3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(8):1400-1409. doi:10.7150/jca.18170 This issue Cite
Research Paper
Downregulation of Mitochondrial Single Stranded DNA Binding Protein (SSBP1) Induces Mitochondrial Dysfunction and Increases the Radiosensitivity in Non-Small Cell Lung Cancer Cells
1. Hubei Key Laboratory of Tumor Biological Behavior, Hubei Cancer Clinical Study Center, Zhongnan Hospital of Wuhan University, Wuhan, China;
2. Department of Radiation and Medical Oncology, Zhongnan Hospital of Wuhan University, Wuhan, China;
3. Department of Radiation Oncology, Hubei Cancer Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China.
* These authors contributed equally to this work.
Received 2016-11-1; Accepted 2017-2-18; Published 2017-5-12
Abstract
Radiotherapy is one of the major therapeutic strategies for human non-small cell lung cancer (NSCLC), but intrinsic radioresistance of cancer cells makes a further improvement of radiotherapy for NSCLC challenging. Mitochondrial function is frequently dysregulated in cancer cells for adaptation to the changes of tumor microenvironment after exposure to radiation. Therefore, targeting mitochondrial biogenesis and bioenergetics is an attractive strategy to sensitize cancer cells to radiation therapy. In this study, we found that downregulation of single-strand DNA-binding protein 1 (SSBP1) in H1299 cells was associated with inducing mitochondrial dysfunction and increasing radiosensitivity to ionizing radiation. Mechanistically, SSBP1 loss induced mitochondrial dysfunction via decreasing mitochondrial DNA copy number and ATP generation, enhancing the mitochondrial-derived ROS accumulation and downregulating key glycolytic enzymes expression. SSBP1 knockdown increased the radiosensitivity of H1299 cells by inducing increased apoptosis, prolonged G2/M phase arrest and defective homologous recombination repair of DNA double-strand breaks. Our findings identified SSBP1 as a radioresistance-related protein, providing potential novel mitochondrial target for sensitizing NSCLC to radiotherapy.
Keywords: SSBP1, NSCLC, radiosensitivity
Introduction
Lung cancer is the most common malignancy and is the leading cause of cancer-related death worldwide [1]. Radiotherapy is one of the major therapeutic strategies in locally advanced [2] and early stage [3] NSCLC. However, intrinsic radioresistance of lung cancer cells, which usually leads to recurrence or metastasis in many lung cancer patients after treatment with radiation [4, 5], makes a further improvement of radiotherapy for NSCLC challenging. As the major energy and metabolite source in cells, mitochondrial function is often dysregulated in cancer, and there is growing interest in understanding how altered mitochondrial function may be targeted to improve radiotherapeutic effects [6]. The modified bioenergetic and biosynthetic states of mitochondria play an eminent role for cancer cells in response to ionizing radiation (IR) [7]. Therefore, identifying novel biomarkers involved in this pathophysiologic process could provide target for enhancing cellular sensitivity to IR and be of great value in the treatment of cancers.
After radiation, many alterations including DNA damages occur in both nuclear DNA (nDNA) and mitochondrial DNA (mtDNA). Among various factors involved in DNA damage response (DDR), single-stranded DNA-binding proteins (SSBs) play essential roles in DNA metabolic processes, including replication, repair and recombination [8, 9]. As one of the compartmentalized SSBs in eukaryotic cells, SSBP1 (also known as mtSSB) is the only known SSBs exists in eukaryotic mitochondria [10]. Distinct from nuclear SSBs, the biological role of SSBP1 is to maintain the structural stability of the mitochondrial genome by binding to single-stranded mtDNA [11]. In addition, SSBP1 can participate in regulation of important physiological and biochemical processes, such as maintenance of mtDNA copy number [12] and changes in metabolic status [13]. Recently, it has been reported that SSBP1 is associated with changes in metabolic status in human cancer cells. The level of SSBP1 protein correlates with the aggressiveness of human osteosarcoma cells, suggesting a link between mtDNA replication and cancer progression [14]. SSBP1 can also suppress epithelial-to-mesenchymal transition and metastasis in triple-negative breast cancer [15]. Apart from that, a study showed that silencing of SSBP1 resulted in an enhancement of etoposide-induced apoptosis in mouse cells [16]. These studies indicate that SSBP1 is not only involved in regulation of mitochondrial function and cell metabolism, but also closely related with tumor progression and cellular response to injury. However, the role of SSBP1 in cellular response to ionizing radiation remains poorly understood. In this study, we show that SSBP1 is a radioresistance-related protein in NSCLC cells for the first time.
Materials and Methods
Cell lines, transfection, plasmids, and regents
The human lung cancer cell line H1299 was obtained from the China center for type culture collection (CCTCC) in Wuhan University. All cells used in this study were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Thermo Fisher, USA), supplemented with 10% fetal bovine serum and incubated under 5% carbon dioxide at 37 °C.
ShRNA duplexes designed against SSBP1 (Gene ID: 6742) with the target sequence (GGCGACAAGCAACAACAATCA) synthesized by Shanghai GenePharma (Shanghai, China), were incorporated into the pU6/GFP/Neo-shRNA vector (GenePharma) to get pU6/GFP/Neo-shRNA-SSBP1. H1299 cells were transfected with pU6/GFP/Neo-shRNA-SSBP1 or the negative control shRNA expression vector using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). SSBP1 low-expressing cells and its negative control cells were selected with 800ug/ml G418 (Biosharp, China) for 5 weeks. The stably-transfected cell lines were named as H1299-sh and H1299-NC, respectively.
X-ray irradiation
X-ray irradiation was performed with an X-ray generator (Primus High-Energy Siemens), emitting at a fixed dose rate of 2 Gy/min (6 Mv X-ray), energy of the X-ray that used to irradiate cells is 0-10 Gy.
