for lung cancer risk prediction model with SNPs was 0.89, and the ROC AUC for lung cancer risk prediction model without SNPs was 0.88. The straight line represented the ROC curve expected by chance alone.
Discussion
In this study, we found that education, BMI, prior diagnosis of COPD, occupational exposure to pesticide, duration of smoking, exposure to heavy cooking emissions, and dietary factors, which included less fish and shrimp, vegetable, soybean products and nuts, and more meat, were all associated with the development of lung cancer. Most importantly, we discovered that, when incorporated with environmental and lifestyle factors, TERT rs2736100 and WWOX rs3764340 were significantly associated with lung cancer.
By using patients' characteristics and epidemiologic, social, and clinical risk factors, several lung cancer risk forecasting models 4-6 have been proposed, but most predictors focused on traditional risk factors, such as age, gender, smoking status, education level, BMI, lung cancer family history, environmental exposure, pneumonia history, and COPD history. The model of Bach et al 4 was developed to determine variation in lung cancer risk among either current or former smokers aged between 55 and 74 years, who were enrolled in a clinical trial for lung cancer prevention. Since this model was developed by using data from individuals with smoking history, it is only applicable to a subset of smokers who are at risk for lung cancer. The initial model by Spitz et al 30 was cross-validated using c statistics (a measure of the discriminative power of the logistic equation) of 0.59, 0.63, and 0.65 in never, former, and current smokers, respectively. It was then improved by adding two markers of DNA repair capacity, which increased the ROC AUC from 0.67 to 0.70 in former smokers and from 0.68 to 0.73 in current smokers31. However, this model requires technical expertise and is not readily available in general practice. Cassidy et al 32 described a lung cancer risk forecasting model, which had an internally validated (cross-validation) ROC AUC of 0.70. By combining patients' socio demographic and clinical records, Iyen-Omofoman et al 33 developed a lung cancer risk forecasting model, which can be used by general practitioners to aid earlier identification of high-risk patients for lung cancer. The Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial) model 5 was developed to determine variation in lung cancer risk among either the general population or ever-smokers; it reported a bootstrap optimism-corrected ROC AUC of 0.857 for the first group and 0.805 for the second group.
In contrast to these traditional risk factors, SNPs are inherited genetic variations that occur during the lifetime of an individual. Inclusion of SNPs may improve the predictive ability of lung cancer risk models. Indeed, the predictive ability of our model was significantly improved by adding SNPs. For example, the ROC AUC of the model with SNP and without SNP was 0.89 and 0.88, respectively. Similarly, by adding a marker SNP (rs663048) in SEZ6L gene, the Liverpool Lung Project risk model did improve the forecasting capability of lung cancer 32,34.
Recently, GWAS studies have identified three susceptibility loci for lung cancer, including the 15q24-25.1 (CHRNA3-5 genes) 14-16, 19, 5p15.33 (TERT-CLPTM1L genes) 17-19, and 6p21.33 (BAT3-MSH5 genes) 19 in European population. In addition, several susceptibility SNPs for lung cancer in the Asian population were also identified. In a GWAS aimed at identifying lung adenocarcinoma susceptibility-related genes, Miki et al.20 found that the TERT rs2736100, and TP63 rs10937405 and rs4488809 were significantly associated with lung adenocarcinoma in Japanese and Korean populations. In another GWAS of lung cancer performed in a Chinese population, Hu et al.21 identified 6 well-replicated SNPs with independent effects and significant lung cancer associations. These SNPs are located in TP63 (rs4488809 at 3q28), TERT-CLPTM1L (rs465498 and rs2736100 at 5p15.33), MIPEP-TNFRSF19 (rs753955 at 13q12.12), and MTMR3-HORMAD2-LIF (rs17728461 and rs36600 at 22q12.2). Moreover, Wu et al. 24 conducted two-stage case-control studies in subjects derived from both Northern and Southern China, and identified 4 novel SNPs (rs2036534 C/T, rs667282 C/T, rs12910984 G/A, and rs6495309 T/C), which were significantly associated with increased lung cancer risk and smoking behavior. In addition, a two-stage case-control study in subjects from Southern and Eastern China showed that 2 tag SNPs (rs3764340 C/G and rs383362 G/T), located in the WWOX gene, were significantly associated with lung cancer risk 23.
