3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(11):1959-1971. doi:10.7150/jca.19364 This issue Cite
Research Paper
Downregulation of ABC Transporters in Non-neoplastic Tissues Confers Better Prognosis for Pancreatic and Colorectal Cancer Patients
1. Department of Biology, Faculty of Medicine in Pilsen, Charles University, Alej Svobody 76, 32300, Pilsen, Czech Republic
2. Biomedical Center, Faculty of Medicine in Pilsen, Charles University, Alej Svobody 76, 32300, Pilsen, Czech Republic
3. Department of Oncology, Faculty of Medicine and Dentistry, Palacky University Olomouc and University Hospital Olomouc, IP Pavlova 6, 77520, Olomouc, Czech Republic
4. Deparment of Surgery, Faculty Hospital and Faculty of Medicine in Pilsen, Charles University, Alej Svobody 80, 30460, Pilsen, Czech Republic
Received 2017-1-26; Accepted 2017-5-18; Published 2017-7-5
Abstract
Transport of a wide variety of substrates, including xenobiotics, is one of the main functions attributed to human ATP-binding cassette (ABC) proteins. Overexpression of ABC genes is considered to be an important mechanism facilitating the development of chemoresistance. Relationships between the expression levels of ABC genes in tumor tissues and established clinicopathological features were extensively studied previously. The current study tested our hypothesis that the expression levels of ABC genes in non-neoplastic (control) tissues also provide important information in relation to the relevant tumor progression. Expression levels of all human ABC genes (48 protein coding and one pseudogene), measured by qRT-PCR, were bioinformatically analyzed. The data originated from four independently collected cohorts covering three types of tumors - breast, colorectal and pancreatic carcinomas. ABC gene expression profiles (signatures) in non-neoplastic tissues (matched to tumor samples from three different tumor types) were characteristically clustered into three main types - those with the vast majority of the genes downregulated, upregulated or heterogeneously regulated. The clusters with mostly downregulated and upregulated genes were shown to possess significant relations to good and poor prognostic markers, respectively, in pancreatic and colorectal cancers. The present findings support the theory that the expression of ABC genes in non-neoplastic tissues can significantly contribute to tumor pathogenesis. Suggested multi-gene panels, consisting of the reduced number of ABC genes, have the potential to be implemented as new prognostic markers, which are especially urgent in pancreatic cancer. The results can also stimulate further primary research in carcinogenesis.
Keywords: ATP-binding cassette, ABC transporters, gene expression, profiles, cancer
Introduction
ATP-binding cassette (ABC) proteins constitute a large family of active transmembrane transporters appearing early in cellular evolution and present in bacterial as well as eukaryotic cells. In humans, 48 ABC protein coding genes and one pseudogene have been described so far and classified into seven subfamilies - ABCA to ABCG - based on amino acid sequence similarities and phylogeny [1]. Transport of a wide range of substrates from inorganic anions, metal ions, peptides, amino acids and sugars to hydrophobic compounds and metabolites such as fatty acids, phospholipids, cholesterol and its derivatives, and vitamins plays a vital role in maintaining cell homeostasis [1, 2]. Substrates are translocated from cytosol out of the cell or inside intracellular organelles such as endoplasmic reticulum, peroxisomes or mitochondria. Notably, some ABC proteins have also been characterized as critical components of the antigen presentation, chromosome segregation, recombination and DNA repair machineries, and regulate or catalyze critical steps of ribosomal protein synthesis [3, 4]. Unfortunately, these important physiological functions of ABC proteins can probably significantly potentiate tumor development in abnormal circumstances such as those happening in cancer cells [5, 6].
Xenobiotics including common anticancer drugs can mimic physiological substrates and are also transported by many members of the ABC family, resulting in drug efflux. This additional function of ABC proteins has led to a significant attention of many research groups worldwide. The overexpression of ABC transporters causes elevation of drug efflux and is considered to be one of the main mechanisms in the development of cancer cell chemoresistance [7, 8]. Individual ABC transporters (mainly ABCB1, ABCC1 and ABCG2) and their mechanisms of drug transportation have been extensively studied for many years. Several inhibitors (modulators) of ABC transporters were developed and entered clinical trials; however, there was limited success [9, 10]. The increased toxicity of drugs administered together with ABC modulators and overlapping function between ABC transporters create high barriers to the further progress in this treatment modality.
Determination of ABC gene expression profiles (clusters, signatures) in relation to tumor characteristics and a search for their clinical relevance were logical directions of the ensuing research. Several research groups showed the importance of ABC gene expression profiles in tumor tissues and their relationships to clinicopathological features. ABC expression profiles in tumor tissues were the main theme of our previous article, where the summary of the relevant literature was presented and the existence of general ABC profiles in tumors was demonstrated [11].
The evidence that underlying genetic alterations of tumor cells and interactions within the tumor microenvironment are equally important to tumor progression was discussed in many recent works [12]. Moreover, it is clear that the metastatic potential is not simply an inherent trait of cancer cells, but is substantially modified by the microenvironment [13]. Based on these facts, we hypothesized that ABC expression profiles in non-neoplastic tissues can also modify tumor progression. Furthermore, ABC profiles in non-neoplastic (control) tissues paired with tumor tissue samples could be classified according to characteristic patterns and possess clinically relevant information. The present work brings evidence in support of this hypothesis.
Materials and methods
Characteristics of datasets
Four datasets - Colorectal I, Colorectal II, Pancreatic, and Breast dataset - containing clinical and laboratory data from oncology patients, who were diagnosed and treated in four hospitals in the Czech Republic (Medicon, Prague; University Hospital Pilsen, Pilsen; Institute of Clinical and Experimental Medicine, Prague; and University Hospital Brno, Brno), were available for our bioinformatic analyses.
The Colorectal I dataset consisted of data from 51 colorectal carcinoma (CC) patients, which were previously characterized in the work of Hlavata et al. (2012) [15]. Gene expression data from paired tumor and non-neoplastic control tissue samples were available for 51 and 50 patients, respectively. These clinical data were considered for statistical testing within this dataset: gender, age, tumor size (pT), lymph node involvement (pN), distant metastases (cM), clinical stage (UICC classification), histological grade, primary tumor localization (colon C18, rectosigmoideum C19, and rectum C20), chemotherapy - first line chemotherapy in adjuvant (cM0) or palliative setting (cM1), grade 3 or 4 toxicity, post-operative radiotherapy, outcome (response to palliative chemotherapy according to RECIST criteria) and disease-free survival (DFS).
