3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(1):57-64. doi:10.7150/jca.16723 This issue Cite
Research Paper
Lentiviral CRISPR/Cas9 vector mediated miR-21 gene editing inhibits the epithelial to mesenchymal transition in ovarian cancer cells
1. Department of Pathology and Laboratory Medicine
2. Center for Cancer Research
3. Department of Physiology
4. Department of Ophthalmology, University of Tennessee Health Science Center, Memphis, TN, USA
5. Henan Agricultural University
6. Henan University of Animal Husbandry and Economy, P.R. China
7. Department of Women's Health Educational System
8. Department of Gynecology, Hokkaido University School of Medicine, Hokkaido University, Sapporo, Japan.
Received 2016-7-4; Accepted 2016-10-29; Published 2017-1-1
Abstract
CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats) mediated genome editing is a powerful approach for loss of function studies. Here we report that lentiviral CRISPR/Cas9 vectors are highly efficient in introducing mutations in the precursor miRNA sequence, thus leading to the loss of miRNA expression and function. We constructed four different lentiviral CRISPR/Cas9 vectors that target different regions of the precursor miR-21 sequence and found that these lentiviral CRISPR/Cas9 miR-21 gRNA vectors induced mutations in the precursor sequences as shown by DNA surveyor mutation assay and Sanger sequencing. Two miR-21 lentiviral CRISPR/Cas9 gRNA vectors were selected to probe miR-21 function in ovarian cancer SKOV3 and OVCAR3 cell lines. Our data demonstrate that disruption of pre-miR-21 sequences leads to reduced cell proliferation, migration and invasion. Moreover, CRISPR/Cas9-mediated miR-21 gene editing sensitizes both SKOV3 and OVCAR3 cells to chemotherapeutic drug treatment. Disruption of miR-21 leads to the inhibition of epithelial to mesenchymal transition (EMT) in both SKOV3 and OVCAR3 cells as evidenced by the upregulation of epithelial cell marker E-cadherin and downregulation of mesenchymal marker genes, vimentin and Snai2. The miR-21 target genes PDCD4 and SPRY2 were upregulated in cells transduced with miR-21gRNAs compared to controls. Our study indicates that lentiviral CRISPR/Cas9-mediated miRNA gene editing is an effective approach to address miRNA function, and disruption of miR-21 inhibits EMT in ovarian cancer cells.
Keywords: miR-21, CRISPR/Cas9, lentiviral vector, ovarian cancer, EMT.
Introduction
MicroRNAs (miRNAs) are a class of non-coding endogenous small RNAs that negatively regulate gene expression at the post-transcriptional level. miRNA genes are transcribed into primary miRNAs by RNA polymerase II in the nucleus and then processed into precursor miRNAs (pre-miRNAs) by the microprocessor complex composed of the RNAase III enzyme Drosha and double-stranded RNA binding protein DGCR8. Mature miRNA is processed in the cytoplasm by RNA induced silencing complex (RISC), which includes RNAase III enzyme Dicer, Ago2, and other RNA binding proteins. miRNAs function by binding 3' untranslated region (3'UTR) of their target genes through complementarity in their seed sequences [1]. Dysregulation of miRNA expression is associated with various human diseases, including cancer and cardiovascular diseases [2, 3].
Gain and loss of function approaches have been utilized to study the roles of miRNAs in vitro and in vivo. The gain of function approach can be easily attained by overexpressing primary miRNAs using plasmid and viral vectors or introducing miRNA mimics. Several loss of function approaches have been used to silence miRNA expression, including plasmid or viral vector based antagomiRNA, miRNAoff and decoymiRNA, or synthesized antisense miRNA inhibitors. miRNA expression is partially silenced using these methods. However, CRISPR/Cas9 is a novel approach for loss of function studies by disrupting miRNA genes through introducing mutations in pre-miRNA sequences and disrupting miRNA expression. To test whether lentiviral CRISPR/Cas9 vector could disrupt miRNA expression, we selected miR-21 for further investigation in ovarian cancer, since it is one of the most consistently upregulated miRNAs in cancer, and contributes to tumor metastasis and chemoresistance. However, how miR-21 contributes to tumor metastasis and chemoresistance in ovarian cancer is still not well-understood. Epithelial to mesenchymal transition (EMT) is associated with tumor metastasis and chemoresistance. Ovarian cancer metastasis is different from other cancers due to tumor spreading through peritoneal fluid and directly invading adjacent organs through routes other than the blood [4]. The role of EMT in ovarian cancer metastasis and chemoresistance has been recently recognized [5-7], and is associated with poor patient survival in ovarian cancer [6, 8, 9]. To determine whether miR-21 contributes to EMT in ovarian cancer cells, miR-21 expression was disrupted by transducing ovarian SKOV3 and OVCAR3 cancer cells with four different miR-21 gRNA lentiviral CRISPR/Cas9 vectors that target different regions of the pre-miRNA sequence. We found that all gRNAs can efficiently introduce mutations in the targeted sequences. We further showed that loss of miR-21 leads to inhibition of cell proliferation, migration, invasion and sensitizes cells to chemotherapy drug treatment. Furthermore, for the first time we showed that miR-21 loss leads to EMT inhibition in ovarian cancer cells.