Lung cancer is a polygenic disease, for which many genetic factors including SNPs appear to play an important role in disease development. But lung cancer risk prediction model include only SNPs from GWAS is not sufficient. Huan Li et al 35 developed a lung cancer risk forecasting model, five single-nucleotide polymorphisms (SNPs) identified in previous GWA or large cohort studies were genotyped in 5068 Chinese case-control subjects. The weighted genetic risk score (wGRS) based on these SNPs was estimated, and the AUC value of it was 0.551. When incorporated with Smoking history, the AUC value increased to 0.639 (0.621-0.652) after adjustment for over-fitting. Which indicate that future studies should focus on establishing a risk assessment model that incorporates both genetic variants and established traditional factors for lung cancer.
Based on these recent findings, we have developed a novel lung cancer risk forecasting model for the Chinese population by incorporating SNPs with environmental and lifestyle factors. We took advantage of a hypothesis-driven candidate gene approach 15,36,37 to identify potentially functional SNPs associated with histologically validated lung cancer. In contrast to genome-wide association (GWA) and quantitative trait locus (QTL) approaches, the candidate gene approach is economical and has rather high statistical power15. Previous studies have suggested that there may be a significant difference in lung cancer susceptibility loci between European and Asian populations. Thus, we selected SNPs identified by GWASs and large scale studies which showed an association with lung cancer risk in the population of Asia. As we just describe that environmental and lifestyle factors are very important during the development of cancers. The AUC of model only include SNPs is about 0.55135. However, the effects of SNPs were always weakened or dismissed when incorporated with several environmental and lifestyle factors. When analyzing the effects of genetic factors, such as SNPs, very few forecasting models of lung cancer included as many environmental and lifestyle factors as did in our model. We found that TERT rs2736100 was still significantly associated with lung cancer when incorporated with many environmental and lifestyle factors. Overall, our data were consist with those from meta-analyses38-40, thus providing evidence that this locus is strongly associated with the development of the disease.
The specific function of TERT rs2736100 in lung cancer is largely unknown. Multiple GWAS studies have suggested its association with leukocyte telomere length (LTL) in different populations41-43. However, the specific mechanism driving the association between rs2736100 C and longer telomeres remains to be determined. A previous bioinformatics analysis suggests that this polymorphism may be located in a regulatory region of the TERT gene44. As compared to the A allele, the C allele of rs2736100 was significantly associated with increased TERT mRNA expression in both normal and lung cancer cells45. Future studies are needed to address how the C allele sequence has a significantly higher capacity for enhancing TERT transcription.
It is also interesting to note that over-sufficient meat consumption was strongly associated with lung cancer risk, even after adjusted for the other confounding factors (Table 4). It is suspected that meat processing, like preservation, cooking and/or processed methods, may increase the exposure to mutagens and carcinogens, such as N-Nitroso compounds, heterocyclic amines and polycyclic aromatic hydrocarbons. Further studies are needed to examine the involvement of these factors in the lung cancer prediction model.