The Colorectal II dataset contained unpublished data from a homogeneous cohort of 60 stage II CC patients (all pN0, cM0). Tumors as well as non-neoplastic tissue samples were available for all of the patients. Information about gender, age, tumor size (pT), histological grade, primary tumor localization (colon C18, rectosigmoideum C19, and rectum C20), oncologic treatment administration, DFS, and OS were considered for statistics. Oncologic treatment was started in high-risk group patients.
The Pancreatic dataset was based on the patients separately published within the study conducted by Mohelnikova-Duchonova et al. (2013) [16], and contained 32 pancreatic adenocarcinoma (PC) tumor tissue samples and 27 paired control non-neoplastic tissue samples. From clinical and prognostic factors, we could evaluate gender, age, histological grade, pTNM stage, angioinvasion (pA), perineural invasion (pP), margin status (R0 versus R1), KRAS mutations in codons 12 and 13 (wild type versus mutant), adjuvant chemotherapy administration, and overall survival (OS) for this dataset.
The Breast dataset, previously discussed within the study conducted by Hlavac et al. (2013) [14], consisted of 68 breast carcinoma (BC) patients. Tumor tissue samples were available for all 68 patients and non-neoplastic tissues for 43 patients. These clinicopathological features were available for statistical analyses: age, menopausal status, tumor size (pT), regional lymph node metastasis (pN), distant metastases (cM), clinical stage (UICC classification), histological type and grade, expression of estrogen receptor (ER), progesterone receptor (PR) and HER2 (v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2; ERBB2), expression of the proliferation marker Ki-67 protein, triple negative breast cancer (TNBC) status, neoadjuvant chemotherapy (NACT) regimen, outcome (response to NACT according to RECIST criteria) and DFS.
Clinical data for all four datasets have been continuously updated with the last update in April 2016 and are provided in the Supporting Information in Table S1. The present study was conducted in accordance with the Declaration of Helsinki and Uniform Requirements for manuscripts submitted to Biomedical journals.
Isolation of total RNA, cDNA preparation and quantitative real-time PCR
Non-neoplastic (control) tissue samples were taken from the macroscopically unaffected resection margins of the resected tissues. The resection margins were microscopically evaluated by an experienced pathologist and only samples free of malignant cells were further analysed. Isolation of total RNA, cDNA preparation and quantitative real-time polymerase chain reactions (qRT-PCR) adhered to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments Guidelines [17], and were precisely described in our previous articles [14-16]. RNA quantity was assessed in duplicates by Quant-iT RNA Assay Kit (Invitrogen, Carlsbad, California) using Infinite M200 multimode reader (Tecan, Männedorf, Switzerland). The quality of RNA was assessed by measurement of RNA Integrity Number (RIN) using Agilent 2100 Bioanalyzer and Agilent RNA 6000 Nano Assay Kit (Agilent Technologies, Santa Clara, California). The same amount and quality of input RNA were applied for each sample within all the studied tumor cohorts. The quality of cDNA in terms of DNA contamination was confirmed by PCR amplification of ubiquitin C discriminating between product from cDNA (190 bp) and from genomic DNA (1,009 bp) as described previously. Prior to qRT-PCR, the preamplification uniformity of cDNA was checked according to the manufacturer's recommendation.
Gene expression profiling
Gene expression of all 49 described members of the human ABC gene family (including the ABCC13 pseudogene) was measured for all four datasets (Colorectal I, Colorectal II, Pancreatic, and Breast dataset). Because the transcript levels of ABCC12 were below the limit of method detection in all samples of the Colorectal I dataset, data from 48 ABC transporters without ABCC12 were included in the bioinformatic analyses, which were performed for the dataset. Similarly, the transcript levels of the genes ABCC13, ABCG4 and ABCG5 were below the limit of method detection in all samples of the Pancreatic dataset and that was the reason why 46 ABC genes were further analyzed for the pancreatic samples. Finally, 28 selected ABC genes (ABCA7, ABCA12, ABCA13, ABCB1, ABCB2, ABCB3, ABCB4, ABCB6, ABCB9, ABCB11, ABCC1, ABCC2, ABCC3, ABCC4, ABCC5, ABCC6, ABCC7, ABCC8, ABCC10, ABCC11, ABCD4, ABCE1, ABCF1, ABCF2, ABCG1, ABCG2, ABCG5, ABCG8), covering all ABC subfamilies, were measured and analyzed for the Colorectal II dataset. When comparing the two colorectal datasets (Colorectal I and Colorectal II), there was no preamplification in the Colorectal II dataset.
Statistics and bioinformatics
Statistical and bioinformatic analyses were performed on the ABC gene expression data obtained from the available non-neoplastic (control) tissues (see Characteristics of datasets). Preprocessing of our gene expression data was performed in Microsoft Excel 2013 and Statistica software (Statsoft, Czech Republic). Raw gene expression data (Ct values) were normalized by the average Ct value of each gene of interest (ABC gene) and then log2-transformed. Statistical analyses (including survival analyses) were conducted with the help of Statistica and the freely available PAST statistical software package [18]. Clusters were determined by tree clustering with Ward's method and then visualized in a heat map manner. With the aim to show comparable characteristics of gene clusters, the median expression value for each ABC gene was calculated. Afterwards one median expression value for the whole cluster, representing all the median expression values of all the ABC genes tested in the cluster, was determined. Clinical data were sorted according to the relevant clusters, visualized in heat maps for an overall review and tested using predominantly non-parametric statistics (the Mann-Whitney U and Kruskal-Wallis tests). Disease-free survival (DFS) was defined as the time elapsed between surgery and disease recurrence or death. Overall survival (OS) was defined as the time elapsed between surgery and the patient´s death from any cause. Median survival was estimated using the Kaplan-Meier method and the difference was tested using the log-rank test. Two-sided p-values of less than 0.05 were considered statistically significant. Monte Carlo values based on the given number of random permutations (default 9999) were also computed, mainly as controls on the asymptotic values. The comprehensive analysis of ABC gene functions was based on multiple searches in several publicly available databases - NCBI Gene, The Weizmann Institute of Science GeneCards, UniProtKB/Swiss-Prot, UCSC Genome Browser, Kyoto Encyclopedia of Genes and Genomes, Genetics Home Reference, and Reactome.