Materials and Methods
Cell culture- The ovarian cancer cell lines SKOV3 and OVCAR3 were obtained from ATCC and cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% FBS (Hyclone; Logan, UT), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen; Carlsbad, CA). HEK293 FT cells were cultured in DMEM supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 1% glutamine, 1% nonessential amino acids, and geneticin at a final concentration of 1 μg/ml.
Lentiviral vector production- The lentiviral CRISPR/Cas9 mediated miR-21 gene editing vectors were constructed by annealing four gRNA oligonucleotide pairs and subcloning them in the BsmII sites of lentiviral vector pLenti CRISPR V2. A control vector was constructed by inserting EGFP gRNA sequences into pLenti CRISPR V2 lentiviral vector. All gRNA sequences were selected from the Human GeCKOv2 CRISPR knockout pooled library [10] and primers for detecting miR-21 mutations in surveyor mutation assay are listed in Table 1. Lentivirus were produced by packaging in 293FT cells as we published previously [11]. Stable cell lines were generated by transducing the SKOV3 and OVCAR3 cells with the lentiviral CRISPR/Cas9 miR-21 gene editing vectors and selected with 5 μg/ml puromycin.
gRNA and primer sequences.
| gRNAs/Primers | Sequences |
|---|---|
| EGFP gRNA | GGGCGAGGAGCTGTTCACCG |
| miR-21 gRNA1 | CTCATGGCAACACCAGTCGA |
| miR-21 gRNA2 | GTCTGATAAGCTACCCGACA |
| miR-21 gRNA3 | ATGTCAGACAGCCCATCGAC |
| miR-21 gRNA4 | ATGTTGACTGTTGAATCTCA |
| miR-21 Forward | GGGGATTTCTTGGTTTGTGAA |
| miR-21 Reverse | ATACAGCTAGAAAAGTCCCTGAAAA |
Surveyor mutation assay- Genomic DNA was extracted from ovarian OVCAR3 cancer cells transduced with miR-21 lentiviral CRISPR/Cas9 and control vectors. PCR was performed by amplifying the mutated region using primers pri-miR-21F and pri-miR-21R as listed in Table 1. PCR product was denatured and reannealed using the PCR program: 95ºC denature for 5min, ramp down to 85ºC at -2ºC/s and then ramp down to 25ºC at -0.1ºC/s; hold at 4ºC. Afterwards, 10 units of T7 endonuclease I was added, incubated at 37ºC for 30 min, and the reaction was stopped by adding 2 ul of 0.25M EDTA and then visualized on a 1.2% agarose gel.
PolyA tailing real time (RT)-PCR- miR-21 expression was detected in ovarian cancer SKOV3 and OVCAR3 cells by polyA tailing real time RT-PCR as we published previously [11]. Briefly, total RNA was extracted from lentiviral CRISPR/Cas9 vector transduced ovarian cancer cells. Following genomic DNA digestion with DNAase, polyA tail was added to total RNA with polyA polymerase and then RT-PCR was performed. Human noncoding RNA U6 was used as a normalization control for the miR-21 expression using the formula: 2 Ct (miR-21-U6). Primers for detecting mature miR-21 and human U6 noncoding RNA as internal control were listed in our previous publication [11].
MTT assay- SKOV3 or OVCAR3 Cells (8000 per well) transduced with lentiviral CRISP/Cas9 for miR-21 editing and control vectors were plated into 96-well plates and cultured for different time points. Thereafter, 10 µl of MTT reagent was added to each well and incubated for ~4 h and then terminated by adding 100 µl detergent reagent to incubate at 22°C in the dark for 2 h. Cell proliferation was assessed by measuring the absorbance at 570 nm wavelength.
Cell migration assay -The cell migration assay was performed using a modified transwell chamber (BD Falcon™, San Jose, CA). These chambers were inserted into 24-well cell culture plates. SKOV3 or OVCAR3 Cells transduced with lentiviral miR-21 gRNA and control vectors (3 × 104) in 300 µl serum-free DMEM were added to the upper chamber. 10% DMEM (serving as the chemoattractant) was added into the lower chamber of each well and incubated for 24 hr. The medium and non-migrated cells in the upper chamber were removed, while the migrated cells in the lower side of the membranes were fixed with methanol and stained with crystal violet. Pictures were taken at 10X magnification, and cell number from at least three different fields were counted.