RNA extraction and Quantitative Real-Time PCR
Total RNA was extracted from the stably-transfected cells using TRIzol reagent (Biosharp, China) according to the manufacturer's protocol. cDNA was synthesized from no more than 5 μg of total RNA by the RecertAidTM First-Strand cDNA Synthesis Kit (Fermentas, Canada) at 42 °C for 10 min, followed by 75 °C for 2 min. Real-time PCR was performed with SYBR Premix Ex Taq TM (Takara, Japan) in a 25 μL reaction volume (12.5μL SYBR Premix Ex Taq, 200mM forward and reverse primers, 10 μL H2O and 1.5μL cDNA template) on a MJ Opticon Monitor Chromo4TM instrument (Bio-Rad, CA). The human GAPDH gene was performed as an internal control. The primers designed and synthesized by Sangon Biotech (Shanghai, China) were as follows: SSBP1 (forward primer 5'-AGCAACTAATGAGATGTGGCGA-3', reverse primer 5'-TTCCCTTCCAAATAAATTCGAG-3'); GAPDH (forward primer 5'-TGGAAGGACTCATGACCACA-3', reverse primer 5'-TTCAGCTCAGGGATGACCTT-3'). The following protocol was used for GAPDH and SSBP1: preincubated at 95 °C for 30 sec followed by 40 cycles of 95 °C for 5 sec, 60 °C for 30 sec and 72 °C for 30 sec. Data were analyzed by the 2(-DeltaDeltaCt) method.
Protein extraction and western blotting analysis
Cultured cells were rinsed twice with pre-cold phosphate-buffered saline (PBS) and mixed with 200 μL lysis buffer (Beyotime Biotechnology, Shanghai, China) containing protease inhibitor cocktail tablets (Roche, Germany) and phosphatase inhibitors (Roche, Germany). After 10 min on the ice, a scraper was used to transfer the cell lysate into an Eppendorf tube. The cells were ultrasonicated 3 times for 10 sec on the ice and then centrifuged at 13,400 g for 15 min, and the supernatants were then collected. The protein concentration was determined with a BCA protein assay kit (Beyotime Biotechnology, Shanghai, China), and then the samples were incubated in boiled water for more than 5 min. These samples were separated by 10 % SDS-PAGE and transferred to PVDF membranes. After blocking with 5% skim milk in TBST, the membranes were incubated with primary antibodies, against human SSBP1 (1:500 dilution; proteintech 12212-1-AP), GAPDH (1:10000 dilution; proteintech 60004-1-Ig), HIF-1α (1:500 dilution; proteintech 20960-1-AP), PDK1 (1:500 dilution; BBI D153265), HK2 (1:500 dilution; BBI D22185), PKM2 (1:500 dilution; BBI D120008), Chk1 (1:1000 dilution; Cell Signaling Technology #2360), p-Chk1 (1:1000 dilution; Cell Signaling Technology #2341), Bax (1:1000 dilution; proteintech 50599-2-Ig), Bcl-2 (1:1000 dilution; proteintech 12789-1-AP), Ku70 (1:1000 dilution; proteintech 10723-1-AP), Ku80 (1:1000 dilution; proteintech 16389-1-AP), Rad51 (1:300 dilution; proteintech 14961-1-AP), BRCA2 (1:300 dilution; proteintech 19791-1-AP), γH2Ax (1:1000 dilution; abcam ab37168) at 4 °C overnight, followed by incubation with Horseradish peroxidase-conjugated secondary antibody diluted at 1:20,000. Specific bands were visualized by ECL (Advansta, USA). Autoradiographs were recorded onto X-Omat AR film (Eastman Kodak Company, Rochester, NY, USA). The density of bands in the resulting film was quantified by the Image J analysis program.
Colony formation assay
Cells were plated in 6-well plate culture plates. After 24h, the cells were irradiated with graded doses (0, 1, 2, 4, 6, 8, 10 Gy) using the X-ray generator (Primus High-Energy Siemens) at a dose rate of 2 Gy/min. After 14 days of incubation, the colonies were fixed and stained with crystal violet (1% crystal violet in absolute alcohol). Those colonies containing more than 50 cells were scored as viable colonies. The data was fit into the single-hit multi-target model, and the survival curve of each group was demonstrated by Graphpad prism 6.0 software (San Diego, CA, USA). Radiobiological parameters, D0, Dq, and SF2 were calculated according to the survival curves.
Transmission electron microscopy (TEM) imaging
Cells were collected and fixed in 2.5% glutaraldehyde and subsequently post-fixed in 1% osmium tetroxide for 3 hours, and then pelleted, dehydrated, infiltrated and embedded. At last, ultrathin sections were cut and stained with uranyl acetate. TEM images were recorded by a Hitachi HT7700 TEM (Tokyo, Japan).
Determination of mtDNA copy number
MtDNA copy number was assessed by quantification of a unique fragment in human mitochondrial genome NC_012920 region relative to a single copy region of the nuclear gene β2M using a real-time PCR (RT-PCR) assay. The detailed method was performed as previously described [17].
Cellular adenosine triphosphate (ATP) concentration detection
The cellular ATP level was detected by the ATP Assay Kit (Beyotime Biotechnology, Shanghai, China) according to the manufacturer's protocol. 100 μL working solution was added to a 10 μL diluted sample, and the luciferase activity was immediately evaluated by Modulus™ II Microplate multifunction tester. ATP content was determined by comparison to a concurrent standard curve and was then normalized by cell number and expressed as μmol/L.
Mitochondria-associated Reaction oxygen species (ROS) measurement
Mitochondria-associated ROS levels were measured by staining cells with MitoSOX Red (Invitrogen, Carlsbad, CA, USA) at 5 μM in the dark for 30 min at 37 °C. Cells were then washed with PBS solution and re-suspended in cold PBS solution for flow cytometry (Beckman Coulter, USA) analysis.