Results
Colorectal I dataset
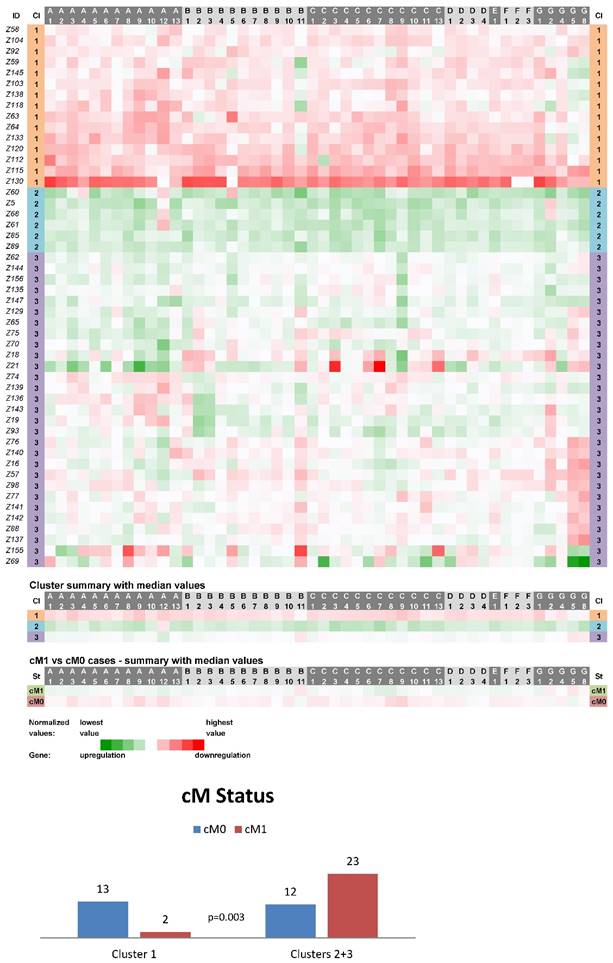
Expression data from 50 non-neoplastic (control) tissue samples paired with CC samples were clustered into three main clusters (CN1-CN3, Fig. 1). The cluster CN1 consisted of 15 cases and its characteristics can be summarized as downregulation of the vast majority of ABC genes (the median number of downregulated genes was 46 out of 48 with the median expression value 0.049 on the logarithmic scale used). Oppositely, upregulation of the vast majority of ABC genes characterized the cluster CN2 containing 6 cases (47 out of 48 genes upregulated with the median expression value -0.090). The rest of the non-neoplastic samples (scattered within the cluster CN3 with N=29) displayed heterogeneous levels of expression of the ABC genes (5 genes downregulated with the median expression value 0.029 and 43 genes upregulated with the median expression value -0.016). Statistical analyses revealed the interesting result that cases positive for distant metastasis (cM1 status) were significantly less prevalent among the cluster CN1 patients (p=0.003, Fig. 1). Moreover, in median value comparisons between all the cM1 versus cM0 cases (N=25 for both groups), regardless of the affiliation to the clusters CN1-CN3, the vast majority of genes showed upregulation within the cM1 group (43 genes) and downregulation within the cM0 group (41 genes; bottom part of the Fig. 1). Survival analyses did not disclose any statistically significant result (median follow-up 65 months).
Colorectal II dataset
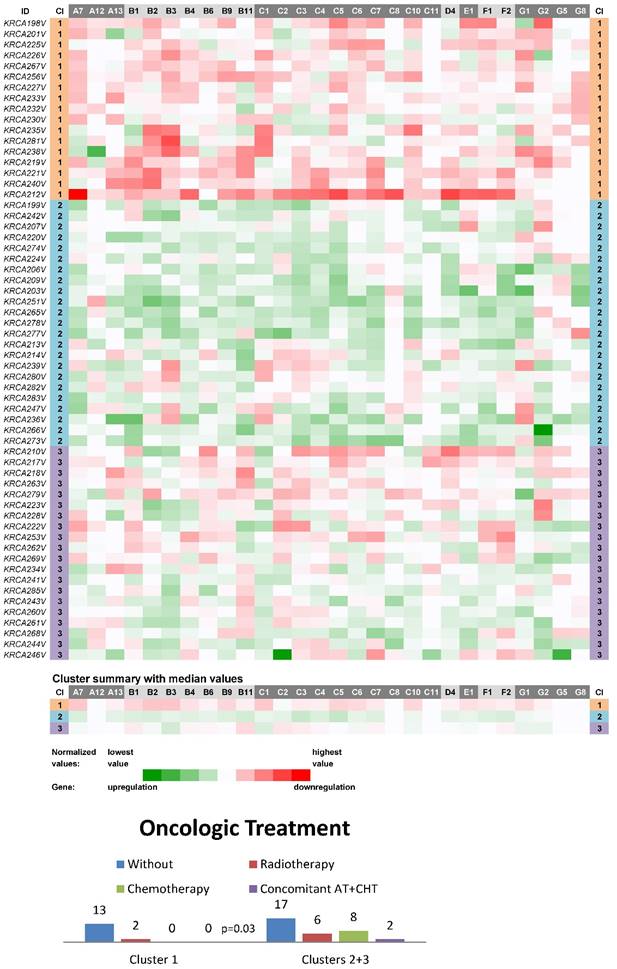
Three main clusters (CIIN1-CIIN3) were also distinguished among non-neoplastic samples from this most homogeneous dataset in terms of clinical stage, which consisted of 60 stage II CC patients (Fig. 2). In a similar manner, the vast majority of ABC genes were downregulated in the cluster CIIN1 (N=17; the median number of downregulated genes was 22 out of 28 with the median expression value 0.036 on the logarithmic scale used), upregulated in CIIN2 (N=23; 24 out of 28 genes upregulated with the median expression value -0.033) and had heterogeneous expression in CIIN3 (N=20; 12 genes upregulated with the median expression value -0.010 and 11 genes downregulated with the median expression value 0.012). Statistical analyses showed this compelling result: patients without the need for oncologic treatment prevailed within the cluster CIIN1 (p=0.03, Fig. 2). Oncologic treatment was started in high-risk group patients in this cohort. Survival analyses, however, showed no significant difference between the clusters (median follow-up 24 months).