Cell invasion assay- SKOV3 and OVCAR3 (5 × 105) cells transduced with lentiviral miR-21 gRNA and control vectors were seeded in serum-free DMEM onto inserts precoated with Matrigel (BD BioCoatTM using 24-well Tumor Invasion System (BD BioSciences, San Jose, CA). DMEM containing 10% FBS was added to the bottom chamber of the invasion system as the chemoattractant. The transwell inserts were stained for 5 mins with hematoxylin and eosin following methanol fixation for 20 mins following overnight incubation. Pictures were taken at 10X magnification. Invaded cells were counted at least in three different fields.
Immunofluorescent staining- To detect EMT marker gene expression, SKOV3 cells transduced with lentiviral miR-21 gRNA and control vectors were fixed onto glass slides for 10 min using 4% PFA, washed three times with 0.1% Tween-20 in PBS (PBST), and incubated with blocking buffer (5% normal goat serum, 3% bovine serum albumin, and 0.1% Triton-X100 in PBS) for 1 hr. The slides were incubated overnight with primary antibodies to E-cadherin and vimentin (1:200 dilution, Cell Signaling, Danvers, MA). After rinsing three times for 5 min with PBST, Alexa 488 or 594 conjugated goat anti-rabbit or mouse (1:200 dilution, Life Technologies) antibodies were added for 1 hr at room temperature. Cell nuclei were counterstained with DAPI (Vector Laboratories, Inc.; Burlingame, CA). Images were captured on a Zeiss LSM700 laser scanning confocal microscope.
Cell apoptosis- Stable SKOV3 and OVCAR3 cancer cell lines established with lentiviral miR-21 gRNA and control vectors were treated with the chemotherapy drug paclitaxel at different doses for 24 hrs. Apoptosis was measured by using a caspase3/7 activity assay kit (Promega). Cell apoptosis was also detected in SKOV3 cells transduced with lentiviral CRISPR/Cas9 mediated miR-21 gRNAs and control vectors using immunofluorescent staining with active-caspase 3 antibody. Cell nuclei were counterstained with propidium iodide.
Western blot- Ovarian cancer cells were collected in RIPA buffer (Thermo Scientific; Rockford, IL) containing 1% Halt Proteinase Inhibitor Cocktail (Thermo Scientific; Rockford, IL). An equal amount of protein (40 µg/lane) was loaded onto 10% SDS-PAGE gels and transferred onto nitrocellulose membranes. The membranes were blocked with 5% non-fat milk for 1 hr and incubated with primary antibodies against PDCD4 (Cell Signaling), GAPDH (Sigma; St. Louis, MO), vimentin, E-cadherin, or snail2 (Cell Signaling).
Statistical analysis- Significant differences were determined from at least two independent experiments performed in triplicate and presented as means ± S.D. using Student's t-test. p < 0.05 was considered significant.
Results
Lentiviral CRISPR/Cas9 is highly efficient in introducing mutations in the precursor miR-21 sequences
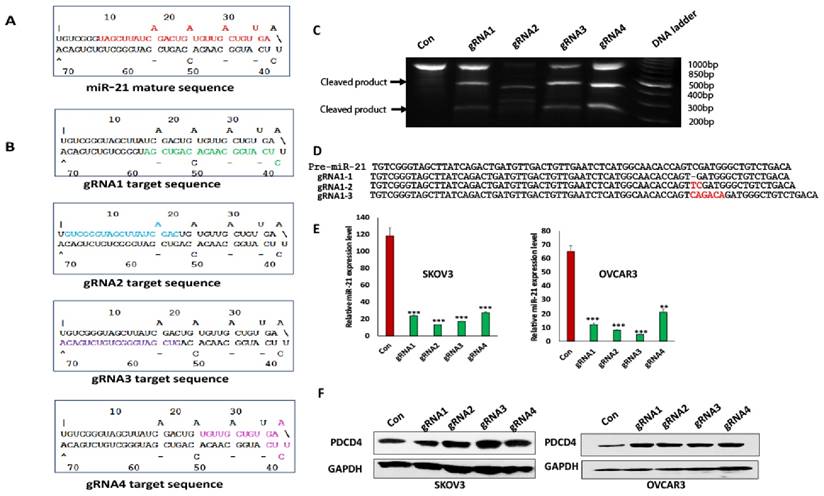
To test whether lentiviral CRISPR/Cas9 vector efficiently disrupts miRNA function, we transduced ovarian cancer OVCAR3 cells with four different miR-21 and control CRISPR/Cas9 lentiviral vectors. Mature miR-21 and all four miR-21 gRNA sequences are indicated in the pre-miR-21 hairpin structure (Fig.1A, B) with miR-21 gRNA1, 2 and 3 targeting the stem of hairpin, while gRNA4 targeting the hairpin loop. Following puromycin selection, we extracted genomic DNA and performed surveyor mutation assay. Compared to control cells, all four lentiviral CRISPR miR-21 gRNA vectors induced mutations in the targeted region (Fig.1C). gRNA targeted region was amplified by PCR and cloned into PCR2.1 vector for sequencing. We found that lentiviral miR-21 CRISPR/Cas9 vector induced mutations, including several base pair deletions and insertions. Sequences from three individual clones from lentiviral CRISPR/Cas9 miR-21 gRNA1 transduced cells are shown in Fig.1D. miR-21 expression was detected with quantitative real-time RT-PCR as we published previously [11] and was significantly reduced with all four gRNA transduced cells compared to control (Fig.1E). The miR-21 target gene PDCD4 was upregulated in ovarian cancer SKOV3 and OVCAR3 cells transduced with all of miR-21 gRNAs compared to control (Fig.1F). Our data demonstrated that lentiviral CRISPR/Cas9 is highly effective in abrogating miR-21 expression by introducing mutations in pre-miRNA hairpin sequences in ovarian cancer cells.