Cell Cycle Analysis
The cell cycle was assessed in cells exposed to 6 Gy of ionizing radiation or without irradiation, and then the cells were incubated for the indicated times. Cells were collected and fixed in suspension in 70% ethanol on ice and then stored at 4 °C overnight. Cells were treated with RNase for 20 min before addition of 5 mg/ml propidium iodide, and analyzed by flow cytometry (Beckman Coulter, USA).
Cell proliferation assay
The stably transfected cells were plated on culture dishes at a density of 5 × 104 cells/cm2 for the indicated times. Cell proliferation was determined by direct counting using a hemocytometer.
Immuno-fluorescence Staining and Imaging
After indicated treatment, cells were fixed with 4% formaldehyde for 15 min and permeabilized with 0.2% Triton X-100 in PBS for 10 minutes at room temperature. After blocked with 5% (w/v) BSA for 2 h, cells were incubated with the primary antibody (γH2AX ser139, dilution 1:400; Millipore Corp., USA) overnight at 4 °C and then washed and incubated with the secondary AF555-conjugated antibody (Millipore Corp., USA) for 1h at 37 °C. Nuclei were stained with DAPI (Sigma, USA) for 5 min at room temperature. At last, images were recorded by a confocal microscope (Leica Microsystems GmbH, Wetzlar, Germany).
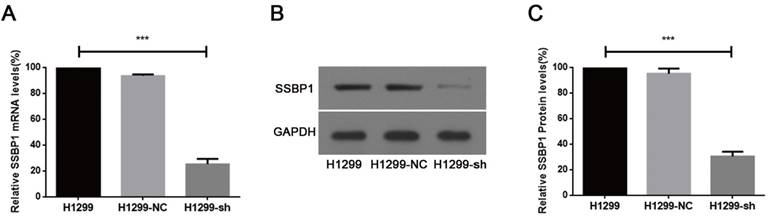
shRNA against SSBP1 decreased SSBP1 mRNA and protein levels. (a) Real-time PCR was performed to determine the relative SSBP1 mRNA levels in H1299, H1299-NC and H1299-sh cells. (b) Western blotting was used to detect the relative SSBP1 protein levels in H1299, H1299-NC and H1299-sh cells. (c) The bands density analysis of western blotting showed the relative SSBP1 protein levels in H1299, H1299-NC and H1299-sh cells. H1299-NC group revealed no significant differences compared with the H1299 group. The bar graph shows the mean ± standard deviation (SD) values of three independent experiments. ***P<0.001.
Statistical analysis
All experiments were performed three times and the data were expressed as mean ± SD. Student's t-test was used to test statistical significance. P < 0.05 was considered to be significant. GraphPad Prism 6 (GraphPad Software, California) software was used as a statistical analysis tool.
Results
Efficient knockdown of SSBP1 in H1299 cells
Real-time PCR and western blotting were used to analyze SSBP1 expression. A comparison of H1299-NC and H1299-sh cells showed that the expression of SSBP1 was reduced in H1299-sh cells at both mRNA (Fig.1A) and protein levels (Fig.1B, 1C). However, no significant change of SSBP1 expression was observed in H1299-NC and H1299 cells. These results indicated that the stable transfection with pU6/GFP/Neo-shRNA-SSBP1 effectively knocked down SSBP1 expression in H1299-sh cells.
SSBP1 downregulation increased radiosensitivity of H1299 cells to IR
To investigate the effect of SSBP1 downregulation on the cell response to radiation, H1299-NC and H1299-sh cells were exposed to different doses of radiation and subjected to a clonogenic assay. Cell clones were counted and radiobiological parameters were described by survival curves for each cell type. As shown in Fig. 2, downregulation of SSBP1 remarkably reduced the cell surviving fraction after exposure to IR. The calculated radiobiological parameters were shown in Table 1. D0, Dq and SF2 values of H1299-sh cells were significantly lower than those of H1299-NC cells. These results suggested that SSBP1 downregulation increased the radiosensitivity of H1299 cells to irradiation.
SSBP1 knockdown increased radiosensitivity of H1299 cells. Each group was irradiated with graded doses of 0, 1, 2, 4, 6, 8, 10 Gy. After 14 days of incubation, the colonies were fixed and stained. Those colonies containing more than 50 cells were scored as viable colonies. These data were fit into the single-hit multi-target model, and surviving curves were demonstrated using GraphPad prism 6.0 software.
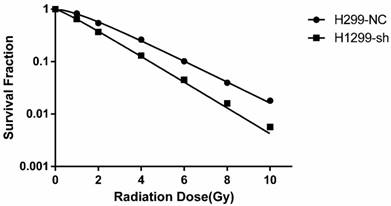
Radiobiological parameters in the different Groups.
| Cell line | D0 | Dq | SF2 |
|---|---|---|---|
| H1299-NC | 2.155±0.131 | 1.120±0.143 | 0.545±0.021 |
| H1299-sh | 1.764±0.094* | 0.356±0.179** | 0.368±0.015** |
D0, the incremental dose required for reducing the fraction of colonies to 37%, indicative of single-event killing; Dq, quasi-threshold dose; SF2, the survival fraction after 2Gy radiation exposure. *P<0.05, **P<0.01.
