3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(10):1258-1264. doi:10.7150/jca.14869 This issue Cite
Review
LncRNAs and Esophageal Squamous Cell Carcinoma - Implications for Pathogenesis and Drug Development
1. Computational Systems Biology Lab, Department of Biochemistry and Molecular Biology, University of Georgia, Athens, GA, USA.
2. Department of Bioinformatics, Shantou University Medical College, Shantou 515041, China.
Received 2016-1-2; Accepted 2016-4-26; Published 2016-6-23
Abstract
LncRNAs are a group of ncRNA species longer than 200 nt, which have fundamental regulatory roles in diverse cellular processes and diseases progression. Esophageal cancer is a serious malignancy with respect to prognosis and mortality rate. It is among the five leading cancer types for the cancer deaths in males of middle age in the United States. In China, esophageal cancer is the fourth most frequently diagnosed cancer and the fourth leading cause of cancer death. The molecular mechanisms of esophageal cancer development are not fully understood, but emerging studies point out that lncRNAs may actively associate with the pathogenesis. In this review, we first provided an introduction of lncRNAs classifications. Then we focused on the recent findings on lncRNA expression and function in esophageal cancer development. Implications for pathogenesis and potential drug developments will also be discussed.
Keywords: LncRNA, Esophageal Squamous Cell Carcinoma, Pathogenesis, Drug Development.
Introduction
In recent years, transcriptomic studies have uncovered an expanding repository of noncoding RNAs (ncRNAs) [1-3]. ncRNAs are generally defined as RNA species that are transcribed but do not code for proteins. ncRNA species longer than 200 nt are named as lncRNAs. The critical regulatory roles of lncRNAs have been linked to diverse biological processes ranging from brain development, stem cell pluripotency and differentiation, to human disease [4-6].
According to their positions relative to closet protein-coding genes, lncRNAs are often classified as: (i) antisense lncRNAs which initiate in the opposite direction from the antisense strand of protein-coding genes; (ii) intronic lncRNAs that are transcribed in either direction within introns of protein-coding genes; (iii) divergent lncRNAs that arise in the opposite direction from the promoter of a protein-coding gene and (iv) long intergenic noncoding RNAs (lincRNAs) that are generated from intergenic regions between protein-coding genes [2]. Since the classification is evolving, lncRNAs can also be grouped according to other criterion, i.e. long intergenic non-coding RNAs (lincRNAs), very long intergenic non-coding RNAs (vlincRNAs), enhancer-associated RNAs (eRNAs), promoter-associated long RNAs (PALRs), natural antisense transcripts (NATs) and circular RNA (circRNAs) [7].
Accumulated investigations indicated that lncRNAs have a broad biological functional repertoire and regulate gene expression at multiple levels. For example, lncRNAs are involved in histone modifications and DNA methylation. Regulation of gene expression by lncRNAs can act both in cis and in trans. In post-transcription level, lncRNAs also modulate miRNAs biogenesis and mRNA alternative splicing [2, 8-10].
Esophageal cancer is a serious malignancy with respect to prognosis and mortality rate. It is among the five leading cancer types for the cancer deaths in males of middle age in the United States. The 5-year relative survival rate of patients with esophageal cancer is less than 20% [11]. In China, esophageal cancer is the fourth most frequently diagnosed cancer and the fourth leading cause of cancer death [12].
Esophageal cancer includes two major histologic types: esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). While the EAC has become the leading cause of esophageal cancer in the United States, ESCC is the predominant histological type in Asians [12, 13]. Thus we will focus on ESCC in this review.
The molecular mechanisms of ESCC development are not fully understood, but emerging studies have pointed out that lncRNAs may actively associate with the pathogenesis. In this review, we will focus on the recent findings on lncRNA expression and function in ESCC development (Table 1, 2 and Figure 1). Implications for pathogenesis and potential drug developments will also be discussed below.
LncRNAs in esophageal squamous cell carcinoma
Although the investigation of lncRNA in ESCC began only a few years ago, multiple evidences indicated that lncRNAs play an important role in the disease progression. First, a few lncRNAs were reported to work as oncogenes or tumor suppressors in several cell based researches; second, the expression pattern of several lncRNAs have also been associated with clinical parameters such as prognosis, stages and chemoradio-therapy resistance in ESCC patient samples. These results suggested lncRNA-based therapeutics have a great potential in ESCC treatment.