Lentiviral CRISPR/Cas9 mediated miR-21 gene editing leads to the inhibition of cell proliferation, migration and invasion in ovarian cancer cells
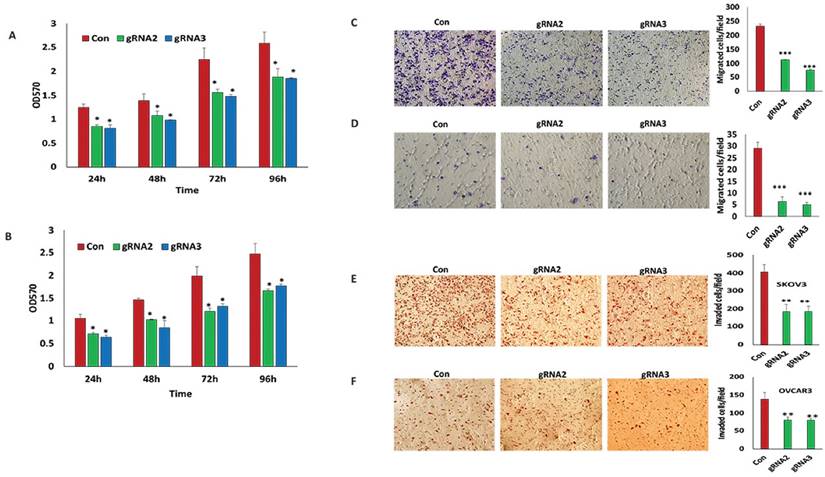
To examine whether lentiviral CRISPR/Cas9 mediated miR-21 gene editing abrogates the oncogenic activity of miR-21 in ovarian cancer cells, we further characterized SKOV3 and OVCAR3 stable cell lines transduced with two miR-21 gRNA lentiviral vectors pLentiCRISPR miR-21gRNA2 and gRNA3. Cell proliferation was determined by MTT assays in SKOV3 and OVCAR3 cells transduced with lentiviral miR-21 gRNAs and control vectors. Loss of miR-21 significantly reduced cell proliferation at all four time points (24, 48, 72 and 96 hr) compared to control transduced cells in both ovarian SKOV3 (Fig.2A) and OVCAR3 cancer cells (Fig.2B). Cell migration determined using transwell plates showed a significant reduction in cell migration in both SKOV3 (Fig.2C) and OVCAR3 (Fig.2D) cells transduced with miR-21 gRNA2 and gRNA3 compared to controls. Cell invasion assessed using matrigel coated transwells also showed a significant reduction in invasiveness of both SKOV3 (Fig.2E) and OVCAR3 cells transduced with miR-21gRNA2 and gRNA3 compared to control transduced cells (Fig. 2F).
Lentiviral CRISPR/Cas9 miR-21vector introduced mutations in the pre-miR-21 sequences. A. miR-21 pre-miRNA hairpin structure. Mature miR-21 sequences were highlighted. B. miR-21 gRNA sequences and locations in the pre-miR-21 hairpin were indicated. C. DNA surveyor mutation assay was performed to examine the mutations induced by lentiviral CRISPR/Cas9. There are three bands in gRNA1, 2, 3 and 4 including two small products cleaved by T7 endonuclease I and one uncleaved large product. In control cells, only one uncleaved band presented. D. Lentiviral CRISPR/Cas9 vector induced mutations were sequenced. E. miR-21 expression in both SKOV3 and OVCAR3 cells transduced with miR-21 different gRNA and control lentiviral vectors was examined by polyA tailing real time RT-PCR(***p<0.001). F. PDCD4 expression in both SKOV3 and OVCAR3 cells transduced with miR-21 different gRNA and control lentiviral vectors was shown by one representative Western blot.