Pancreatic dataset
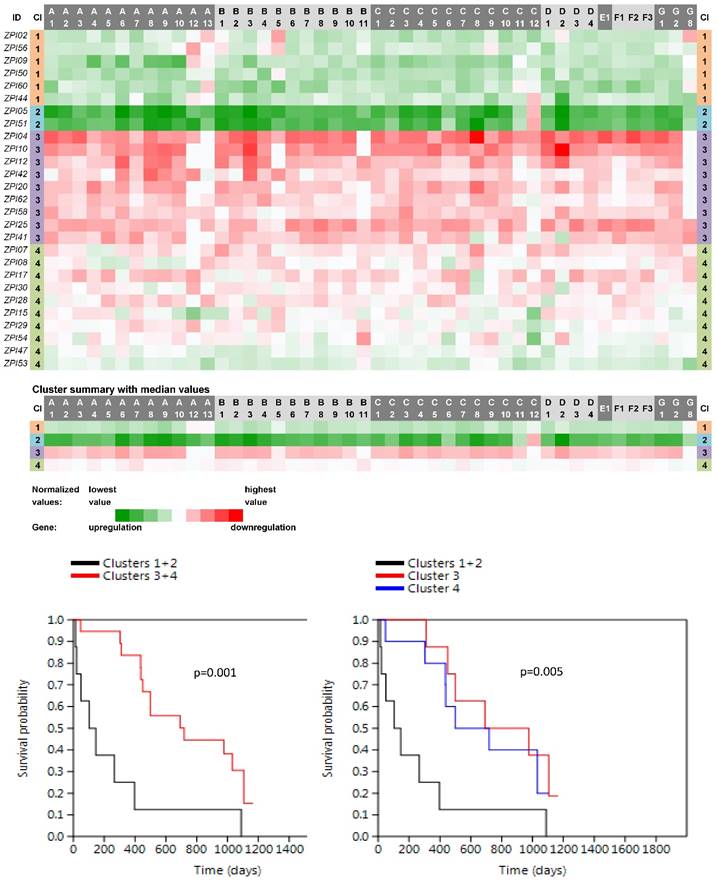
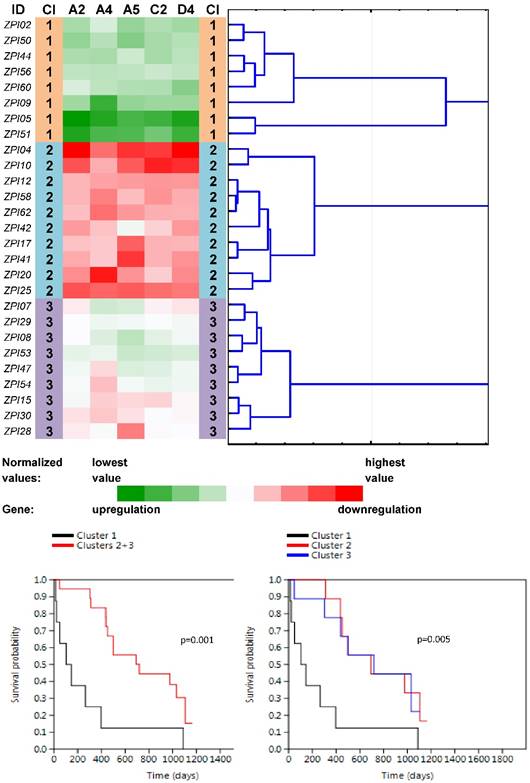
Four main clusters (PN1-PN4) were revealed between 27 non-neoplastic tissue samples related to PC samples (Fig. 3). The vast majority of ABC genes showed upregulation within the cluster PN1 (N=6; median number of upregulated genes was 44 out of 46 with the median expression value -0.083 on the logarithmic scale used) and even stronger upregulation within the cluster PN2 (N=2; 45 out of 46 genes upregulated with the median expression value -0.252). Contrastingly, the vast majority of genes showed downregulation within PN3 (N=9; 42 out of 46 genes downregulated with the median expression value 0.078). The rest of the cases with heterogeneous levels of expression were assigned to the cluster PN4 (N=10; 8 genes only slightly upregulated with the median expression value -0.005 and 38 genes downregulated with the median expression value 0.011). Survival analyses revealed the important result - patients belonging to the clusters PN1 and PN2 had significantly shorter OS compared to patients within PN3 and PN4 (the Log rank test, p=0.001, Fig. 3).
Breast dataset
Forty-three non-neoplastic tissue samples paired with BC samples were divided into three main clusters (BN1-BN3; heat map visualization of the tree clustering is provided in Fig. S1 in the Supporting Information). Cluster BN1 contained 11 samples and can be characterized as having the vast majority of ABC genes downregulated (the median number of downregulated genes was 48 out of 49 with the median expression value 0.045 on the logarithmic scale used). Cluster BN2, representing the majority of samples, contained 29 samples and the vast majority of genes were upregulated (48 out of 49 genes upregulated with the median expression value -0.052). Only three samples were included into the cluster BN3; however, the vast majority of ABC genes were strongly downregulated (48 out of 49 genes downregulated with the median expression value 0.316). No statistically significant relationships were revealed between these three main ABC gene expression profiles in non-neoplastic tissues and clinicopathological features (including DFS) of our BC patients.
Heat maps showing results of the tree clustering (Ward's method, Euclidean distances) of the expression levels of 48 human ABC transporters in non-neoplastic (control) tissues of colorectal carcinoma patients (Colorectal I cohort) and cM Status histograms; Cl, cluster
Heat maps showing results of the tree clustering (Ward's method, Euclidean distances) of the expression levels of 28 human ABC transporters in non-neoplastic (control) tissues of stage II colorectal carcinoma patients (Colorectal II cohort) and Oncologic Treatment histograms; Cl, cluster
Reduced ABC gene profiles suggested as new prognostic markers
In order to find ABC gene profiles more suitable for use in routine laboratory praxis, we reduced the number of ABC genes, while the statistical significance of the discovered relationships was maintained. Normalized expression values for each of the analyzed ABC genes were sorted from the smallest to the largest. Relevant patients were divided into two mutually exclusive groups - one containing only the negative values and the other one positive values. For the following analyses of the individual ABC genes, upregulation of the relevant ABC gene was represented by the group containing only the negative values and downregulation by the group with only positive values. We were aware of the inaccuracy caused by the cases with the values close to zero, which had median expression; however, it was acceptable at that stage of the ABC gene selection process. Afterwards, statistical analyses comparing the two established groups (upregulation versus downregulation) for each ABC gene individually were performed for the relevant clinicopathological features (cM status for Colorectal I dataset and OS for Pancreatic dataset).
Heat maps showing results of the tree clustering (Ward's method, Euclidean distances) of the expression levels of 46 human ABC transporters in non-neoplastic (control) tissues of pancreatic adenocarcinoma patients and survival analyses (the Kaplan-Meier plots, the Log rank test p-values); Cl, cluster
Twenty-two genes showed individually significant differences (p<0.05) in cM status and 10 genes (ABCA3, ABCA5, ABCB8, ABCB10, ABCC1, ABCC6, ABCC7, ABCC8, ABCC10, ABCF1) an even more significant difference (p<0.01) within the Colorectal I dataset when comparing downregulated versus upregulated cases (results of the statistical analyses are disclosed in Figs. S2, S3, S4, S5 and S6 in the Supporting Information). Similarly, 5 genes (ABCA2, ABCA4, ABCA5, ABCC2, and ABCD4) were revealed to have significantly different OS between downregulated and upregulated cases within the Pancreatic dataset (individual survival analyses are presented in Fig. S7 in the Supporting Information).
