3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(13):2554-2560. doi:10.7150/jca.20031 This issue Cite
Research Paper
A Multi-Center Prospective Study to Validate an Algorithm Using Urine and Plasma Biomarkers for Predicting Gleason ≥3+4 Prostate Cancer on Biopsy
1. NeoGenomics Laboratories, Irvine, CA;
2. Odense University Hospital, Odense, Denmark;
3. University of California, Irvine, CA;
4. Comprehensive Urology, Royal Oak, MI;
5. Carolina Urologic Research Center, Myrtle Beach, SC.
Received 2017-3-11; Accepted 2017-5-28; Published 2017-8-2
Abstract
Background: Unnecessary biopsies and overdiagnosis of prostate cancer (PCa) remain a serious healthcare problem. We have previously shown that urine- and plasma-based prostate-specific biomarkers when combined can predict high grade prostate cancer (PCa). To further validate this test, we performed a prospective multicenter study recruiting patients from community-based practices.
Patients and Methods: Urine and plasma samples from 2528 men were tested prospectively. Results were correlated with biopsy findings, if a biopsy was performed as deemed necessary by the practicing urologist. Of the 2528 patients, biopsy was performed on only 524 (21%) patients.
Results: Of the 524 patients, Gleason≥3+4 PCa was found in 161 (31%) and Gleason ≥4+3 was found in 62 (12%) of the patients.
The urine/plasma biomarkers algorithm showed sensitivity and specificity of 75% and 69% for predicting Gleason ≥3+4. However, upon incorporating prostate size and prior history of biopsy in the algorithm, we achieved a sensitivity between 97% and 86% and a specificity between 36% and 57%, dependent on the used cut-off point. Sensitivity for predicting PCa Gleason ≥4+3 was between 96% and 99% and specificity between 59% and 37%, dependent on the cut-off point. Diagnosis of Gleason ≥3+4 was missed in 1% to 3% of tested patients and of Gleason ≥4+3 in 0.2% to 1%.
Conclusion: This test when integrated with prostate volume and the prior prostate biopsy enhance the sensitivity and specificity for predicting the presence of high grade prostate cancer with negative predictive value (NPV) of 90% to 97% for Gleason ≥3+4 and between 98% to 99% for Gleason ≥4+3.
Keywords: prostate cancer, high grade, prediction, biomarkers, urine, plasma, biopsy, Gleason.
Introduction
Over diagnosis and overtreatment of prostate cancer are significant concerns in the US healthcare system. More than one million prostate biopsies are performed each year in the US, with an attendant positive biopsy rate of ~40%1,2. Biopsies can lead not only to patient discomfort and additional healthcare costs, but also to significant complications in a minority of patients, including death3-5. Furthermore approximately 40-50% of patients with newly diagnosed PCa may have indolent disease and many of these patients may be best suited for an active surveillance strategy6.
PSA testing remains the predominant testing modality for determining whether a patient considers a biopsy or not, despite a range of published controversial3,7,8 recommendations. Recently, new versions of measuring PSA or its isoforms are being introduced and showed significant improvement in predicting high grade prostate cancer, therefore reducing the need for biopsies9-11. The four kallikrein (4K) measures 4 isoforms of PSA, then combines the results with age, digital rectal exam (DRE) findings, and history of prior prostate Bx and generates a score (4KScore) for predicting the presence of aggressive (Gleaso≥7) PCa. In a prospective study in patients recommended for biopsy, the 4KScore showed an AUC of 0.8210.
Another biomarker assay called PHI (Prostate Health index) uses a formula that incorporates pro-PSA, fPSA, and PSA for predicting PCa. The AUC was 0.70 in patients with PSA between 2 and 10 ng/ml and negative DRE9.
A recent study used a urine exosome-derived normalized RNA levels of PCA3 and ERG genes in combination with multiple clinical parameters (PSA level, age, race, and family history of PCA) reported distinguishing prostate cancer Gleason ≥7 from benign disease with AUA of 77% in the training set and 73% in the testing set12.
We recently reported the development of a combined urine/plasma assay which measures the expression levels of 10 different genes in urine and plasma by incorporating an algorithm that includes serum PSA (sPSA) and age, allowing for the prediction biopsy proven PCa Gleason ≥3+4)13,14
Here we report the performance of this urine/plasma test in randomly selected patients visiting urology practices for a spectrum of prostate issues.