Lentiviral CRISPR/Cas9 mediated miR-21 gene editing leads to reduced cell survival, migration and invasion. A, B. Cell proliferation was examined in SKOV3 and OVCAR3 cells transduced with miR-21gRNA2, gRNA3 and control lentiviral vectors using MTT assay (*p<0.05). C, D. Migration of SKOV3 and OVCAR3 cells transduced with lentiviral CRISPR/Cas9 miR-21 gRNA2, gRNA3 and control vectors. Cells were stained using Crystal Violet and imaged by microscopy, respectively (**p<0.01). E,F. Invaded SKOV3 and OVCAR3 cells transduced with miR-21 gRNA2, gRNA3 and control lentiviral vectors were stained with hematoxylin/eosin and imaged by microscopy (**p<0.01).
Lentiviral CRISPR/Cas9 mediated miR-21 gene editing sensitizes cell response to chemotherapy drug treatment
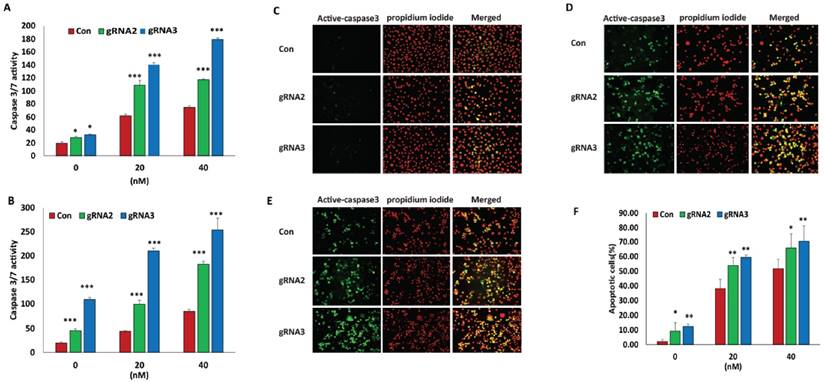
We previously found that high miR-21 expression contributes to the chemoresistance of various cancer cells [12]. To examine whether lentiviral CRISPR/Cas9 mediated miR-21 gene editing sensitizes ovarian cancer cells to the response of chemotherapy drugs, we treated both ovarian SKOV3 and OVCAR3 cancer cells transduced with lentiviral miR-21 gRNA2 and gRNA3 vectors at different doses of the chemotherapy drug, paclitaxel. Disrupting miR-21 expression in ovarian SKOV3 or OVCAR3 cancer cells transduced with both lentiviral miR-21 gRNA2 and gRNA3 vectors not only significantly induced basal cell apoptosis, but also sensitized cells to the induction of apoptosis is response to chemotherapy drug treatment (Fig.3A, B). We also performed immunofluorescent staining to examine apoptosis in SKOV3 cells transduced with lentiviral miR-21 gRNAs and control vectors using active-caspase3 antibody, which consistently showed that miR-21 loss sensitized cells to their response to the chemotherapy drug paclitaxel at different doses (Fig.3C, D, E, and F). Our data indicate that disruption of miR-21 expression enhanced the efficacy of chemotherapy drug treatment in ovarian cancer cells.