Tree clustering (Ward's method, Euclidean distances), heat map visualization and statistical analyses (clusters versus cM status histograms, the Kruskal-Wallis test p-value) of gene expression levels of the ten ABC transporters panel in non-neoplastic (control) tissues of colorectal carcinoma patients (Colorectal I cohort); Cl, cluster
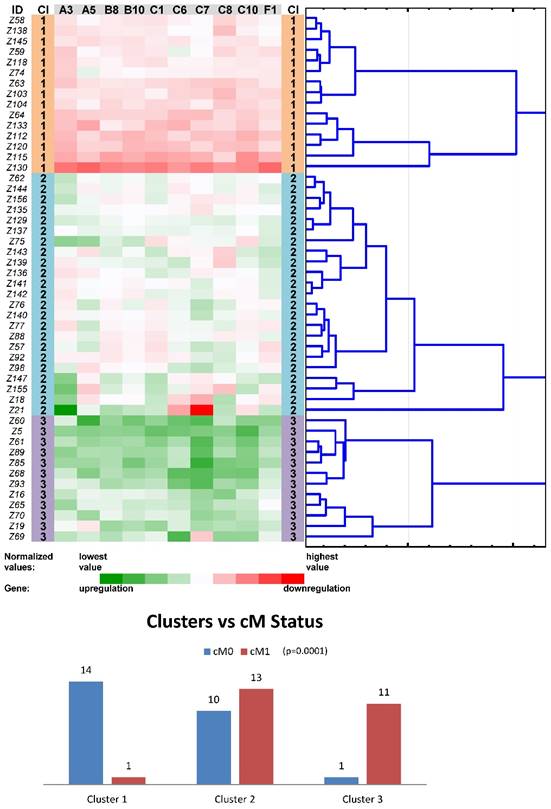
Another round of tree clustering analyses followed by statistical tests were done with the two reduced panels of individually significant ABC genes (for the Colorectal I and Pancreatic datasets), in a similar way as before with the whole set of human ABC genes (Figs. 4 and 5). Both panels (ABCA3, ABCA5, ABCB8, ABCB10, ABCC1, ABCC6, ABCC7, ABCC8, ABCC10, and ABCF1 for CC patients and ABCA2, ABCA4, ABCA5, ABCC2, and ABCD4 for PC patients) were shown to cluster non-neoplastic samples into three main clusters - with mostly downregulated, upregulated or heterogeneously expressed genes. Most importantly, both panels also retained the same prognostic relevance, which was shown for the whole set of human ABC genes before (p=0.001 for the cM status in the Colorectal I dataset and p=0.001 for the OS in the Pancreatic dataset; Figs. 4 and 5). Notably, a trend for better OS in the patients within the downregulated cluster with a reduced number of ABC genes could be distinguished also for the Colorectal I cohort. However, this result was without statistical significance and a larger cohort and longer follow-up is needed to prove this possible relationship to OS in colorectal cancer patients (see Fig. S8 in the Supporting Information).
Discussion
In the current article, we presented the evidence that the gene expression values of all human ABC genes in non-neoplastic (control) tissues, taken together with tumor tissues during surgery treatment, can be characteristically clustered into three main profiles (signatures). These main profiles were characterized by the downregulation, upregulation or heterogeneous deregulation of the vast majority of ABC genes. Furthermore, the downregulated and upregulated main profiles were significantly related to cM status and OS in colorectal and pancreatic cancer, respectively. The suggested multi-gene panels, consisting of the reduced number of ABC genes, could be easily implemented in clinical practice as new prognostic markers. These findings support the theory that the expression of ABC genes in non-neoplastic tissues can contribute to tumor progression generally.
Tree clustering (Ward's method, Euclidean distances), heat map visualization and survival analyses (the Kaplan-Meier plots, the Log rank test p-values) of gene expression levels of the five ABC transporters panel in non-neoplastic (control) tissues of pancreatic carcinoma patients; Cl, cluster
The framework for our theory was mainly based on the results of the previous studies, which explored ABC expression profiles in tumor tissues and compared them to the relevant non-neoplastic tissues. Using oligonucleotide microarrays, Park et al. (2006) [19], analyzed the expression profiles of all human ABC genes in a cohort of 21 breast cancer patients who underwent neoadjuvant chemotherapy. They compared the ABC profiles between two groups of pretreatment tumor samples according to the patients' pathological response - residual disease versus complete response. Several ABC transporters showed differential expression between the two groups and the feasibility of developing a multigene predictor model of pathological response to neoadjuvant chemotherapy using gene expression profiles of ABC transporters was evaluated. Their results suggested that several ABC transporters in human breast cancer cells may affect the clinical response to neoadjuvant chemotherapy, and transcriptional profiling of these genes may be useful to predict the pathological response.
Similarly, the relative expression levels of all ABC genes in 51 tumor samples originating from acute myeloid leukemia (AML) patients were analyzed with the use of TaqMan custom arrays in the study described by Marzac et al. [20]. Patients were divided into two extreme groups, one very sensitive and the other very resistant to the administered chemotherapy. The prognostic impact of six ABC genes, selected in the first part of the study, was than evaluated on a cohort of 281 AML patients. Interestingly, the strongest prognostic factor was the number of genes overexpressed among the three ABC genes - ABCB1, ABCG1, and ABCG2. Therefore, ABC transporters may cooperate to promote chemoresistance rather than overexpression of single transporters being responsible, and modulation of multiple transporters might be required to increase intracellular drug accumulation and to induce chemotherapeutic eradication of leukemic cells in AML.
Previous studies originating from our laboratories also explored transcript levels of all human ABCs in several tumors (BC, CC, and PC) in comparison with paired non-neoplastic (control) tissues [14-16]. In the case of our BC cohort, comparable to the cohort published by Park et al. [19], ABC gene transcript levels were determined in post-treatment tumors together with non-neoplastic tissues from patients treated by neoadjuvant chemotherapy. Specimens (tumor and paired non-neoplastic tissues) from our CC patient cohort I were collected before the first line of treatment by a 5-fluorouracil (5-FU)-containing regimen, and from our PC patients immediately after surgical treatment. Several significant connections between individual ABC genes and clinicopathological features were revealed. We recently proposed the existence of ABC gene expression profiles common to more types of tumors based on a comprehensive bioinformatic analysis of our results concerning ABC gene expression profiles in different tumor tissues together with a literature review [11].