Examples of the involvement of lncRNAs in esophageal squamous cell carcinoma.
| LncRNAs | Targeting Genes | Function | PMID |
|---|---|---|---|
| MALAT1 | miR-101/miR-217 | Silencing of MALAT1 by miRNAs inhibits proliferation, migration and invasion | 25538231 |
| HOTAIR | LAMB3, JAM2, MX1, and OAS-1 | Mediating multiple downstream DNA methylation changes | 24022190 |
| HOTAIR | WIF-1 | Wnt/β-catenin signaling pathway | 24118380 |
| linc-POU3F3 | POU3F3 | CpG methylation | 24631494 |
| TUG1 | PRC2 | Proliferation and migration | 25366138 |
| ANRIL | p15,TGFβ1 | Proliferation | 24747824 |
| SOX2-OT | SOX2 | Cell cycle distribution, neural differentiation | 24105929 |
Examples of lncRNAs as diagnostic tools in esophageal squamous cell carcinoma.
| LncRNAs | Expression in cancer | Prognosis or stages | PMID |
|---|---|---|---|
| UCA1 | Up | Poor prognosis | 25550835 |
| HOTAIR | Up | Poor prognosis | 25157956 |
| SPRY4-IT1 | Up | Poor prognosis and advanced stages | 24810925 |
| CBR3-AS1 | Up | Advanced clinical stage and lymph node metastasis | 24337686 |
| FOXCUT | Up | Poor prognosis, advanced stages and metastasis | 25031703 |
| TUSC7 | Down | Poor prognosis | 23680400 |
Known mechanisms of action of lncRNA in esophageal squamous cell carcinoma. (A) LncRNAs can bind with miRNAs. On the one hand, lncRNAs work as 'miRNA sponge' to sequester miRNAs. On the other hand, these miRNAs can compete with other lncRNAs interaction targets. Thus, these interactions significantly influence both miRNAs and lncRNAs downsteam targets. (B) Linc-POU3F3 recruits EZH2 to form the PRC2 complex, which then activates DNA Methylation of POU3F3. Thus it influences the transcription of downstream genes, such as Notch signaling. Similar epigenetic control mechanism has been identified for HOTAIR, TUG and ANRIL in ESCC.
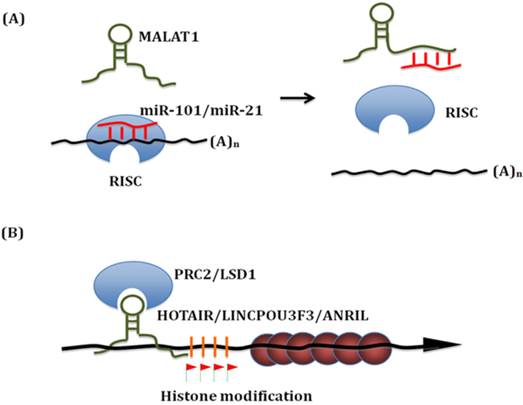
LncRNAs as oncogenes or tumor suppressors
(1). MALAT1
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1, also known as NEAT2 (nuclear-enriched abundant transcript 2)) is a highly conserved nuclear lncRNA. It is an established oncogene across many cancer types including lung cancer [14, 15], colorectal cancer [16], bladder cancer [17] and liver cancer [18]. It is usually over-expressed in cancers compared with matched normal tissues. MALAT1 is a versatile regulator since it can employ a variety of molecular mechanisms to become active with several downstream genes to finally induce migration, metastasis phenotype. For example, MALAT1 interacts with hnRNP C and B-MYB, to precisely control cell cycle [19, 20].
The proposed mechanisms of MALAT1 action include alternative splicing control and gene expression regulation. Interestingly, Wang et al. found that MALAT1 interacts with miR-101 and miR-217 to inhibit proliferation, migration and invasion in ESCC [10]. MALAT1 functions as a competing endogenous RNAs (ceRNA) to sequester miR-101 and miR-217. On the other hand, these miRNAs can also compete with other MALAT1 interaction targets. Thus, MALAT1-miR-101/miR-217 interactions significantly modulate each other's expression and influence both miRNAs and lncRNAs downstream targets. This finding elucidated a novel regulation mechanism for MALAT1 and demonstrated the involvement of MALAT1 in ESCC pathogenesis.