Materials and Methods
Study Design and Patients
Randomly selected patients from 8 different Urology practice sites were recruited for this study between July 2014 and December 2015. Inclusion criteria included any prostate problem or lower urinary tract symptoms (LUTS) (frequent or difficult urination, pain during urination, blood in urine) or elevated PSA. Exclusion criteria included prior diagnosis of PCa ≥7 (accepted 3+3), receiving any medication influencing PSA level, prostate DRE within 24 hours, ejaculation within 24 hours, receiving any cancer-related therapy, or prior prostatectomy. The primary outcome of the study was predicting biopsy results of Gleason ≥3+4 as determined in community-based practice using our test. The goal of the test is to reduce the rate of unnecessary biopsies. We assumed prevalence of Gleason ≥3+4 at 25% and power at 90%. Performing prostate biopsy was not a condition for recruitment, but consenting to provide biopsy results if performed was required. Urine and blood samples were collected from each patient without a required DRE or prostate massage and sent for analysis. Results of testing were not used to determine if biopsy is needed. All sites were asked at 3 months post-testing to provide biopsy reports, if performed. All biopsies reviewed information required a minimum of 10 cores. All work was performed after obtaining institutional review board (IRB) approval (Western IRB).
Urine and Plasma Processing
The urine sample from each patient was concentrated and plasma was separated and RNA from plasma and urine was extracted as previously described13,14. Briefly, urine samples were concentrated by centrifugation using Amcion Ultra-15 Centrifugal Filter Units with 3 kDa membrane (Millipore, Billerica, MA). Total nucleic acid was extracted from urine and plasma.
Quantitative Reverse Transcription-Polymerase Chain Reaction
Quantitative reverse transcription real time polymerase chain reaction (qRT-PCR) was performed as previously detailed13, 14. Quantitative qRT/PCR was performed to measure the levels of PDLIM5, HSPD1, IMPDH2, PCA3, TMPRSS2, ERG, UAP1, PTEN, AR, GAPDH and B2M RNA in urine and plasma.
Statistical Analysis
Sensitivity and specificity were calculated using standard formula. Comparisons between groups were performed using the Wilcoxon rank sum or Kruskal-Wallis tests for continuous variables and the Chi-Square test for categorical variables. Two-sided statistical tests were used in all analyses. Statistical significance considered at P-value <0.05, but correction for false detection rate (FDR) due to multiple testing is also used and expressed as q-value. Relationships among groups were investigated using Fisher's exact test for categorical variables. For developing the new model univariate and multivariate logistic regression analysis was performed.
Results
Prediction high grade PCa using biomarkers only
Between July 2014 and December 2015, a total of 2528 patients were enrolled in this study. Treating physicians elected to perform biopsies within three months on only 524 (21%) of these patients based on their clinical judgment and current standard practice. All enrolled patients were randomly selected. All enrolled patients were referred to urologists because of lower urinary tract symptoms. However, low level of PSA was not an exclusion criterion. Patients with prior biopsy and diagnosis of PCa Gleason 6 were accepted since the goal of this study is to predict high grade cancer. We excluded patients with prior diagnosis of PCa Gleason ≥7, receiving any medication influencing PSA level, prostate DRE within 24 hours, ejaculation within 24 hours, receiving any cancer-related therapy, or prior prostatectomy. PSA in these 524 patients was <4 ng/ml in 78 (15%) and >10 ng/ml in 63 (12%) of these patients (Table 1). The median age was 65 years (range: 40-89). The median prostate size was 40 g (range: 3.8-251 g). The majority of patients (404, 77%) were caucasian. African American were 15% (N=80). Nineteen percent (N=99) of these patients had prior biopsy and 4% (N=22) of all patients had prior diagnosis of PCa Gleason 3+3.
Patients characteristics.