A novel perspective on this theme was brought by our theory that the ABC gene expression profiles in non-neoplastic tissues can also bear clinically important information in relation to tumor progression. The results in support of this theory were addressed in the present article.
Notably, with the use of the selected clustering methodology expression data from non-neoplastic tissues were divided into similar three main clusters independently in four cohorts, which represented three types of tumors (BC, CC and PC). Downregulation, upregulation or heterogeneous deregulation of the vast majority of ABC genes were proposed as the characteristics of the main types of clusters. Clinical stage independence of the determined clusters was demonstrated by the similar results of the clustering within our Colorectal II cohort, which was the most homogeneous in terms of clinical stages and consisted of only stage II CC patients. Moreover, statistical analyses revealed significant relationships between the main clusters and cM status in colorectal cancer and OS in pancreatic cancer. In line with these findings, oncologic treatment was started significantly less frequently among the patients with the downregulation of the vast majority of ABC genes in the homogeneous Colorectal II cohort. Importantly, the administration of oncologic treatment was limited to the high-risk patients in that cohort. In our opinion, all the presented results support the theory that was proposed at the beginning. Therefore, the presence of downregulation and upregulation of the vast majority of ABC genes in control tissues was stated as a promising positive and negative prognostic marker, respectively, for the two types of gastrointestinal tumors, but not for breast carcinoma.
In addition, selected multi-gene panels with a reduced number of ABC genes were suggested for a possible testing in clinical laboratory practice. There is still a lack of efficient prognostic markers in CC and especially in PC. Because PC represents one of the most deadly tumors, the effective prognostic stratification of patients and subsequent therapy modifications are of major importance worldwide. The mechanisms underlying the relationships between ABC genes in non-neoplastic tissues and tumor progression are still unknown and to reveal them can be a new and exciting issue for the primary research into the principles of carcinogenesis.
The current study was a pioneering one and has several limitations. The distance from the removed control tissues to the tumor tissues can represent an important factor. Due to the common practices in tumor surgery, the largest distance between tumor and paired control tissues was in CC samples, the shortest in BC and the median in PC. The number of patients in the Pancreatic dataset and the length of BC and CC patient follow-up were further limitations, which need to be solved by the subsequent studies. However, the possible importance and benefit of the presented results would satisfy the publication and enables quicker validation or modification of the results by other research groups.
We selected ABC profiles with a reduced number of genes, while maintaining the statistical significance for the discovered relationships, in order to find an affordable assay suitable for use in majority of routine clinical laboratories. For CC samples the reduced panel includes 10 genes (ABCA3, ABCA5, ABCB8, ABCB10, ABCC1, ABCC6, ABCC7, ABCC8, ABCC10, and ABCF1) and for PC samples 5 genes (ABCA2, ABCA4, ABCA5, ABCC2, and ABCD4). Importantly, both panels need further validation on larger cohorts of CC and PC patients. Using clustering methodology adopted in this study, the cut-off values for determination of downregulation or upregulation can be easily specified by each laboratory individually, becoming more precise with the increasing number of cases involved in the panel validation. The advantage of this approach is also in the independence of the laboratory equipment used for the gene expression measurements.
The physiological functions of the ABC genes, included into the two assay panels, are mostly still poorly understood; however, connections to cancer were shown for all of them. The ABCA3 protein (the first member of the panel for CC patients) transports phospholipids into the lamellar bodies where they interact with surfactant proteins to form surfactant. The ABCA3 protein also appears to be involved in the formation of normal lamellar bodies and may be involved in development of resistance to xenobiotics and engulfment during programmed cell death (if not specifically stated, information about gene functions was retrieved from the publicly available databases; see Materials and Methods). The protein encoded by the ABCA5 gene, located in the 17q24 chromosome region in a cluster together with ABCA6, ABCA8, ABCA9 and ABCA10 genes, is probably involved in lipid transport including cholesterol efflux. Induction of ABCA5, together with ABCB1, appeared to be correlated with the differentiation state of human colon tumors, and may have a role in tumor development [21]. It has been speculated that ABCB8, like other mitochondrial transporters - ABCB6, ABCB7, and ABCB10, is involved in the transport of heme, phospholipids, and/or peptides. ABCB8 is able to form complexes with succinate dehydrogenase, nucleotide translocators, inorganic phosphate carriers, and ATP synthase in mitochondrial ATP-sensitive K+ channels, which are probably involved in the protection of cells against oxidative stress [22]. Similarly, ABCB10 may mediate critical mitochondrial transport functions related to heme biosynthesis; however, recent evidence points toward ABCB10 as an important player in the protection from oxidative stress in mammals [23]. ABCC1 (MRP1) mediates ATP-dependent export of glutathione and glutathione conjugates, leukotriene C4, estradiol-17-beta-o-glucuronide, methotrexate, antiviral drugs and other xenobiotics from the cytoplasm and confers resistance to anticancer drugs. Some studies suggest that ABCC6 (MRP6) stimulates the release of adenosine triphosphate (ATP) from cells through an unknown mechanism. Moreover, ABCC6 may participate directly in the active transport of drugs into subcellular organelles or influence drug distribution indirectly; isoform 2 inhibits TNF-alpha-mediated apoptosis through blocking one or more caspases. Chloride channels, encoded by the ABCC7 (official symbol CFTR) gene, are a family of anion-selective channels involved in the regulation of the excitability of neurons, skeletal, cardiac and smooth muscle, cell volume regulation, transepithelial salt transport and the acidification of intra- and extracellular compartments. Surprisingly, an important role of CFTR protein in gastrointestinal cancer development was suggested by several recent works [24, 25] and also by our previous results [11]. The ABCC8 protein functions as a modulator of ATP-sensitive potassium channels and insulin release, and together with ABCD2 was recently suggested to play a putative role in breast cancer progression and prognosis of patients [14, 26]. ABCC10 is ATP-dependent transporter probably involved in cellular detoxification through lipophilic anion extrusion. Recent data identify roles for ABCC10 in breast and colorectal cancer pathogenesis and docetaxel resistance [27, 28]. The ABCF1 protein may be regulated by tumor necrosis factor-alpha and play a role in enhancement of protein synthesis and the inflammation process.
ABCA2 (the first member of the panel for PC patients) is highly expressed in brain tissue and may play a role in macrophage lipid metabolism and neural development. Together it and ABCA9, ABCA10, ABCC9, ABCG2 and SLC16A14 present novel putative markers of epithelial ovarian cancer progression [29]. ABCA4 is expressed almost exclusively in retinal photoreceptor cells, indicating the gene product mediates transport of essential molecules across the photoreceptor cell membrane. Its connection to cancer is limited to a few reports to date [30]. ABCA5 was included into the both assay panels and discussed above. ABCC2 (MRP2) mediates hepatobiliary excretion of numerous organic anions and anticancer drugs such as vinblastine or cisplatin belong to its substrates. Vinette et al. [31] reported that ABCC2 expression was upregulated by adenosine 5'-triphosphate (ATP) in colorectal cancer cells and enhanced their survival when exposed to chemotherapeutic drugs. The ABCD4 protein may be involved in intracellular processing of vitamin B12 (cobalamin) and there are only a very few reports connecting it to cancer [15, 32].