(2). HOTAIR1
HOX transcript antisense RNA (HOTAIR) is located within the Homeobox C (HOXC) loci on chromosome 12. It is co-expressed with HOXC gene cluster and highly expressed in multiple tumors. HOTAIR interacts with both polycomb repressive complex 2 (PRC2) and lysine specific demethylase 1 (LSD1) complexes to reprogram chromatin states. These two protein complexes recruit a variety of histone methylase and demethylase to deregulate multiple downstream genes including a set of metastasis-suppressing genes [21, 22].
Does HOTAIR function in ESCC and employ a similar mechanism in ESCC? Both in ESCC cell lines and in vivo mouse xenograft model, Li et al. found that blocking of HOTAIR down-regulates cell migration and cell metastasis. They analyzed the methylation level profiles in ESCC. The profiling data suggested that HOTAIR indeed reprogrammed the gene expression profile of ESCC cells [23]. In ESCC cells, one of the key down-steam targets is wnt inhibitory factor 1(WIF-1). HOTAIR was found to induce its histone H3K27 methylation in the promoter region thereby decreasing WIF-1 expression, and finally activated the Wnt/β-catenin signaling pathway [24]. This newly identified HOTAIR/WIF-1 axis mediates the signal transduction from chromatin change to phenotype change. Genetic studies also identified an ESCC susceptibility SNP rs920778 which locates within the HOTAIR intronic enhancer region. This allelic regulation of rs920778 on HOTAIR expression and ESCC risk clearly demonstrates the vital role of HOTAIR in ESCC progression [25].
(3). Linc-POU3F3
Li et al. recently screened 13 candidate lincRNAs in 182 pairs of ESCC tissue samples and found that linc-POU3F3 was significantly up-regulated in ESCC than nearby normal tissues. Linc-POU3F3 was predominately expressed in the nucleus and associated with the enhancer of zeste homolog 2 (EZH2), a key component of the polycomb repressive complex 2 (PRC2). Stable over-expression of linc-POU3F3 induced hypermethylated CpG islands of POU3F3 and reduced the expression of POU3F3 mRNA; whereas knockdown of linc-POU3F3 reduced the methylation at these sites and increased the levels of POU3F3 mRNA. In ESCC cells, up-regulation of linc-POU3F3 increased proliferation and ability to form colonies. ESCC cells with knockdown of linc-POU3F3 formed xenograft tumors more slowly in mice than control ESCC cells [26].
(4). TUG1
Taurine up-regulated 1(TUG1) was first identified in developing retinal cells [27]. TUG1 is known to bind the methylated and un-methylated Prc2 together with lncRNAs MALAT1/NEAT2, which works as a scaffold to mediate gene activation programs [2, 28]. In human non-small cell lung cancer, the expression of TUG1 was significantly low regulated compared with the normal control. TUG1 expression was induced by p53, which further modulates HOXB7 via epigenetic control [29]. In contrast to non-small cell lung cancer, Xu et al. provided clear evidences on TUG1 function in ESCC. In a cohort of 62 patients, TUG1 was significantly over-expressed in ESCC tissues compared with paired adjacent normal tissues. Silencing via siRNA block of TUG1 inhibits proliferation, migration, and cell cycle in ESCC cells [30]. Is TUG1 a potential oncogene or tumor suppressor? The inconsistency may indicate an alternative mechanism related with TUG1 in ESCC cells.
(5). ANRIL
CDKN2B antisense RNA 1(CDKN2B-AS1, also known as ANRIL, p15AS) is located within the CDKN2B-CDKN2A gene cluster at chromosome 9p21. ANRIL is transcribed in the opposite direction from this gene cluster. Similar to HOTAIR1, ANRIL can redirect protein complex-1 (PRC1) and -2 (PRC2) to specific genomic loci, thus inhibiting gene expression, regulating cancer invasion and metastasis thereby. This locus has been associated with cardiovascular disease and several cancers [31]. It is abnormally expressed in gastric cancer [32], primary myelofibrosis [33] and ESCC [34]. In human gastric cancer, E2F1 activates ANRIL expression, which then binds to the promoter region of miR-99a/miR-449a together with PRC2. Silencing of miR-449a expression leads to the release of E2F1, which forms a positive feedback loop and continues to promote gastric cancer cell proliferation [32]. In ESCC, inhibition of ANRIL was found to promote the expression of p15 (INK4b) and transforming growth factor β1 (TGFβ1), and block of ANRIL in ESCC cell lines reduces cellular proliferation [34].