SSBP1 downregulation induced mitochondrial dysfunction
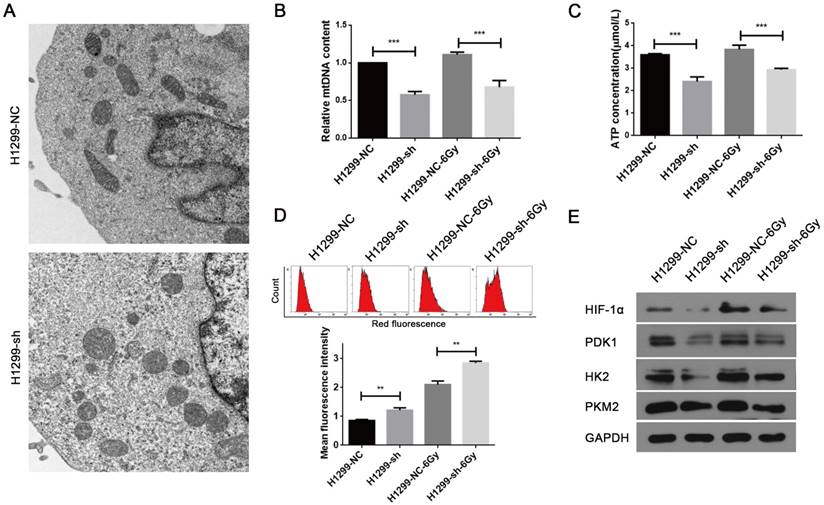
The major physiological functions of mitochondria include the production of ATP by oxidative phosphorylation, generation and detoxification of reactive oxygen species, involvement in some forms of apoptosis, regulation of cytoplasmic and mitochondrial matrix calcium, synthesis and catabolism of metabolites [18]. Thus, abnormality in any of these processes can lead to mitochondrial dysfunction. As SSBP1 is essential for mtDNA replication [19], we hypothesized that SSBP1 downregulation would affect the mtDNA copy number and then induce the alteration in mitochondrial morphology and function. Firstly, mitochondrial morphology was observed with transmission electron microscopy. As shown in Fig. 3A, mitochondria of H1299-NC cells were oval or round with complete membrane structures and longitudinally arranged ridges that were tidy and compact. Whereas mitochondria of H1299-sh cells were swollen and damaged with somewhat complete outer membranes, disorganized or even invisible ridge structures, and barely visible inner structures. Then, the mtDNA copy number was measured by real-time PCR and results showed that SSBP1 knockdown decreased the mtDNA copy number in H1299 cells. (Fig. 3B). Next, ATP production was investigated and the result indicated that the knockdown of SSBP1 led to a lower ATP production (Fig. 3C). Additionally, after exposure to 6Gy IR, both the mtDNA copy number and the ATP concentration of H1299-sh cells were still significantly lower than those of H1299-NC cells (Fig. 3B and Fig. 3C). Furthermore, mitochondrial-generated ROS was measured using cell-permeating probe mitoSOX. As shown in Fig.3D, whether the cells were exposed to IR or not, SSBP1 knockdown led to a higher level of mitoSOX Red fluorescence, which implied that suppression of SSBP1 could enhance the ROS accumulation in mitochondrial. Finally, the expressions of mitochondria metabolism-related proteins, including hypoxia-inducible factor 1α (HIF-1α), pyruvate dehydrogenase kinase 1 (PDK1), hexokinase 2 (HK2) and pyruvate kinase M2 (PKM2), were assessed by western blotting (Fig. 3E). The results showed that SSBP1 knockdown significantly decreased protein levels of HIF-1α, PDK1, HK2, and PKM2 in the absence of IR. When the cells were exposed to IR, the levels of these proteins in H1299-sh were still downregulated compared with H1299-NC cells. Taken together, these results indicated that SSBP1 was required for maintaining mitochondrial morphology and function and knockdown of SSBP1 induced mitochondrial dysfunction.
SSBP1 knockdown induced mitochondrial dysfunction of H1299 cells. (a) Mitochondrial morphology was observed with transmission electron microscopy in H1299-NC and H1299-sh cells (magnification, ×5,000). (b) Real-time PCR was determined to analyze the mtDNA copy number in H1299-NC and H1299-sh groups, without radiation or 30 min after 6Gy. (c) Intracellular ATP concentrations in the H1299-NC and H1299-sh cells were measured by the ATP Assay Kit, without radiation or 30 min after 6Gy. (d) Mitochondrial ROS (mROS) production levels were measured by MitoSOX Red fluorescence intensity, without radiation or 30 min after 6Gy. (upper image: mROS measured by flow cytometry; lower chart: mROS levels were depictured as a bar chart) (e) The effects of SSBP1 knockdown on the expressions of HIF-1α, PDK1, HK2, PKM2, without radiation or 24 h after 6Gy. **P<0.01, ***P<0.001.