| Age | 65 (40-89) |
| Prostate size (gm) | 40 (3.8-251) |
| Gleason | |
| 3+3 | 109 (21%) |
| 3+4 | 99 (19%) |
| ≥4+3 | 62 (12%) |
| sPSA (ng/ml) | |
| ≤4 | 78 (15%) |
| 4 to 10 | 383 (73%) |
| >10 | 63 (12%) |
| Race | |
| Caucasian | 404 (77%) |
| African American | 80 (15%) |
| Hispanic | 7 (1%) |
| Asian | 1 (0.2%) |
| Missing | 32 (6%) |
| Family History | |
| Yes | 84 (16%) |
| No | 299 (57%) |
| Unknown | 141 (27%) |
| Prior Biopsy | |
| Yes, Negative | 77 (15%) |
| Yes, Positive | 22 (4%) |
| No | 425 (81%) |
| DRE | |
| Normal | 347 (66%) |
| Abnormal | 44 (8%) |
| Unknown | 133 (25%) |
| Stage | |
| T1a | 14 (3%) |
| T1b | 56 (11%) |
| T1c | 154 (29%) |
| T2a | 10 (2%) |
| T2b | 6 (1%) |
| T2c | 9 (2%) |
Patients who underwent biopsy included 270 (51%) patients with cancer. Biopsy data showed 109 (21%) patients with Gleason 3+3, 99 (19%) with Gleason 3+4, and 62 (12%) with Gleason ≥4+3 PCa and the rest were negative. Cases with Gleason 3+4 are defined by having major cancer pattern as grade 3 and minor component as grade 3 and the opposite is true for Gleason 4+3. Using the urine/plasma biomarkers algorithm without any modification, we predicted the presence of cancer in 234 patients. However, PCa Gleason ≥3+4 was reported on biopsy in 40 patients that were missed by the urine/plasma test. Of these 40 patients, only 11 had PCa Gleason ≥4+3. The sensitivity and specificity of the test was at 75% and 69% (respectively) for predicting Gleason ≥3+4 (Table 2), but they were 82% and 61% for predicting Gleason ≥4+3 (Table 3). If the test recommendations were followed, there was a risk of missing or delaying diagnosis of Gleason ≥3+4 in 8% or missing/delaying diagnosis of Gleason ≥4+3 in 2% of patients. The negative predictive value of the urine/plasma test was at 86% for patients with Gleason ≥3+4 (Table 2) and 96% for patients with Gleason ≥4+3 (Table 3). The prevalence of Gleason ≥3+4 in this group of patients was 31%, which is higher than the reported by other studies10.
To rule out a bias in the patient population who underwent biopsy procedure, we compared the score between those with biopsy and those without biopsy. There was no significant difference in the overall score between the two groups (P=0.36, Kruskal-Wallis test). However, overall, 25% of patients were predicted by the test to have Gleason ≥3+4.
Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) for predicting Gleason ≥ 3+4.
| Estimated Value | 95% Confidence Interval | ||
|---|---|---|---|
| Lower Limit | Upper Limit | ||
| Sensitivity | 75% | 68% | 81% |
| Specificity | 69% | 64% | 74% |
| PPV | 52% | 45% | 58% |
| NPV | 86% | 82% | 90% |
Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) for predicting Gleason ≥ 4+3.
| Estimated Value | 95% Confidence Interval | ||
|---|---|---|---|
| Lower Limit | Upper Limit | ||
| Sensitivity | 82% | 70% | 90% |
| Specificity | 61% | 56% | 65% |
| PPV | 22% | 17% | 28% |
| NPV | 96% | 93% | 98% |
Incorporating clinical information with biomarkers in predicting biopsy high grade PCa
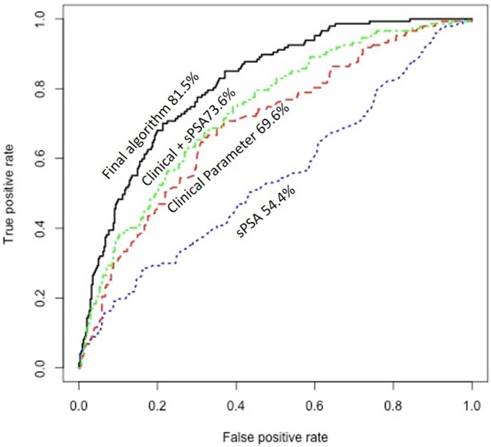
Since prior studies have shown that clinical information may significantly improve prediction of the presence of PCa15, we explored the potential of using available clinical information on improving the prediction of the presence of aggressive PCa. This included race, DRE data, history of prior biopsy, family history, and prostate size. Only 489 patients had the most complete information on these clinical variables. Univariate analysis showed that prostate size associated significantly with the presence of aggressive PCa (P=0.00001/q=0.0004). More specifically, patients with aggressive PCa tend to have lower prostate size. In addition, history of prior prostate biopsy associated directly with the presence of aggressive PCa (P=0.003/q=0.02). We combined urine/plasma biomarkers algorithm with prostate size in a decision tree and dichotomized patients into two groups: one includes high proportion of patients with aggressive PCa and a second group that includes high proportion of patients with non-aggressive PCa. We further refined the prediction in each group using logistic models based on history of prior biopsy along with urine/plasma biomarkers and developed two independent algorithms: one for prediction of biopsy results in the group with high proportion of patients with aggressive PCa and one for prediction of biopsy results in the second group with high proportion of patients with non-aggressive PCa. The scores obtained from the two logistic models, when incorporated together in analyzing patients, showed significantly higher sensitivity and specificity dependent on the cut-off used (Table 4) (Fig. 1). As shown in Figure 1, area under the curve (AUC) of sPSA was at 54.4%. Combining the clinical information (age, prostate size, and history of prior biopsy) improved the AUC to 73.6%, but our final algorithm that also incorporated the urine and plasma biomarkers significantly improved the AUC to 81.5%.