(6). SOX2-OT
SOX2 overlapping transcript (SOX2-OT) locates in 3q26.33 locus, which has been demonstrated in vertebrate development [35]. Importantly, the established pluripotency state modulator, SOX2 located within the third intron of SOX2OT. Recently Shahryari et al. examined the expression pattern of SOX2OT and SOX2 in paired tumor/non-tumor samples of ESCC and NTERA2 (NT2), a human embryonal carcinoma (EC) stem cell line. In both circumstances, they found SOX2OT was correlated with SOX2, and co-upregulated in tumor samples or ESCC cells. Furthermore, they showed that blocking SOX2OT significantly altered the cell cycle, and SOX2OT was reduced upon the neural differentiation in NT2 Cell Line. Collectively, these evidences suggested lncRNA SOX2OT can work with its cousin transcriptional factor SOX2, to module cancer stem cell differentiation [36].
LncRNAs as diagnostic tools
Accumulated evidences indicate that lncRNA is usually expressed in specific cells or tissues. This unique feature suggests that their expression patterns could successfully be used for accurate cancer diagnostics and subtype classification. Increasing studies have found the profilings of multiple lncRNAs are correlated with prognosis, stages and chemoradio-therapy resistance in ESCC.
(1). MALAT-1
MALAT-1 is known as a biomarker for lung cancer and hepatocellular carcinoma. Upregulation of MALAT-1 predicts metastasis, limited survival and poor prognosis [15, 37]. In a recent investigation, Yao et al. found the upregulation of MALAT-1 was also associated with a decreased survival rate in patients with esophageal squamous cell carcinoma. Inhibition of MALAT-1 affected multiple cellular functions such as apoptosis, migration/invasion, colony formation and cell cycle arrest [38].
(2). HOTAIR
Since HOTAIR could recruit epigenetic protein complex to induce genome-wide reprogramming chromatin status and deregulate multiple downstream genes, it is involved in the development and progression of many types of cancers. Recently, a meta-analysis including nineteen studies with a total of 2,033 patients suggested that over-expression of HOTAIR can serve as a biomarker for poor prognosis in different types of cancers in both Asian and Western countries. Specifically, high HOTAIR expression was significantly associated with TNM stage in esophageal carcinoma [39]. Up-regulation of the long non-coding RNA HOTAIR also predicted the cancer metastasis and poor prognosis [23, 40].
(3). UCA1
LncRNA UCA1 is upregulated in bladder cancer [41, 42], breast cancer [43], tongue squamous cell carcinomas [44], and ESCC [45]. Higher UCA1 expression correlated with an advanced clinical stage and a poorer prognosis [45]. Its downstream targets include cAMP response element-binding protein (CREB) [46], p27 (Kip1) [43] and hexokinase 2 (HK2) [47] that modulate a variety of cellular processes including cell cycle and glycolysis.
(4). TUSC7
Previously, low expression of TUSC7 (also known as LOC285194) was associated with poor prognosis in colorectal cancer. Consistent with this finding, Tong et al. recently found that among patients treated with preoperative chemo-radiotherapy followed by surgical resection, the pathological complete response rate was significantly higher in patient groups with high LOC285194 expression, than patient groups with low LOC285194 expression. This indicated that lncRNA LOC285194 is a tumor suppressor associated with chemo-radiotherapy resistance and poor prognosis in ESCC [48].
(5). SPRY4-IT1
In melanoma, high expression of SPRY4-IT1 inhibits apoptosis and promotes invasion [49, 50]. SPRY4-IT1 is upregulated in ESCC tissues compared to the adjacent non-tumor tissues. Higher SPRY4-IT1 expression predicts advanced clinical stage and poorer prognosis [51].
(6). CBR3-AS1
Previously Cui et al. found that CBR3-AS1 was significantly elevated in human CaPs compared with normal tissues and benign prostatic hyperplasia (BPH). Inhibition of PlncRNA-1 led to blocking of proliferation and induction of apoptosis in CaP cell lines LNCaP and LNCaP-AI [52]. CBR3-AS1 is significantly higher in human ESCC and correlates with advanced clinical stage and lymph node metastasis [53].
(7). FOXCUT
FOXCUT and adjacent coding genes, the Fork head box C1 (FOXC1) gene, are over-expressed in both oral squamous cell carcinoma and ESCC [54, 55]. Notably, up-regulated FOXCUT and FOXC1 expression levels were correlated with advanced lymph node classification, metastasis and worse prognosis [55].