SSBP1 downregulation decreased the proportion of cells in the S phase, but increased the proportion of cells in the G2/M phase and IR-induced G2/M phase arrest in H1299 cells
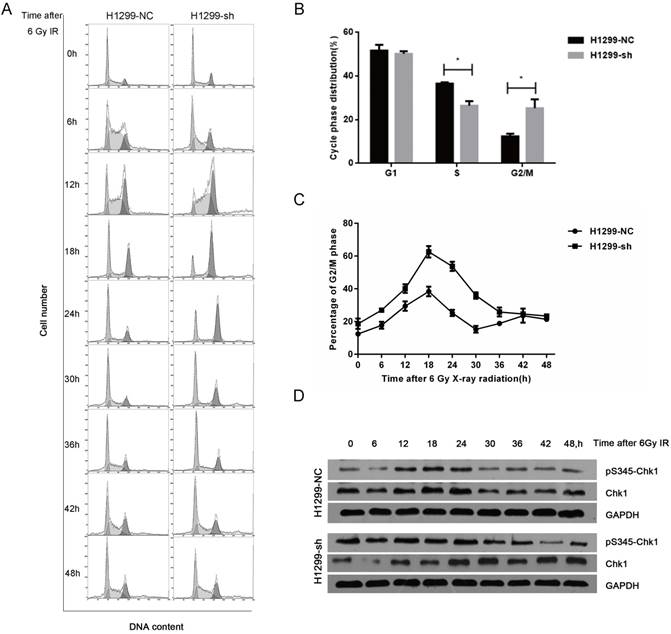
As cell cycle distribution is one of the key factors affecting radiosensitivity, we next investigated the SSBP1 knockdown-mediated effect on the cell cycle distribution with flow cytometry. In absence of radiation exposure, the proportion of cells in the S phase in H1299-sh cells was significantly lower than that in H1299-NC cells, while the percentage of cells in G2/M phase cells showed the opposite trend (Fig. 4B). However, SSBP1 depletion had no significant effect on the proportion of cells in the G1 phase. After exposure to 6 Gy of IR, although the G2/M arrest reached a peak in both H1299-NC and H1299-sh cells at 18 hours, the proportion of H1299-sh cells arrested in the G2/M phase was significantly higher (Fig. 4C). More importantly, the kinetic response was different between these two groups. In H1299-NC cells, the G2/M peak gradually decreased from 18 h after IR and returned to basal levels at approximately 30 h. However, the G2/M arrest in H1299-sh cells was maintained at a high level until 42 h after IR (Fig. 4C). These results suggested that SSBP1 knockdown prolonged G2/M arrest after IR exposure in H1299 cells (Fig. 4A and 4C). To investigate the molecular mechanisms of prolonged G2/M arrest after IR exposure in SSBP1-downregulation cells, we measured Chk1 protein and its phosphorylation levels at Ser345. As shown in Fig. 4D, phosphorylation levels of Chk1 at Ser345 remained higher till 36 h after IR exposure in H1299-sh cells. In contrast, the levels in H1299-NC cells returned to normal levels at about 30h after IR exposure. These results indicated that IR- induced prolonged G2/M arrest after SSBP1 knockdown was likely due to the sustained phosphorylation of Chk1 at Ser345 in H1299 cells.
SSBP1 knockdown prolonged IR-induced G2/M phase arrest by activating sustained phosphorylation of Chk1 at Ser345. (a) H1299-NC and H1299-sh cells were irradiated with 6Gy X-ray and recovered for indicated times. (b) The changes of cell cycle phase distribution after SSBP1 knockdown. (c) The proportion of cells in G2/M phases at indicated time points after exposure to 6 Gy radiation. (d) The effects of SSBP1 knockdown on Chk1 and pS435-Chk1 protein levels, without radiation or 24 h after 6Gy. *P<0.05
SSBP1 knockdown inhibited proliferation and induced apoptosis in H1299 cells. (a) H1299-sh cells exhibited profoundly decelerated growth kinetics compared to H1299-NC cells. (b) The effects of SSBP1 knockdown on cell apoptosis proteins levels (Bax, Bcl-2), without radiation or 24 h after 6Gy. (c) The bands density were analyzed using Image J and then the ratios of Bax/Bcl-2 were presented as bar graph. *P<0.05, **P<0.01, ***P<0.001.
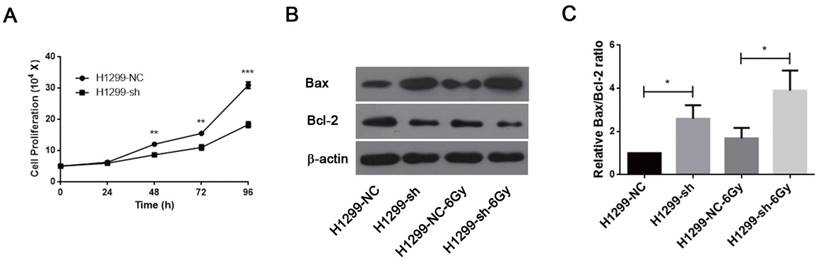
SSBP1 was involved in controlling proliferation and apoptosis of H1299 cells
Downregulation of SSBP1 inhibited the proliferation of H1299 cells (Fig. 5A). IR is known to induce DNA damage. If DNA damage repair is insufficient after irradiation, cells may undergo apoptosis and/or necrosis. Since SSBP1 downregulation sensitized H1299 cells to irradiation, we further investigated the effect of SSBP1 knockdown on cell apoptosis by measuring expressions of Bax and Bcl-2 proteins with western blotting. As showed in Fig 5B and 5C, the proportion of Bax/Bcl-2 increased in the absence of SSBP1 with or without exposure to radiation. These results suggested that the depletion of SSBP1 led to inhibition of proliferation and induced apoptosis in H1299 cells.