Lentiviral CRISPR/Cas9 mediated miR-21 gene editing leads to the inhibition of EMT in ovarian cancer cells
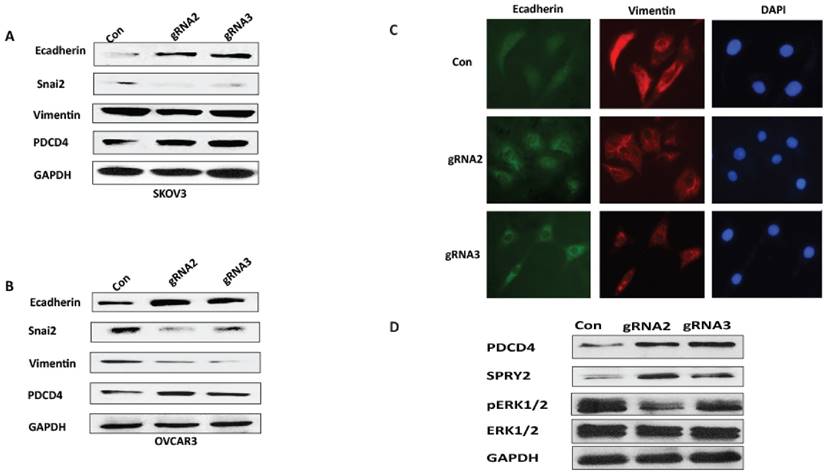
Whether miR-21 contributes to EMT in ovarian cancer is unknown. Therefore, we analyzed the expression of EMT-associated marker genes, including E-cadherin, Vimentin and Snai2, in both SKOV3 and OVCAR3 cells transduced with miR-21 gRNA2 and gRNA3 lentiviral vectors. Compared to control cells, disruption of miR-21 results in upregulation of E-cadherin expression and downregulation of Snai2 and Vimentin in both SKOV3 and OVCAR3 cells. Furthermore, expression of the well-known miR-21 target gene PDCD4 was upregulated in both SKOV3 and OVCAR3 cells transduced with lentiviral miR-21 gRNAs compared to controls (Fig.4A, B). In addition, we also performed immunostaining for the EMT markers, E-cadherin and vimentin, in SKOV3 cells transduced with miR-21 gRNAs and control lentiviral vectors. E-cadherin was upregulated whereas vimentin was downregulated in SKOV3 transduced with miR-21 gRNA vectors comparted to control cells (Fig.4C). We also found that SPRY2 is a target gene of miR-21 in ovarian cancer, which was upregulated in SKOV3 cells transduced with miR-21gRNA2 and 3 lentiviral vectors compared to control cells. The phosphorylation of ERK1/2, which is downstream of cellular survival signaling, was attenuated in SKOV3 cells transduced with lentiviral miR-21 gRNAs compared to control cells (Fig.4D). Taken together these data show that lentiviral CRISPR/Cas9 mediated miR-21 gene editing leads to inhibition of EMT in ovarian cancer cells.
Lentiviral CRISPR/Cas9 mediated miR-21 gene editing sensitized ovarian cancer cells to chemotherapy drug treatment. A, B. Cell apoptosis induced by the chemotherapy drug paclitaxel was examined in SKOV3 and OVCAR3 transduced with miR-21 gRNA2, gRNA3 and control lentiviral vectors by measuring Caspase3/7 activity (*p<0.05,***p<0.001). C, D, E. Apoptosis in SKOV3 cells transduced with control, gRNA2 and 3 lentiviral vectors was detected by immunofluorescence staining using active-caspase3 antibody with 0(C), 20(D) and 40nM(E) of paclitaxel treatment. F. Quantitation of apoptosis of SKOV3 cells transduced with control, gRNA2 and gRNA3 lentiviral vectors. Data were shown by normalizing apoptotic cells versus total cell nuclei stained with propidium iodide. At least apoptotic cells were imaged with a Nikon fluorescent microscope and counted using image J software from three different fields under 200X magnification (*p<0.05, **p<0.01).
Lentiviral CRISPR/Cas9 mediated miR-21gene editing leads to inhibition of EMT.A, B. EMT marker gene expression was examined in SKOV3 or OVCAR3 cells transduced with lentiviral CRISPR/Cas9 miR-21 gRNA2 and gRNA3 vectors using Western blot. C. SKOV3 cells transduced with miR-21gRNA and control lentiviral vectors were immunostained with EMT marker gene E-cadherin and Vimentin. D. miR-21 targeted gene SPRY2 expression was upregulated while downstream phospho-ERK1/2 was attenuated in SKOV3 cells transduced with miR-21gRNA2 and gRNA3 compared to control cells.
Discussion
The lentiviral CRISPR/Cas9 vector provides a powerful approach for disrupting not only gene expression, but also non-coding RNAs including miRNA genes. A previous study showed that a plasmid based CRISPR/Cas9 was efficient in knocking down miRNAs [13]. In this study we showed that lentiviral CRISPR/Cas9 vector is highly effective in disrupting miRNA expression. Targeting several different regions of pre-miR-21, including the stem and hairpin loop, reduced miR-21 expression in ovarian cancer cells as shown by our results with four different gRNA sequences. We analyzed the newly mutated sequences of pre-miR-21 by folding RNA secondary structure and found that the mutated sequences of pre-miR-21 are still capable of forming secondary hairpin structures (Supplementary Figure 1). Interestingly, the new hairpin structure of miR-21 is relatively stable based on predicted energy compared with wild-type miR-21 hairpin structure. There are few base pair mismatches between 5' and 3' stem of pre-miR-21 hairpin structure. However, whether this newly formed hairpin leads to new off-targets by generating some novel miRNAs will require further investigation. Our data demonstrate that miR-21 expression is significantly reduced, although low residual miR-21 expression still remains. This may reflect a small subpopulation of untransduced cells that remain in the cell cultures. The residual miR-21 expression can be depleted by further selecting through subcloning. We also found that the lentiviral CRISPR/Cas9 mediated gene editing induced different mutations including deletion and insertions in the pre-miRNA sequences of miR-21. We selected two different miR-21 gRNAs (gRNA2 and 3) that target the stem regions of pre-miR-21 hairpin for functional studies in ovarian cancer cells. However, disruption of miR-21 expression using two different gRNAs leads to similar results, indicating that lentiviral CRISPR/Cas9 mediated miR-21 is fully functional by targeting pre-miR-21 hairpin sequences.