Sensitivity and specificity after integrating prostate size and history of prior biopsy into the biomarkers algorithm and analyzing the group with high proportion of non-aggressive patient in a separate algorithm (Logistic 1) and the group with the high proportion of aggressive patients in a second algorithm (Logistic 2). The sensitivity of specificity at various cut-off points are shown.
| Logistic 2 | ||||||
| Logistic 1 | Sensitivity | 0.10 | 0.15 | 0.20 | 0.25 | 0.30 |
| 0.10 | 0.973 | 0.952 | 0.946 | 0.925 | 0.905 | |
| 0.15 | 0.946 | 0.925 | 0.918 | 0.898 | 0.878 | |
| 0.20 | 0.912 | 0.891 | 0.884 | 0.864 | 0.844 | |
| 0.25 | 0.878 | 0.857 | 0.850 | 0.830 | 0.810 | |
| 0.30 | 0.857 | 0.837 | 0.830 | 0.810 | 0.789 | |
| Logistic 2 | ||||||
| Logistic 1 | Specificity | 0.10 | 0.15 | 0.20 | 0.25 | 0.30 |
| 0.10 | 0.363 | 0.377 | 0.401 | 0.447 | 0.465 | |
| 0.15 | 0.442 | 0.456 | 0.480 | 0.526 | 0.544 | |
| 0.20 | 0.506 | 0.520 | 0.544 | 0.591 | 0.608 | |
| 0.25 | 0.544 | 0.558 | 0.582 | 0.629 | 0.646 | |
| 0.30 | 0.570 | 0.585 | 0.608 | 0.655 | 0.673 | |
Receiver operating characteristic (ROC) curve showing the area under the curve (AUC) of serum PSA (sPSA), clinical information, combined clinical information with sPSA, and the final algorithm that incorporate the urine and plasma biomarkers with sPSA and clinical information.
Predicted and actual data for selected three cut-off points with overall sensitivity, specificity, PPV and NPV for predicting patients with PCa ≥3+4 and patients with PCa Gleason ≥4+3. The number of cases missed by the algorithm were 16, 2, and 3 for Gleason 3+4, 4+3, and ≥4+3, respectively, using low cut-off; 9, 1, and 3 for Gleason 3+4, 4+3, and ≥4+3, respectively, using standard cut-off; and 3, 0, and 1 for Gleason 3+4, 4+3, and ≥4+3, respectively, using high cut-off.
| Cut-off | Neg | 3+3 | 3+4 | 4+3 | >4+3 | Total Missed | Sensitivity | Specificity | NPV | PPV | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≥3+4 | ≥4+3 | ≥3+4 | ≥4+3 | ≥3+4 | ≥4+3 | ≥3+4 | ≥4+3 | ||||||||
| Actual | 239 | 103 | 92 | 29 | 26 | ||||||||||
| Predicted | Low | 93 | 54 | 76 | 27 | 23 | 21 (4.3%) | 86% | 96% | 57% | 59% | 90% | 98% | 46% | 46% |
| Standard | 106 | 63 | 83 | 28 | 23 | 13 (2.7%) | 91% | 97% | 51% | 44% | 93% | 98% | 44% | 52% | |
| High | 148 | 70 | 89 | 29 | 25 | 4 (0.8%) | 97% | 99% | 36% | 37% | 97% | 99% | 40% | 40% | |
For clinically practical approach, we selected three cut-off point: standard, high and low. These cut-off points show sensitivity at 91%, 97%, and 86% and specificity at 51%, 36%, and 57%, respectively for predicting PCa Gleason ≥ 3+4 (Table 5). However, the sensitivity is significantly higher for predicting the presence of PCa Gleason ≥4+3 (Table 5).
Discussion
This prospective multicenter study showed a high sensitivity and specificity, negative predictive value for predicting Gleason ≥7 using biomarkers in urine and peripheral blood plasma for the prediction of prostate biopsy findings. The purpose of the test is to reduce unnecessary biopsies and to select patients who would benefit from the diagnosis of a significant PCa. The assay was offered as an option to community-based practicing urologists without the condition of performing biopsy. Treating urologists determined to perform biopsy based on their clinical judgment. The study is designed to reduce the rate of unnecessary biopsies in urology community practices. Since the purpose of performing a biopsy in a specific patient is to find out if this patient has Gleason grade ≥3+4 as determined by community-based practitioner, we designed the study to collect samples from these patients and correlate test results with biopsy results in these patients. This will allow us to determine how many of these patients will avoid performing biopsy if this test is performed.