SSBP1 downregulation led to an increased level of DNA damage and impaired the homologous recombination repair of DSBs induced by IR
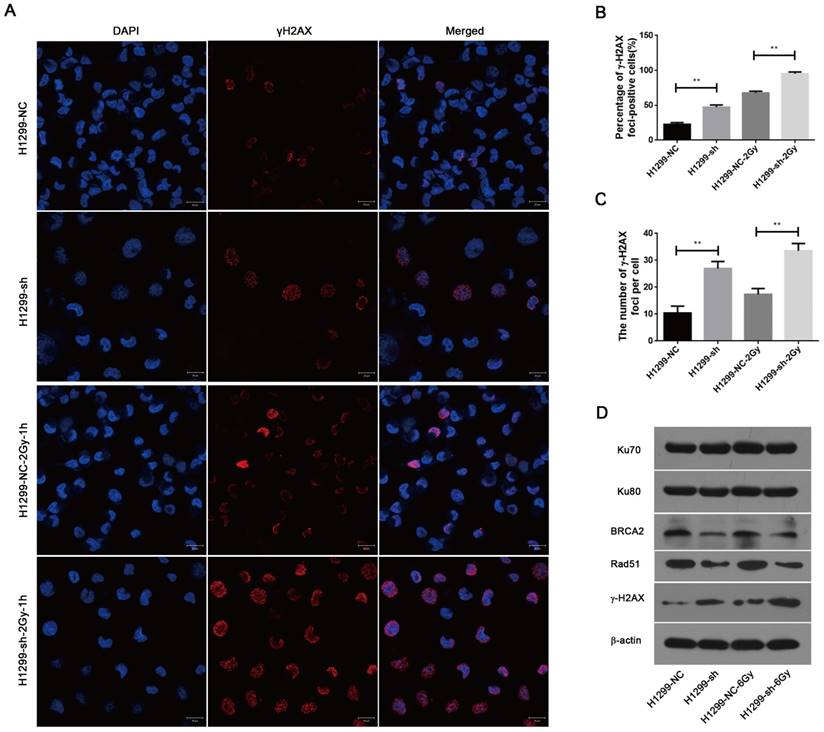
To investigate the effect of SSBP1 knockdown on DNA damage, immunofluorescence was used to assess the changes in phospho-γH2AX. As shown in Fig. 6A, 6B and 6C, γH2AX foci significantly increased in H1299-sh cells compared with those in H1299-NC cells. In addition, one hour after exposure to 2 Gy IR, the number of γH2AX foci in H1299-sh cells was still higher than that in H1299-NC cells. This implied that the SSBP1 downregulation led to an increased level of DNA damage. In eukaryotes cells, DSBs are repaired by two basic approaches, homologous recombination (HR) repair and non-homologous end joining (NHEJ). To explore the effect of SSBP1 knockdown on DSBs repair pathways in H1299 cells, the expression of DNA damage repair proteins (Ku70, Ku80, Rad51 and BRCA2) were assessed by western blotting. Among these DNA repair factors, BRCA2 and Rad51 are important proteins involved in HR repair, while Ku70 and Ku80 are essential for NHEJ. Our results revealed that SSBP1 knockdown reduced the expressions of Rad51 and BRCA2, whereas SSBP1 loss showed no effect on Ku70 and Ku80 protein levels (Fig. 6D). After exposure to IR, Rad51 and BRCA2 protein levels decreased when SSBP1 was downregulated; however, the protein levels of Ku70 and Ku80 remained stable (Fig. 6D). These data indicated that the depletion of SSBP1 induced an increased level of DNA damage and impaired the homologous recombination repair of DSBs in H1299 cells.
Discussion
In the current study, we demonstrated that SSBP1 downregulation led to a significant decrease in the mtDNA copy number, which induced mitochondrial dysfunction, and increased the radiosensitivity of human NSCLC cells to ionizing radiation. Several studies have supported a role of mtDNA defects in modulating radiosensitivity in human fibroblast and tumor cells [20-22], but the upstream regulators of mtDNA alteration remain poorly understood. Here, our data led to the identification of SSBP1 as a novel mitochondrial biomarker for sensitizing human NSCLC to radiotherapy.
In this study, we investigated the clonogenic survival of NSCLC cells after knockdown of SSBP1 and demonstrated that downregulation of SSBP1 increased radiosensitivity in H1299 cells. Then, we provided evidence that SSBP1 was not only required for maintaining the integrity and normal morphology of mitochondria, but also involved in modulating mitochondrial functions in NSCLC cells. It has previously been reported that depletion of SSBP1 resulted in a significant decrease in mtDNA level. Moreover, mtDNA depletion was revealed to impair the ATP synthase activity [23] and to cause more ROS production [24, 25]. Similarly, our data demonstrated that the decrease of mtDNA copy number and ATP production induced by SSBP1 knockdown was associated with an increase in mitochondrial ROS generation in H1299 cells. Recent study shows that enhanced aerobic glycolysis was discovered in acquired radioresistance of tumor cells by long-term exposure to fractionated radiation [26]. In addition, as the upstream regulator of PDK1, HK2 and PKM2 [27, 28], HIF-1α has been reported to promote radiation resistance in vivo and in vitro [29]. Here, we demonstrated that, whether the cells were exposed to radiation or not, the protein levels of HIF-1α, PKM2, PDK1 and HK2 were downregulated in the absence of SSBP1, suggesting that SSBP1 knockdown attenuated aerobic glycolysis in H1299 cells. Collectively, we consider that downregulation of SSBP1 increases the radiosensitivity of H1299 cells probably via inducing mitochondrial dysfunction. Nevertheless, the exact mechanism about how mitochondrial dysfunction regulates radiosensitivity deserves further study. And related hypotheses are also under investigation in our laboratory.
SSBP1 knockdown induced an increased level of DNA damage and impaired the HR repair of DSBs. (a) Confocal microscopy recorded the images in 4 groups as follows: H1299-NC, H1299-sh, H1299-NC-2Gy-1h and H1299-sh-2Gy-1h. (b) The percentages of γH2AX foci-positive cells in these four groups were obtained by analyzing 50 randomly selected cells in each group. (c) The numbers of H2AX foci per cell in these four groups were obtained by analyzing 50 randomly selected cells in each group. (d) The effects of SSBP1 knockdown on double-strand breaks repair proteins (Ku78, Ku80, Rad51, BRCA2, γH2AX), without radiation or 24 h after 6Gy. **P<0.01
Classical radiation biology suggests that the changes in the cell cycle distribution are one of the key factors regulating radioresistance. The G1 phase and early S phase are the most radioresistant phases of the cell cycle, while the G2/M phase is the most radiosensitive phase [30]. In this study, we found that SSBP1 knockdown had no significant effect on the proportion of cells in the G1 phase, but significantly decreased the number of cells in the S phase, whereas it increased the number of cells in G2/M phase arrest, indicating that changes in cell cycle distribution might be a factor underlying increased radiosensitivity after SSBP1 knockdown. Furthermore, our results showed that downregulation of SSBP1 prolonged IR-induced G2/M arrest. IR-induced G2/M arrest is generally considered to promote cell survival and reduce the probability of genomic alterations. However, it is reported that enhanced radiosensitivity and G2/M arrest were observed in radioresistant esophageal cancer cells by knocking down replication protein A (RPA) [31], which implies that the definite contribution of G2/M arrest to cell survival after radiation remains unclear. Interestingly, both RPA and SSBP1 are members of single-stranded DNA-binding proteins. Activation of Chk1 regulates the G2/M arrest in response to DNA damage, and phosphorylation of Chk1 on S345 is regarded as an indicator of Chk1 activation [32]. In this paper, we found that Chk1 phosphorylation in SSBP1 knockdown cells was elevated and sustained till later time points after exposure to IR. Thus, IR-induced prolonged G2/M arrest after knocking down SSBP1 may be mediated via the sustained phosphorylation of Chk1 on S345.