We further verified the function of miR-21 in both ovarian cancer SKOV3 and OVCAR3 cells by characterizing stable cell lines transduced with lentiviral CRISPR/Cas9 miR-21 gRNA2 and 3 vectors. Consistent with the oncogenic properties of miR-21 reported in ovarian cancer [14, 15], CRISPR/Cas9 mediated miR-21gene editing leads to reduced cell proliferation, migration and invasion in ovarian cancer cells. Meanwhile, disruption of miR-21 expression enhanced the sensitivity of cells to the apoptotic activity of the chemotherapy drug, paclitaxel. Our study suggests that lentiviral CRISPR/Cas9 is a highly effective method to assess miRNA function.
In addition, our studies for the first time showed that loss of miR-21 leads to the inhibition of EMT in ovarian cancer cells. Our finding is consistent with several other studies showing that miR-21 promotes EMT in different cancers including breast cancer [16], cholangiocarcinoma [17], prostate cancer [18], and renal cancer [19]. Although it is unclear how miR-21 regulates EMT in ovarian cancer cells, it is possible that miR-21 may directly or indirectly regulate EMT through suppressing the expression of target genes or downstream pathways of these genes. For example, miR-21 was previously reported to target SPRY2 in glioma and colon cancers [20-22]. We found that miR-21 also targeted SPRY2 in ovarian cancer cells, which was a negative regulator of ERK1/2 cell survival pathway as shown in Figure 4D. Therefore, disruption of miR-21 inhibited EMT at least by targeting SPRY2 and subsequently attenuating ERK1/2 pathway in ovarian cancer cells. Previous studies showed that ERK1/2 is a positive regulator of EMT in various human cancer [23, 24]. TGFβ also induces EMT in a variety of human cancers. SMAD7 is another target gene of miR-21 and is a negative regulator of TGFβ pathway, which may be an additional pathway for miR-21 to promote EMT in ovarian cancer cells.
In conclusion, our study showed that the use of lentiviral CRISPR/Cas9 vectors is highly effective in disrupting miRNA expression in ovarian cancer cells. Introducing mutations in the pre-miRNA hairpin sequences, including stem and hairpin loop, disrupted miRNA expression using CRISPR/Cas9 system. We found that lentiviral CRISPR/Cas9 vector mediated miR-21 gene editing inhibited cell proliferation, migration and invasion. Furthermore, we showed for the first time that disruption of miR-21 resulted in inhibition of EMT in ovarian cancer cells, suggesting a novel role of miR-21 in contributing to tumor metastasis and chemoresistance.
Supplementary Material
Supplementary figure 1.
Acknowledgements
This study was partially supported by a grant from American Heart Association (15GRNT25000015) and West Cancer Center to Yue, J.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Du P, Wang L, Sliz P, Gregory RI. A Biogenesis Step Upstream of Microprocessor Controls miR-17 approximately 92 Expression. Cell. 2015;162:885-99
2. Piletic K, Kunej T. MicroRNA epigenetic signatures in human disease. Arch Toxicol. 2016;90:2405-19
3. Voglova K, Bezakova J, Herichova I. Micro RNAs: an arguable appraisal in medicine. Endocr Regul. 2016;50:106-24
4. Yang D, Sun Y, Hu L, Zheng H, Ji P, Pecot CV. et al. Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer Cell. 2013;23:186-99
5. Lili LN, Matyunina LV, Walker LD, Wells SL, Benigno BB, McDonald JF. Molecular profiling supports the role of epithelial-to-mesenchymal transition (EMT) in ovarian cancer metastasis. J Ovarian Res. 2013;6:49
6. Ahmed N, Abubaker K, Findlay J, Quinn M. Epithelial mesenchymal transition and cancer stem cell-like phenotypes facilitate chemoresistance in recurrent ovarian cancer. Curr Cancer Drug Targets. 2010;10:268-78
7. Yan H, Sun Y. Evaluation of the mechanism of epithelial-mesenchymal transition in human ovarian cancer stem cells transfected with a WW domain-containing oxidoreductase gene. Oncol Lett. 2014;8:426-30