LncRNA therapeutics
As our understanding of lncRNAs in ESCC pathogenesis has accumulated rapidly, it also opens up the possibility to use them as anti-cancer therapeutic targets. SiRNAs, antisense oligonucleotides (ASOs) as well as ribozymes or deoxyribozymes, can be designed to silence endogenous lncRNAs. Conversely, synthetically modified lncRNA mimics or plasmid delivery may be used to alleviate beneficial lncRNAs in vivo [56, 57]. Although lncRNA-based therapeutic drugs are still in preclinical studies, the potential use of lncRNAs for therapies is tremendous and promising. For example, MALAT1 was found to be highly expressed in a variety of cancers, and was involved in cancer development. In a MALAT1 knockout lung tumor model, Diederichs and co-workers demonstrated that blocking MALAT1 with antisense oligonucleotides prevented metastasis formation after tumor implantation [14]. Thus, targeting MALAT1 provides a promising therapeutic approach to prevent cancer metastasis.
Perspective and Discussion
In the past decades, considerable interests have focused on the vital role of lncRNA in cancers and other diseases. In this review, we outline several novel lncRNA players in ESCC initiation, development, metastasis and their potential use in clinical applications. As of November, 2015, there are ~300 experimentally verified eukaryotic lncRNAs collected in the lncRNAdb v2.0 database [58]. Among them, only a small proportion has been examined for their biological function in ESCC. We proposed that these functional lncRNAs should be carefully examined combined with microarray and RNA-seq based profiling data. Furthermore, lncRNA genes are prevalent in eukaryotic genomes; therefore, it is likely that other ESCC associated lncRNAs will continually be found in the near future.
In this review, we identified that lncRNAs mainly adopt two molecular mechanisms in ESCC. Firstly and mainly, they act as a signal and guide to recruit chromatin regulator PRC1/2 complex to modulate nearby or distant downstream genes. Another case can be seen in the MALAT example, where they work as decoys to titer the endogenous miRNAs. Besides chromatin regulator and decoy mechanism, lncRNAs actually possesses a rich palette of regulatory routes and are considered as a versatile regulator, therefore there will be a large chance that more and more novel molecular mechanisms will surely be identified in ESCC. For example, traditionally, lncRNAs were thought to play the regulatory function of these transcripts in the nucleus. But a recent study found that lncRNAs are located in all subcellular locations. For example, the ribosome is enriched with lncRNAs H19 and TUG1[59]. Considering TUG1 is highly expressed in ESCC samples and is a key modulator in ESCC pathogenesis, it would be interesting to find alternative molecular mechanisms in ESCC.
Regarding the clinical use of lncRNA-targeted strategies, there are still unsolved technical barriers currently. These include relative large size, complex secondary structure and possible degradation by nucleases which hamper the efficient design, synthesis and delivery of functional therapeutic agents to targeting sites. Furthermore, safety evaluation of possible toxicity and unexpected side effects should be taken into consideration in the development of lncRNA-based therapeutics. Nevertheless, a detailed deciphering of the crucial role of lncRNAs in ESCC tumorigenesis will eventually facilitate the design of new generation of lncRNA therapeutics, which will complement traditional therapy to have profound clinical implications.