Bax functions as a pro-apoptotic protein, whereas Bcl-2 functions as an anti-apoptotic protein. The Bax/Bcl-2 ratio is an indicator of cell apoptosis. Recent studies have suggested that silencing of SSBP1 induced a significant apoptosis to apoptotic stimuli by etoposide [16]. Our study showed that SSBP1 knockdown inhibited the cell proliferation and increased the proportion of cells undergoing spontaneous and radiation-induced apoptosis. Thus, the inhibition of proliferation induced by knocking down SSBP1 might be mediated by decreasing ATP production and increasing spontaneous apoptosis. Moreover, the increased apoptosis induced by depletion of SSBP1 was probably related to increased radiosensitivity in H1299 cells. However, the exact mechanism underlying these processes requires further study.
As γH2AX is a highly specific and sensitive molecular marker for monitoring DSBs initiation and resolution [33], the number of γH2AX foci can be used as a representation of DNA damage and DNA repair capacity. We found that phosphorylation of histone γH2AX was enhanced by knocking down SSBP1 and that combination of SSBP1 depletion with IR induced an increased level of DNA damages, indicating that SSBP1 knockdown impaired DNA damage repair ability in NSCLC cells after radiation. HR repair and NHEJ are two basic approaches for higher eukaryotes to process DSBs. HR repair is predominantly error-free using the sister chromatid as template to retrieve lost sequence information, thus is restricted to S- and G2-phase of the cell cycle, while NHEJ is considered as the prevalent DSB repair pathway in whole cell cycle [34]. In the present study, we found that SSBP1 knockdown compromised the repair kinetics of DSBs after radiation by reducing the HR repair proteins expression (Rad51, BRCA2) while maintaining stable expression of NHEJ repair proteins (Ku70, Ku80). Furthermore, combined with our data showing that downregulation of SSBP1 prolonged IR-induced G2/M phase arrest in H1299 cells, it is suggested that those DSBs were not sufficiently repaired when cells were arrested in G2/M phase due to the defective HR repair induced by SSBP1 knocking down. All these results indicate that SSBP1 knockdown increases the radiosensitivity of NSCLC cells by inducing both prolonged G2/M phase arrest and defective HR repair capability.
In conclusion, our results demonstrate that SSBP1 downregulation induces mitochondrial dysfunction and increases radiosensitivity in NSCLC cells. Therefore, our study indicates that SSBP1 may be a promising target for the radiotherapy of NSCLC. Moreover, SSBP1 depletion might have the potential for improving radiotherapy efficacy.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (NO. 81172129 and NO. 81472798).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7-30
2. Eberhardt WE, De Ruysscher D, Weder W, Le Pechoux C, De Leyn P, Hoffmann H. et al. 2nd ESMO Consensus Conference in Lung Cancer: locally advanced stage III non-small-cell lung cancer. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2015;26:1573-88
3. Vansteenkiste J, De Ruysscher D, Eberhardt WE, Lim E, Senan S, Felip E. et al. Early and locally advanced non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2013;24(Suppl 6):vi89-98
4. Garg S, Gielda BT, Kiel K, Turian JV, Fidler MJ, Batus M. et al. Patterns of locoregional failure in stage III non-small cell lung cancer treated with definitive chemoradiation therapy. Pract Radiat Oncol. 2014;4:342-8
5. Willers H, Azzoli CG, Santivasi WL, Xia F. Basic mechanisms of therapeutic resistance to radiation and chemotherapy in lung cancer. Cancer J. 2013;19:200-7
6. van Gisbergen MW, Voets AM, Starmans MH, de Coo IF, Yadak R, Hoffmann RF. et al. How do changes in the mtDNA and mitochondrial dysfunction influence cancer and cancer therapy? Challenges, opportunities and models. Mutat Res Rev Mutat Res. 2015;764:16-30
7. Bol V, Bol A, Bouzin C, Labar D, Lee JA, Janssens G. et al. Reprogramming of tumor metabolism by targeting mitochondria improves tumor response to irradiation. Acta oncologica. 2015;54:266-74
8. Chase JW, Williams KR. Single-stranded DNA binding proteins required for DNA replication. Annu Rev Biochem. 1986;55:103-36
9. Ashton NW, Bolderson E, Cubeddu L, O'Byrne KJ, Richard DJ. Human single-stranded DNA binding proteins are essential for maintaining genomic stability. BMC Mol Biol. 2013;14:9
10. Kaguni LS. DNA polymerase gamma, the mitochondrial replicase. Annu Rev Biochem. 2004;73:293-320
11. Broderick S, Rehmet K, Concannon C, Nasheuer HP. Eukaryotic single-stranded DNA binding proteins: central factors in genome stability. Subcell Biochem. 2010;50:143-63
12. Ruhanen H, Borrie S, Szabadkai G, Tyynismaa H, Jones AW, Kang D. et al. Mitochondrial single-stranded DNA binding protein is required for maintenance of mitochondrial DNA and 7S DNA but is not required for mitochondrial nucleoid organisation. Biochim Biophys Acta. 2010;1803:931-9
13. Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an Organizer/Mobilizer of Genome Maintenance Complexes. Critical reviews in biochemistry and molecular biology. 2008;43:289-318
14. Shapovalov Y, Hoffman D, Zuch D, de Mesy Bentley KL, Eliseev RA. Mitochondrial dysfunction in cancer cells due to aberrant mitochondrial replication. J Biol Chem. 2011;286:22331-8
15. Jiang HL, Sun HF, Gao SP, Li LD, Huang S, Hu X. et al. SSBP1 Suppresses TGFbeta-Driven Epithelial-to-Mesenchymal Transition and Metastasis in Triple-Negative Breast Cancer by Regulating Mitochondrial Retrograde Signaling. Cancer Res. 2016;76:952-64
16. Arakaki N, Nishihama T, Kohda A, Owaki H, Kuramoto Y, Abe R. et al. Regulation of mitochondrial morphology and cell survival by Mitogenin I and mitochondrial single-stranded DNA binding protein. Biochim Biophys Acta. 2006;1760:1364-72
17. Xu H, He W, Jiang HG, Zhao H, Peng XH, Wei YH. et al. Prognostic value of mitochondrial DNA content and G10398A polymorphism in non-small cell lung cancer. Oncol Rep. 2013;30:3006-12
18. Brand MD, Nicholls DG. Assessing mitochondrial dysfunction in cells. The Biochemical journal. 2011;435:297-312
19. Oliveira MT, Kaguni LS. Functional roles of the N- and C-terminal regions of the human mitochondrial single-stranded DNA-binding protein. PLoS One. 2010;5:e15379
20. Yamazaki H, Yoshida K, Yoshioka Y, Isohashi F, Ozeki S, Koizumi M. et al. Impact of mitochondrial DNA on hypoxic radiation sensitivity in human fibroblast cells and osteosarcoma cell lines. Oncology reports. 2008;19:1545-9
21. Yoshioka Y, Yamazaki H, Yoshida K, Ozeki S, Inoue T, Yoneda M. et al. Impact of mitochondrial DNA on radiation sensitivity of transformed human fibroblast cells: clonogenic survival, micronucleus formation and cellular ATP level. Radiation research. 2004;162:143-7
22. Cloos CR, Daniels DH, Kalen A, Matthews K, Du J, Goswami PC. et al. Mitochondrial DNA depletion induces radioresistance by suppressing G2 checkpoint activation in human pancreatic cancer cells. Radiation research. 2009;171:581-7
23. Buchet K, Godinot C. Functional F1-ATPase essential in maintaining growth and membrane potential of human mitochondrial DNA-depleted rho degrees cells. The Journal of biological chemistry. 1998;273:22983-9
24. Indo HP, Davidson M, Yen HC, Suenaga S, Tomita K, Nishii T. et al. Evidence of ROS generation by mitochondria in cells with impaired electron transport chain and mitochondrial DNA damage. Mitochondrion. 2007;7:106-18
25. Miranda S, Foncea R, Guerrero J, Leighton F. Oxidative stress and upregulation of mitochondrial biogenesis genes in mitochondrial DNA-depleted HeLa cells. Biochemical and biophysical research communications. 1999;258:44-9
26. Shimura T, Kakuda S, Ochiai Y, Nakagawa H, Kuwahara Y, Takai Y. et al. Acquired radioresistance of human tumor cells by DNA-PK/AKT/GSK3beta-mediated cyclin D1 overexpression. Oncogene. 2010;29:4826-37
27. Sun Q, Chen X, Ma J, Peng H, Wang F, Zha X. et al. Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4129-34
28. Kim JW, Dang CV. Cancer's molecular sweet tooth and the Warburg effect. Cancer research. 2006;66:8927-30
29. Zhang M, Qiu Q, Li Z, Sachdeva M, Min H, Cardona DM. et al. HIF-1 Alpha Regulates the Response of Primary Sarcomas to Radiation Therapy through a Cell Autonomous Mechanism. Radiation research. 2015;183:594-609
30. Pawlik TM, Keyomarsi K. Role of cell cycle in mediating sensitivity to radiotherapy. International journal of radiation oncology, biology, physics. 2004;59:928-42
31. Di Z, Sanyuan S, Hong L, Dahai Y. Enhanced radiosensitivity and G2/M arrest were observed in radioresistant esophageal cancer cells by knocking down RPA expression. Cell Biochem Biophys. 2014;70:887-91
32. Borst GR, McLaughlin M, Kyula JN, Neijenhuis S, Khan A, Good J. et al. Targeted radiosensitization by the Chk1 inhibitor SAR-020106. International journal of radiation oncology, biology, physics. 2013;85:1110-8
33. Sak A, Stuschke M. Use of gammaH2AX and other biomarkers of double-strand breaks during radiotherapy. Semin Radiat Oncol. 2010;20:223-31
34. Mladenov E, Magin S, Soni A, Iliakis G. DNA double-strand-break repair in higher eukaryotes and its role in genomic instability and cancer: Cell cycle and proliferation-dependent regulation. Semin Cancer Biol. 2016;37-38:51-64
Author contact
![]() Corresponding author: Fuxiang Zhou, MD, PhD, Department of Radiation Oncology and Medical Oncology, Zhongnan Hospital of Wuhan University, 169 Donghu Road, Wuhan 430071, China. Phone: 86-27-67813155, Email: fxzhouwhucom
Corresponding author: Fuxiang Zhou, MD, PhD, Department of Radiation Oncology and Medical Oncology, Zhongnan Hospital of Wuhan University, 169 Donghu Road, Wuhan 430071, China. Phone: 86-27-67813155, Email: fxzhouwhucom

 Global reach, higher impact
Global reach, higher impact