8. Helleman J, Smid M, Jansen MP, van der Burg ME, Berns EM. Pathway analysis of gene lists associated with platinum-based chemotherapy resistance in ovarian cancer: the big picture. Gynecol Oncol. 2010;117:170-6
9. Yoshida S, Furukawa N, Haruta S, Tanase Y, Kanayama S, Noguchi T. et al. Expression profiles of genes involved in poor prognosis of epithelial ovarian carcinoma: a review. Int J Gynecol Cancer. 2009;19:992-7
10. Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783-4
11. Yue J, Sheng Y, Ren A, Penmatsa S. A miR-21 hairpin structure-based gene knockdown vector. Biochem Biophys Res Commun. 2010;394:667-72
12. Yang CH, Yue J, Fan M, Pfeffer LM. IFN induces miR-21 through a signal transducer and activator of transcription 3-dependent pathway as a suppressive negative feedback on IFN-induced apoptosis. Cancer Res. 2010;70:8108-16
13. Chang H, Yi B, Ma R, Zhang X, Zhao H, Xi Y. CRISPR/cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Scientific reports. 2016;6:22312
14. Chan JK, Blansit K, Kiet T, Sherman A, Wong G, Earle C. et al. The inhibition of miR-21 promotes apoptosis and chemosensitivity in ovarian cancer. Gynecol Oncol. 2014;132:739-44
15. Liu S, Fang Y, Shen H, Xu W, Li H. Berberine sensitizes ovarian cancer cells to cisplatin through miR-21/PDCD4 axis. Acta Biochim Biophys Sin (Shanghai). 2013;45:756-62
16. Wu ZH, Tao ZH, Zhang J, Li T, Ni C, Xie J. et al. MiRNA-21 induces epithelial to mesenchymal transition and gemcitabine resistance via the PTEN/AKT pathway in breast cancer. Tumour Biol. 2015
17. Liu Z, Jin ZY, Liu CH, Xie F, Lin XS, Huang Q. MicroRNA-21 regulates biological behavior by inducing EMT in human cholangiocarcinoma. Int J Clin Exp Pathol. 2015;8:4684-94
18. Coppola V, Musumeci M, Patrizii M, Cannistraci A, Addario A, Maugeri-Sacca M. et al. BTG2 loss and miR-21 upregulation contribute to prostate cell transformation by inducing luminal markers expression and epithelial-mesenchymal transition. Oncogene. 2013;32:1843-53
19. Cao J, Liu J, Xu R, Zhu X, Liu L, Zhao X. MicroRNA-21 stimulates epithelial-to-mesenchymal transition and tumorigenesis in clear cell renal cells. Mol Med Rep. 2016;13:75-82
20. Frey MR, Carraro G, Batra RK, Polk DB, Warburton D. Sprouty keeps bowel kinases regular in colon cancer, while miR-21 targets Sprouty. Cancer Biol Ther. 2011;11:122-4
21. Feng YH, Wu CL, Shiau AL, Lee JC, Chang JG, Lu PJ. et al. MicroRNA-21-mediated regulation of Sprouty2 protein expression enhances the cytotoxic effect of 5-fluorouracil and metformin in colon cancer cells. Int J Mol Med. 2012;29:920-6
22. Kwak HJ, Kim YJ, Chun KR, Woo YM, Park SJ, Jeong JA. et al. Downregulation of Spry2 by miR-21 triggers malignancy in human gliomas. Oncogene. 2011;30:2433-42
23. Xie L, Law BK, Chytil AM, Brown KA, Aakre ME, Moses HL. Activation of the Erk pathway is required for TGF-beta1-induced EMT in vitro. Neoplasia. 2004;6:603-10
24. Strippoli R, Loureiro J, Moreno V, Benedicto I, Perez Lozano ML, Barreiro O. et al. Caveolin-1 deficiency induces a MEK-ERK1/2-Snail-1-dependent epithelial-mesenchymal transition and fibrosis during peritoneal dialysis. EMBO Mol Med. 2015;7:102-23
Author contact
![]() Corresponding authors: Dr. Junming Yue, University of Tennessee Health Science Center, 19 S. Manassas St., Rm. 266, Memphis, TN 38163; Fax: 901-448-3910; Phone: 901-448-2091; Email: jyueedu. Or Dr. Peixin Dong, Department of Women's Health Educational System, Hokkaido University School of Medicine, Hokkaido University, Sapporo, Japan. Email: dpx1cncom.
Corresponding authors: Dr. Junming Yue, University of Tennessee Health Science Center, 19 S. Manassas St., Rm. 266, Memphis, TN 38163; Fax: 901-448-3910; Phone: 901-448-2091; Email: jyueedu. Or Dr. Peixin Dong, Department of Women's Health Educational System, Hokkaido University School of Medicine, Hokkaido University, Sapporo, Japan. Email: dpx1cncom.

 Global reach, higher impact
Global reach, higher impact