Acknowledgements
This work was supported by the National High Technology Research and Development Program of China (863 Program, 2014AA021102) and by the Department of Education, Guangdong Government under the Top-tier University Development Scheme for Research and Control of Infectious Diseases (2015017) and the National Natural Science Foundation of China (No. 61502292).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X. et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503-10
2. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145-66
3. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904-14
4. Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB. et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433-45
5. Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012;13:528-41
6. Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3-7
7. Di Gesualdo F, Capaccioli S, Lulli M. A pathophysiological view of the long non-coding RNA world. Oncotarget. 2014;5:10976-96
8. Li Q, Su Z, Xu X, Liu G, Song X, Wang R. et al. AS1DHRS4, a head-to-head natural antisense transcript, silences the DHRS4 gene cluster in cis and trans. Proc Natl Acad Sci U S A. 2012;109:14110-5
9. Soreq L, Guffanti A, Salomonis N, Simchovitz A, Israel Z, Bergman H. et al. Long non-coding RNA and alternative splicing modulations in Parkinson's leukocytes identified by RNA sequencing. PLoS Comput Biol. 2014;10:e1003517
10. Wang X, Li M, Wang Z, Han S, Tang X, Ge Y. et al. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 2015;290:3925-35
11. Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9-29
12. Lin Y, Totsuka Y, He Y, Kikuchi S, Qiao Y, Ueda J. et al. Epidemiology of esophageal cancer in Japan and China. J Epidemiol. 2013;23:233-42
13. Napier KJ, Scheerer M, Misra S. Esophageal cancer: A Review of epidemiology, pathogenesis, staging workup and treatment modalities. World J Gastrointest Oncol. 2014;6:112-20
14. Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G. et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180-9
15. Schmidt LH, Spieker T, Koschmieder S, Schaffers S, Humberg J, Jungen D. et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6:1984-92
16. Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z. et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014;111:736-48
17. Zhang X, Wu D, Chen L, Li X, Yang J, Fan D. et al. RAID: a comprehensive resource for human RNA-associated (RNA-RNA/RNA-protein) interaction. RNA. 2014;20:989-93
18. Wang J, Wang H, Zhang Y, Zhen N, Zhang L, Qiao Y. et al. Mutual inhibition between YAP and SRSF1 maintains long non-coding RNA, Malat1-induced tumourigenesis in liver cancer. Cell Signal. 2014;26:1048-59
19. Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X. et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9:e1003368
20. Yang F, Yi F, Han X, Du Q, Liang Z. MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Lett. 2013;587:3175-81
21. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071-6
22. Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F. et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689-93
23. Li X, Wu Z, Mei Q, Li X, Guo M, Fu X. et al. Long non-coding RNA HOTAIR, a driver of malignancy, predicts negative prognosis and exhibits oncogenic activity in oesophageal squamous cell carcinoma. Br J Cancer. 2013;109:2266-78
24. Ge XS, Ma HJ, Zheng XH, Ruan HL, Liao XY, Xue WQ. et al. HOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates Wnt pathway. Cancer Sci. 2013;104:1675-82
25. Song Y, Li L, Ou Y, Gao Z, Li E, Li X. et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509:91-5
26. Li W, Zheng J, Deng J, You Y, Wu H, Li N. et al. Increased levels of the long intergenic non-protein coding RNA POU3F3 promote DNA methylation in esophageal squamous cell carcinoma cells. Gastroenterology. 2014;146:1714-26 e5
27. Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol. 2005;15:501-12
28. Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD. et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773-88
29. Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH. et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5:e1243
30. Xu Y, Wang J, Qiu M, Xu L, Li M, Jiang F. et al. Upregulation of the long noncoding RNA TUG1 promotes proliferation and migration of esophageal squamous cell carcinoma. Tumour Biol. 2015;36:1643-51
31. Congrains A, Kamide K, Ohishi M, Rakugi H. ANRIL: Molecular Mechanisms and Implications in Human Health. Int J Mol Sci. 2013;14:1278-92
32. Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L. et al. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5:2276-92
33. Pennucci V, Zini R, Norfo R, Guglielmelli P, Bianchi E, Salati S. et al. Abnormal expression patterns of WT1-as, MEG3 and ANRIL long non-coding RNAs in CD34+ cells from patients with primary myelofibrosis and their clinical correlations. Leuk Lymphoma. 2015;56:492-6