From the above-described plethora of cellular functions it seems that the prognostic significance of the common dysregulation profiles (up or downregulation) revealed by the present study in non-tumor tissues most likely cannot be simply attributed to a few functions such as drug or lipid transport. We also do not know the timeline of these changes and a clear link to carcinogenesis is still missing. Andersen and coworkers [33, 34] reported that changes in the levels of ABC transporters were early events in the adenoma-carcinoma sequence leading to CC. It would definitely be interesting to track the changes defined in the current article in some pre-neoplastic material as intestinal polyps or tissues from patients with pancreatitis. Such changes if found there could bear predictive information and help with earlier diagnosis of carcinogenesis, which is urgently needed, especially for PC. Moreover, the genetic background of these changes could add another dimension, because genetic testing based on phenotype would be more cost and labor effective. Such studies are now in the scope of our next research.
In conclusion, results in support of the theory that deregulation of the expression of ABC genes in non-neoplastic tissues, matched to tumor samples, can bear clinically relevant information in relation to tumor progression were shown. ABC gene expression profiles with dominating downregulation or upregulation patterns were suggested to be potent prognostic markers in two different types of gastrointestinal tumors and are awaiting further validation.
Supplementary Material
Supplementary figures and tables.
Abbreviations
ABC: ATP-binding cassette; BC: breast carcinoma; CC: colorectal carcinoma; cM: distant metastases; DFS: disease-free survival; ER: estrogen receptor; NACT: neoadjuvant chemotherapy; OS: overall survival; PC: pancreatic adenocarcinoma; pN: regional lymph node metastasis; PR: progesterone receptor; pT: tumor size; qRT-PCR: quantitative real-time polymerase chain reaction; TNBC: triple negative breast cancer
Acknowledgements
This work was supported by the Ministry of Health of the Czech Republic [Conceptual Development of Research Organization -Faculty Hospital in Pilsen - FNPl, grant no. 00669806; and grant no. AZV 15-25618A]; the Czech Science Foundation [project no. P303/12/G163]; and the National Sustainability Program I (NPU I) provided by the Ministry of Education Youth and Sports of the Czech Republic [project no. LO1503].
Ethical approval
All procedures involving human participants were in accordance with the ethical standards of the institutional and national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Competing Interests
The authors declare no competing financial interests.
References
1. Vasiliou V, Vasiliou K, Nebert D. Human ATP-binding cassette (ABC) transporter family. Hum Genomics. 2009;3:281-90
2. Falasca M, Linton K. Investigational ABC transporter inhibitors. Expert Opin Investig Drugs. 2012;21:657-66 doi: 10.1517/13543784.2012.679339
3. Dean M, Rzhetsky A, Allikmets R. The Human ATP-Binding Cassette (ABC) Transporter Superfamily. Genome Res. 2001;11:1156-66
4. Hopfner K. Invited review: Architectures and mechanisms of ATP binding cassette proteins. Biopolymers. 2016;105:492-504 doi: 10.1002/bip.22843
5. Aye I, Singh A, Keelan J. Transport of lipids by ABC proteins: Interactions and implications for cellular toxicity, viability and function. Chem Biol Interact. 2009;180:327-39 doi: 10.1016/j.cbi.2009.04.012
6. Lee B, Taylor M, Robinet P, Smith J, Schweitzer J, Sehayek E, Falzarano S, Magi-Galluzzi C, Klein E, Ting A. Dysregulation of Cholesterol Homeostasis in Human Prostate Cancer through Loss of ABCA1. Cancer Res. 2013;73:1211-8 doi: 10.1158/0008-5472.CAN-12-3128
7. Wu Q, Yang Z, Nie Y, Shi Y, Fan D. Multi-drug resistance in cancer chemotherapeutics: Mechanisms and lab approaches. Cancer Lett. 2014;347:159-66 doi: 10.1016/j.canlet.2014.03.013
8. Kachalaki S, Ebrahimi M, Mohamed Khosroshahi L, Mohammadinejad S, Baradaran B. Cancer chemoresistance; biochemical and molecular aspects: a brief overview. Eur J Pharm Sci. 2016;89:20-30 doi: 10.1016/j.ejps.2016.03.025
9. Shukla S, Ohnuma SV, Ambudkar S. Improving Cancer Chemotherapy with Modulators of ABC Drug Transporters. Curr Drug Targets. 2011;12:621-30
10. Kathawala R, Gupta P, Ashby C, Chen Z. The modulation of ABC transporter-mediated multidrug resistance in cancer: A review of the past decade. Drug Resist Updat. 2015;18:1-17 doi: 10.1016/j.drup.2014.11.002
11. Dvorak P, Pesta M, Soucek P. ABC gene expression profiles have clinical importance and possibly form a new hallmark of cancer. Tumor Biol. 2017;39:1-16 doi: 10.1177/1010428317699800
12. Hu Y, Yu X, Xu G, Liu S. Metastasis: an early event in cancer progression. J Cancer Res Clin Oncol. 2016 [Epub ahead of print]. doi:10.1007/s00432-016-2279-0
13. Alderton G. Tumour microenvironment: Driving relapse. Nat Rev Cancer. 2015;15:195. doi: 10.1038/nrc3935
14. Hlaváč V, Brynychová V, Václavíková R, Ehrlichová M, Vrána D, Pecha V, Koževnikovová R, Trnková M, Gatěk J, Kopperová D, Gut I, Souček P. The expression profile of ATP-binding cassette transporter genes in breast carcinoma. Pharmacogenomics. 2013;14:515-29 doi: 10.2217/pgs.13.26
15. Hlavata I, Mohelnikova-Duchonova B, Vaclavikova R, Liska V, Pitule P, Novak P, Bruha J, Vycital O, Holubec L, Treska V, Vodicka P, Soucek P. The role of ABC transporters in progression and clinical outcome of colorectal cancer. Mutagenesis. 2012;27:187-96 doi: 10.1093/mutage/ger075
16. Mohelnikova-Duchonova B, Brynychova V, Oliverius M, Honsova E, Kala Z, Muckova K, Soucek P. Differences in Transcript Levels of ABC Transporters Between Pancreatic Adenocarcinoma and Nonneoplastic Tissues. Pancreas. 2013;42:707-16 doi: 10.1097/MPA.0b013e318279b861
17. Bustin S, Benes V, Garson J, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl M, Shipley G, Vandesompele J, Wittwer C. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin Chem. 2009;55:611-22 doi: 10.1373/clinchem.2008.112797
18. Hammer Ø, Harper D, Ryan P. PAST: Paleontological Statistics Software Package for Education and Data Analysis [Internet]; Palaeontologia Electronica; 2001 [cited 2016 Oct 16]. http://palaeo-electronica.org/2001_1/past/issue1_01.htm.