This study had several limitations. First, we could not include all SNPs that were identified in the Chinese population or other Asian countries populations. Additional susceptibility loci for lung cancer remain to be discovered. It is possible that rare variants with high penetrance would explain the remaining heritability. Combining these remaining SNPs would result in an ethnicity-specific classification of lung cancer risk. Second, our assessment model was an internal validation, and thus external validation needs to be performed in future studies. We will continue to recruit study subjects in this ongoing project. With the increment in statistic power, we will be able to validate our risk models using different approaches, including the integrated discrimination improvement index or change in c statistics. Thirdly, this study is retrospective. Our data provided evidence that lung cancer is associated with the education, BMI, prior diagnosis of COPD, occupational exposure to pesticide, duration of smoking, exposure to heavy cooking emission, less fish and shrimp, vegetable, soybean products and nuts. Meat consumption was not convincing, probably due to relatively small sample size. It cannot be excluded that some of the findings, although statistically significant, are probably owing to type 1 error. Thus, they need to be validated by a large-scale prospective study. Fourthly, in the screening of the model, we include the patients with family history, which might lead to bias. So a future study will be needed to include more subjects so that patients who have family history will be excluded from the analysis or appropriately be adjusted in the modeling. Finally, the association between lung cancer and WWOX rs3764340 was statistically significant in the multivariate logistic regression model (Tables 3-4), but not in the univariate analysis (Table 2). A further interaction analysis with other factors involved in the model showed that WWOX rs3764340 was also related to TERT rs2736100 (p=0.017) and cooking emissions (p=0.008). The Spearman's correlation coefficient was not 0. Thus, Future studies are needed to address the role of the gene-gene or gene-environment interaction in this prediction model.
In conclusion, our lung cancer risk forecasting model has demonstrated high discrimination capability to distinguish between high- and low-risk individuals. Most importantly, our study discovered that TERT rs2736100 and WWOX rs3764340 are associated with lung cancer risk when combined with environmental and lifestyle factors. Future large cohort studies are needed to validate this model in Chinese populations. The final lung cancer risk forecasting model will be used to discriminate individuals with high risk of developing lung cancer
Acknowledgements
We thank all individuals who participated in this study. We specially thank Li Deng for assistance in sample collection and preparation, and Lina Jin and Hua He for data and statistical analysis. This work was supported by Jilin Provincial Science and Technology Department Grant (3D512J243428), the National Natural Science Foundation of China grants (#81501962) to X.W; California Institute of Regenerative Medicine (CIRM) grant (#RT2-01942), Jilin International Collaboration Grant (#20120720), the National Natural Science Foundation of China grants (#81272294 and #31430021) to J.F. Hu; the National Natural Science Foundation of China grants (#81071920 and #81372835) and Jilin Science and Technique Program grant (#11ZDGG003) to W. Li.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ferlay J, Soerjomataram I, Dikshit R. et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E386
2. Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277-300
3. Marugame T, Hirabayashi Y. Comparison of time trends in lung cancer mortality (1990-2006) in the world, from the WHO Mortality Database. Japanese journal of clinical oncology. 2009;39:696-697