34. Chen D, Zhang Z, Mao C, Zhou Y, Yu L, Yin Y. et al. ANRIL inhibits p15(INK4b) through the TGFbeta1 signaling pathway in human esophageal squamous cell carcinoma. Cell Immunol. 2014;289:91-6
35. Amaral PP, Neyt C, Wilkins SJ, Askarian-Amiri ME, Sunkin SM, Perkins AC. et al. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA. 2009;15:2013-27
36. Shahryari A, Rafiee MR, Fouani Y, Oliae NA, Samaei NM, Shafiee M. et al. Two novel splice variants of SOX2OT, SOX2OT-S1, and SOX2OT-S2 are coupregulated with SOX2 and OCT4 in esophageal squamous cell carcinoma. Stem Cells. 2014;32:126-34
37. Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM. et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031-41
38. Yao W, Bai Y, Li Y, Guo L, Zeng P, Wang Y. et al. Upregulation of MALAT-1 and its association with survival rate and the effect on cell cycle and migration in patients with esophageal squamous cell carcinoma. Tumour Biol. 2015
39. Zhang S, Chen S, Yang G, Gu F, Li M, Zhong B. et al. Long noncoding RNA HOTAIR as an independent prognostic marker in cancer: a meta-analysis. PLoS One. 2014;9:e105538
40. Chen FJ, Sun M, Li SQ, Wu QQ, Ji L, Liu ZL. et al. Upregulation of the long non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinog. 2013;52:908-15
41. Wang F, Li X, Xie X, Zhao L, Chen W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 2008;582:1919-27
42. Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW, Li MQ. et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res. 2006;12:4851-8
43. Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M. et al. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis. 2014;5:e1008
44. Fang Z, Wu L, Wang L, Yang Y, Meng Y, Yang H. Increased expression of the long non-coding RNA UCA1 in tongue squamous cell carcinomas: a possible correlation with cancer metastasis. Oral Surg Oral Med Oral Pathol Oral Radiol. 2014;117:89-95
45. Li JY, Ma X, Zhang CB. Overexpression of long non-coding RNA UCA1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:7938-44
46. Yang C, Li X, Wang Y, Zhao L, Chen W. Long non-coding RNA UCA1 regulated cell cycle distribution via CREB through PI3-K dependent pathway in bladder carcinoma cells. Gene. 2012;496:8-16
47. Li Z, Li X, Wu S, Xue M, Chen W. Long non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase 2 through the mTOR-STAT3/microRNA143 pathway. Cancer Sci. 2014;105:951-5
48. Tong YS, Zhou XL, Wang XW, Wu QQ, Yang TX, Lv J. et al. Association of decreased expression of long non-coding RNA LOC285194 with chemoradiotherapy resistance and poor prognosis in esophageal squamous cell carcinoma. J Transl Med. 2014;12:233
49. Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS. et al. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852-62
50. Mazar J, Zhao W, Khalil AM, Lee B, Shelley J, Govindarajan SS. et al. The functional characterization of long noncoding RNA SPRY4-IT1 in human melanoma cells. Oncotarget. 2014;5:8959-69
51. Xie HW, Wu QQ, Zhu B, Chen FJ, Ji L, Li SQ. et al. Long noncoding RNA SPRY4-IT1 is upregulated in esophageal squamous cell carcinoma and associated with poor prognosis. Tumour Biol. 2014;35:7743-54
52. Cui Z, Ren S, Lu J, Wang F, Xu W, Sun Y. et al. The prostate cancer-up-regulated long noncoding RNA PlncRNA-1 modulates apoptosis and proliferation through reciprocal regulation of androgen receptor. Urol Oncol. 2013;31:1117-23
53. Wang CM, Wu QQ, Li SQ, Chen FJ, Tuo L, Xie HW. et al. Upregulation of the long non-coding RNA PlncRNA-1 promotes esophageal squamous carcinoma cell proliferation and correlates with advanced clinical stage. Dig Dis Sci. 2014;59:591-7
54. Kong XP, Yao J, Luo W, Feng FK, Ma JT, Ren YP. et al. The expression and functional role of a FOXC1 related mRNA-lncRNA pair in oral squamous cell carcinoma. Mol Cell Biochem. 2014;394:177-86
55. Pan F, Yao J, Chen Y, Zhou C, Geng P, Mao H. et al. A novel long non-coding RNA FOXCUT and mRNA FOXC1 pair promote progression and predict poor prognosis in esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:2838-49
56. Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov. 2013;12:433-46
57. Mizrahi A, Czerniak A, Levy T, Amiur S, Gallula J, Matouk I. et al. Development of targeted therapy for ovarian cancer mediated by a plasmid expressing diphtheria toxin under the control of H19 regulatory sequences. J Transl Med. 2009;7:69
58. Quek XC, Thomson DW, Maag JL, Bartonicek N, Signal B, Clark MB. et al. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015;43:D168-73
59. van Heesch S, van Iterson M, Jacobi J, Boymans S, Essers PB, de Bruijn E. et al. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 2014;15:R6
Author contact
![]() Corresponding author: Jianzhen Xu. Ph.D. E-mail: jzxu01edu.cn or xujz0451com Phone: +86-754-88900491.
Corresponding author: Jianzhen Xu. Ph.D. E-mail: jzxu01edu.cn or xujz0451com Phone: +86-754-88900491.

 Global reach, higher impact
Global reach, higher impact