19. Park S, Shimizu C, Shimoyama T, Takeda M, Ando M, Kohno T, Katsumata N, Kang Y, Nishio K, Fujiwara Y. Gene expression profiling of ATP-binding cassette (ABC) transporters as a predictor of the pathologic response to neoadjuvant chemotherapy in breast cancer patients. Breast Cancer Res Treat. 2006;99:9-17
20. Marzac C, Garrido E, Tang R, Fava F, Hirsch P, De Benedictis C, Corre E, Lapusan S, Lallemand J, Marie J, Jacquet E, Legrand O. ATP Binding Cassette transporters associated with chemoresistance: transcriptional profiling in extreme cohorts and their prognostic impact in a cohort of 281 acute myeloid leukemia patients. Haematologica. 2011;96:1293-301 doi: 10.3324/haematol.2010.031823
21. Ohtsuki S, Kamoi M, Watanabe Y, Suzuki H, Hori S, Terasaki A. Correlation of Induction of ATP Binding Cassette Transporter A5 (ABCA5) and ABCB1 mRNAs with Differentiation State of Human Colon Tumor. Biol Pharm Bull. 2007;30:1144-6
22. Sachrajda I, Ratajewski M. Mithramycin A suppresses expression of the human melanoma-associated gene ABCB8. Mol Genet Genomics. 2011;285:57-65 doi: 10.1007/s00438-010-0586-8
23. Liesa M, Qiu W, Shirihai O. Mitochondrial ABC transporters function: The role of ABCB10 (ABC-me) as a novel player in cellular handling of reactive oxygen species. Biochim Biophys Acta. 2012;1823:1945-57 doi: 10.1016/j.bbamcr.2012.07.013
24. Than B, Linnekamp J, Starr T, Largaespada D, Rod A, Zhang Y, Bruner V, Abrahante J, Schumann A, Luczak T, Niemczyk A, O'Sullivan M. et al. CFTR is a tumor suppressor gene in murine and human intestinal cancer. Oncogene. 2016;35:4179-87 doi: 10.1038/onc.2015.483
25. Xu J, Lin L, Yong M, Dong X, Yu T, Hu L. Adenovirus-mediated overexpression of cystic fibrosis transmembrane conductance regulator enhances invasiveness and motility of serous ovarian cancer cells. Mol Med Rep. 2016;13:265-72 doi: 10.3892/mmr.2015.4509
26. Soucek P, Hlavac V, Elsnerova K, Vaclavikova R, Kozevnikovova R, Raus K. Whole exome sequencing analysis of ABCC8 and ABCD2 genes associating with clinical course of breast carcinoma. Physiol Res. 2015;64(Suppl 4):S549-S557
27. Domanitskaya N, Wangari-Talbot J, Jacobs J, Peiffer E, Mahdaviyeh Y, Paulose C, Malofeeva E, Foster K, Cai K, Zhou Y, Egleston B, Hopper-Borge E. Abcc10 status affects mammary tumour growth, metastasis, and docetaxel treatment response. Br J Cancer. 2014;111:696-707 doi: 10.1038/bjc.2014.326
28. Krizkova V, Dubova M, Susova S, Vycital O, Bruha J, Skala M, Liska V, Daum O, Soucek P. Protein expression of ATP-binding cassette transporters ABCC10 and ABCC11 associates with survival of colorectal cancer patients. Cancer Chemother Pharmacol. 2016;78:595-603 doi: 10.1007/s00280-016-3114-7
29. Elsnerova K, Mohelnikova-Duchonova B, Cerovska E, Ehrlichova M, Gut I, Rob L, Skapa P, Hruda M, Bartakova A, Bouda J, Vodicka P, Soucek P. et al. Gene expression of membrane transporters: Importance for prognosis and progression of ovarian carcinoma. Oncol Rep. 2016;35:2159-70 doi: 10.3892/or.2016.4599
30. Nymoen D, Holth A, Hetland Falkenthal T, Tropé C, Davidson B. CIAPIN1 and ABCA13 are markers of poor survival in metastatic ovarian serous carcinoma. Mol Cancer. 2015;14:44. doi: 10.1186/s12943-015-0317-1
31. Vinette V, Placet M, Arguin G, Gendron F. Multidrug Resistance-Associated Protein 2 Expression Is Upregulated by Adenosine 5'-Triphosphate in Colorectal Cancer Cells and Enhances Their Survival to Chemotherapeutic Drugs. PLoS One. 2015;10:e0136080. doi: 10.1371/journal.pone.0136080
32. Heimerl S, Bosserhoff A, Langmann T, Ecker J, Schmitz G. Mapping ATP-binding cassette transporter gene expression profiles in melanocytes and melanoma cells. Melanoma Res. 2007;17:265-73
33. Andersen V, Vogel U, Godiksen S, Frenzel FB, Sæbø M, Hamfjord J, Kure E, Vogel LK. Low ABCB1 gene expression is an early event in colorectal carcinogenesis. PLoS One. 2013;8:e72119. doi: 10.1371/journal.pone.0072119
34. Andersen V, Vogel LK, Kopp TI, Sæbø M, Nonboe AW, Hamfjord J, Kure EH, Vogel U. High ABCC2 and low ABCG2 gene expression are early events in the colorectal adenoma-carcinoma sequence. PLoS One. 2015;10:e0119255. doi: 10.1371/journal.pone.0119255
Author contact
![]() Corresponding author: Pavel Dvorak, MSc.; Charles University, Faculty of Medicine in Pilsen, Department of Biology, Alej Svobody 76, 323 00, Pilsen, Czech Republic; Tel.: +420 377 593 263, Email: Pavel.Dvorakcuni.cz
Corresponding author: Pavel Dvorak, MSc.; Charles University, Faculty of Medicine in Pilsen, Department of Biology, Alej Svobody 76, 323 00, Pilsen, Czech Republic; Tel.: +420 377 593 263, Email: Pavel.Dvorakcuni.cz

 Global reach, higher impact
Global reach, higher impact