4. Spahn L, Siligan C, Bachmaier R, Schmid JA, Aryee DN, Kovar H. Homotypic and heterotypic interactions of EWS, FLI1 and their oncogenic fusion protein. Oncogene. 2003;22:6819-6829
5. Tammemagi CM, Pinsky PF, Caporaso NE. et al. Lung cancer risk prediction: Prostate, Lung, Colorectal And Ovarian Cancer Screening Trial models and validation. Journal of the National Cancer Institute. 2011;103:1058-1068
6. Cassidy A, Myles JP, van Tongeren M. et al. The LLP risk model: an individual risk prediction model for lung cancer. Br J Cancer. 2008;98:270-276
7. Young RP, Hopkins RJ, Hay BA. et al. Lung cancer susceptibility model based on age, family history and genetic variants. PLoS One. 2009;4:e5302
8. Takezaki T, Hirose K, Inoue M. et al. Dietary factors and lung cancer risk in Japanese: with special reference to fish consumption and adenocarcinomas. Br J Cancer. 2001;84:1199-1206
9. Chiu YL, Wang XR, Qiu H, Yu IT. Risk factors for lung cancer: a case-control study in Hong Kong women. Cancer causes & control: CCC. 2010;21:777-785
10. Villeneuve PJ, Parent ME, Sahni V, Johnson KC, Canadian Cancer Registries Epidemiology Research G. Occupational exposure to diesel and gasoline emissions and lung cancer in Canadian men. Environmental research. 2011;111:727-735
11. Andreotti G, Hou L, Beane Freeman LE. et al. Body mass index, agricultural pesticide use, and cancer incidence in the Agricultural Health Study cohort. Cancer causes & control: CCC. 2010;21:1759-1775
12. Leon DA, Thomas P, Hutchings S. Lung cancer among newspaper printers exposed to ink mist: a study of trade union members in Manchester, England. Occupational and environmental medicine. 1994;51:87-94
13. Pepe MS, Janes HE. Gauging the performance of SNPs, biomarkers, and clinical factors for predicting risk of breast cancer. Journal of the National Cancer Institute. 2008;100:978-979
14. Hung RJ, McKay JD, Gaborieau V. et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633-637
15. Amos CI, Wu X, Broderick P. et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nature genetics. 2008;40:616-622
16. Thorgeirsson TE, Geller F, Sulem P. et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638-642
17. McKay JD, Hung RJ, Gaborieau V. et al. Lung cancer susceptibility locus at 5p15.33. Nature genetics. 2008;40:1404-1406
18. Wang Y, Broderick P, Webb E. et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nature genetics. 2008;40:1407-1409
19. Broderick P, Wang Y, Vijayakrishnan J. et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Research. 2009;69:6633-6641
20. Miki D, Kubo M, Takahashi A. et al. Variation in TP63 is associated with lung adenocarcinoma susceptibility in Japanese and Korean populations. Nature genetics. 2010;42:893-896
21. Hu Z, Wu C, Shi Y. et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nature genetics. 2011;43:792-796
22. Hsiung CA, Lan Q, Hong YC. et al. The 5p15.33 locus is associated with risk of lung adenocarcinoma in never-smoking females in Asia. PLoS genetics. 2010:6
23. Huang D, Qiu F, Yang L. et al. The polymorphisms and haplotypes of WWOX gene are associated with the risk of lung cancer in southern and eastern Chinese populations. Molecular carcinogenesis. 2013;52(Suppl 1):E19-27
24. Wu C, Hu Z, Yu D. et al. Genetic variants on chromosome 15q25 associated with lung cancer risk in Chinese populations. Cancer Research. 2009;69:5065-5072
25. Metzger R, Vallbohmer D, Muller-Tidow C. et al. Increased human telomerase reverse transcriptase (hTERT) mRNA expression but not telomerase activity is related to survival in curatively resected non-small cell lung cancer. Anticancer Research. 2009;29:1157-1162
26. Nishio Y, Nakanishi K, Ozeki Y. et al. Telomere length, telomerase activity, and expressions of human telomerase mRNA component (hTERC) and human telomerase reverse transcriptase (hTERT) mRNA in pulmonary neuroendocrine tumors. Japanese Journal of Clinical Oncology. 2007;37:16-22
27. Fernandez-Garcia I, Ortiz-de-Solorzano C, Montuenga LM. Telomeres and telomerase in lung cancer. Journal of Thoracic Oncology. 2008;3:1085-1088
28. Dietary guidelines and the Food Guide Pagoda. The Chinese Nutrition Society. Journal of the American Dietetic Association. 2000;100:886-887
29. Sauer S, Gelfand DH, Boussicault F. et al. Facile method for automated genotyping of single nucleotide polymorphisms by mass spectrometry. Nucleic Acids Research. 2002;30:e22-e22
30. Spitz MR, Hong WK, Amos CI. et al. A risk model for prediction of lung cancer. Journal of the National Cancer Institute. 2007;99:715-726
31. Spitz MR, Etzel CJ, Dong Q. et al. An expanded risk prediction model for lung cancer. Cancer prevention research. 2008;1:250-254
32. Cassidy A, Duffy SW, Myles JP, Liloglou T, Field JK. Lung cancer risk prediction: a tool for early detection. International journal of cancer Journal international du cancer. 2007;120:1-6
33. Iyen-Omofoman B, Tata LJ, Baldwin DR, Smith CJ, Hubbard RB. Using socio-demographic and early clinical features in general practice to identify people with lung cancer earlier. Thorax. 2013;68:451-459
34. Raji OY, Agbaje OF, Duffy SW, Cassidy A, Field JK. Incorporation of a genetic factor into an epidemiologic model for prediction of individual risk of lung cancer: the Liverpool Lung Project. Cancer prevention research. 2010;3:664-669
35. Li H, Yang L, Zhao X. et al. Prediction of lung cancer risk in a Chinese population using a multifactorial genetic model. BMC Medical Genetics. 2012;13:118
36. Clark AG. The role of haplotypes in candidate gene studies. Genetic epidemiology. 2004;27:321-333
37. Leng S, Stidley CA, Liu Y. et al. Genetic determinants for promoter hypermethylation in the lungs of smokers: a candidate gene-based study. Cancer Research. 2012;72:707-715
38. Nie W, Zang YS, Chen JQ. et al. TERT rs2736100 polymorphism contributes to lung cancer risk: a meta-analysis including 49,869 cases and 73,464 controls. Tumor Biol. 2014;35:5569-5574
39. Yang JH, Jiao SC. Increased lung cancer risk associated with the TERT rs2736100 polymorphism: an updated meta-analysis. Tumor Biol. 2014;35:5763-5769
40. Wang HM, Zhang XY, Jin B. TERT Genetic Polymorphism rs2736100 Was Associated with Lung Cancer: A Meta-Analysis Based on 14,492 Subjects. Genet Test Mol Bioma. 2013;12:937-941
41. Codd V, Nelson CP, Albrecht E. et al. Identification of seven loci affecting mean telomere length and their association with disease. Nat Genet. 2013;45:422e427
42. Lan Q, Cawthon R, Gao Y. et al. Longer telomere length in peripheral white blood cells is associated with risk of lung cancer and the rs2736100 (CLPTM1L-TERT ) polymorphism in a prospective cohort study among women in China. PLoS One. 2013;8:e59230
43. Machiela MJ, Hsiung CA, Shu XO. et al. Genetic variants associated with longer telomere length are associated with increased lung cancer risk among never-smoking women in Asia: a report from the female lung cancer consortium in Asia. Int J Cancer. 2015;137:311e319
44. Zou P, Gu A, Ji G. et al. The TERT rs2736100 polymorphism and cancer risk: a meta-analysis based on 25 case-control studies. BMC Cancer. 2012;12:7
45. Wei R, Cao L, Pu H. et al. TERT polymorphism rs2736100-C is associated with EGFR mutation-positive non-small cell lung cancer. Clin Cancer Res. 2015;21:5173
Author contact
![]() Corresponding authors: Wei Li, M.D., Ph.D., Cancer Center, First Affiliated Hospital, Jilin University, Changchun, Jilin 130061, P.R. China, e-mail: jdyylwcom or Ji-Fan Hu, M.D., Ph.D., Palo Alto Veterans Institute for Research, Palo Alto, CA. 94304, USA, Tel: 650-493-5000, x63175, Fax: 650-725-7085, e-mail:Jifanedu.
Corresponding authors: Wei Li, M.D., Ph.D., Cancer Center, First Affiliated Hospital, Jilin University, Changchun, Jilin 130061, P.R. China, e-mail: jdyylwcom or Ji-Fan Hu, M.D., Ph.D., Palo Alto Veterans Institute for Research, Palo Alto, CA. 94304, USA, Tel: 650-493-5000, x63175, Fax: 650-725-7085, e-mail:Jifanedu.

 Global reach, higher impact
Global reach, higher impact