Per the design of the trial, information on biopsy was collected at three months of performing the test. This showed that 21% of the enrolled patients underwent prostate biopsy. This most likely reflects the rate of performing biopsy in general practice at this time. Interestingly, of these patients only 51.5% had prostate cancer on biopsy, but more importantly only 31% had PCa Gleason ≥3+4, which suggests that 69% of the biopsied patients had no cancer or indolent/clinically insignificant cancer. This is significant issue in routine clinical practice, since performing biopsy is not without consequences of bleeding and infection.
Toward this goal, we developed a test that combines biomarkers in urine and peripheral blood plasma. These biomarkers have been previously described12,13. They include: PDLIM5, HSPD1, IMPDH2, PCA3, TMPRSS2, ERG, UAP1, PTEN, AR, GAPDH and B2M in addition to the serum PSA (sPSA) protein and age. These biomarkers were previously validated by studying their levels in patients with confirmed PCa and by comparing their levels with the levels in patients with benign prostate hyperplasia as well as with the levels in patients after prostatectomy12.
We first tested the performance of the algorithm as it was previously described without any modification. The urine/plasma original algorithm recommended performing biopsy to confirm or rule out PCa Gleason ≥3+4 in 45% of the patients. It accurately detected the presence or absence of prostate cancer in 70.8% of patients. Recommended unnecessary biopsy in 21.5% of patients. It accurately predicted the presence of PCa Gleason ≥3+4 in 23% of all patients. However, 7.6% of patients were misclassified as they do not need biopsy or lack the presence of PCa, but actually had PCa Gleason ≥3+4. The algorithm missed 2% of the patients with PCa Gleason ≥4+3. The original algorithm had NPV of 96% for Gleason ≥4+3.
This study proved that the urine/plasma biomarkers are reliable and show reproducible results in predicting high grade PCa. However, it misses the diagnosis of significant number of patients. To improve the performance of this test, we took advantage of some clinical parameters and incorporated these parameters in the prediction model. While this was not pre-planned in the study, but clinical parameters are constant and not part of the laboratory testing, therefore, including them in a final analysis may improve the prediction without influencing the testing. Univariate analysis showed that prostate size and history of prior biopsy are highly predictive of the presence of aggressive PCa. Therefore, we incorporated these two clinical variables in our biomarkers model and modified the current algorithm. If clinical data was not available for a specific patient, a median equivalent was used and the score was calculated. Incorporating clinical information in the algorithm lead to significant improvement in the sensitivity and NPV (Fig 1).
We selected three cut-off points: standard, low and high. As demonstrated in table 5, the high cut-off point provides very high sensitivity and NPV but the standard cut-off is also very practical with NPV at 93% for PCa Gleason ≥3+4 and 98% for ≥4+3. With the standard cut-off point, 38% of patients will avoid performing biopsy and only 2.7% of patients with Gleason ≥3+4 would have a delayed diagnosis. Choosing which cut-off point to use for determining if biopsy should be performed should be based on other clinical findings and discussion with the patient. For example, a patient over the age of 70, might tolerate higher risk and accept low cut-off point as compared to a patient at younger age or to a patient with strong family history.
The results of this study show that this urine/plasma test is reliable in reducing unnecessary biopsies and highly reliable in ruling out high grade (Gleason ≥4+3) prostate cancer. Most likely the levels of the analyzed biomarkers in peripheral blood plasma and urine reflect the expected higher levels of turnover of tumor cells in more aggressive or higher grade PCa. Tumor cells with higher turnover pour more of their RNA/DNA into blood and urine. Therefore, when properly measured, they can be used for predicting the aggressiveness of the cancer. This test is not totally relying on measuring PSA or PSA isoforms. Therefore, it can be combined with other tests such as 4KScore or PHI for managing and screening patients with potential prostate cancer. The prevalence of PCa Gleason ≥3+4 in our series was at 31%, slightly higher than that reported in the 4k stud.10 If we adjust the prevalence of Gleason ≥3+4 to 23%, the NPV of our urine/plasma test changes and will become even higher, but based on our data, most likely the prevalence of high grade PCa is higher than 23% and our data is more representative of todays practice.
This test could be very useful in monitoring elderly man with elevated PSA avoiding performing a biopsy. Some of these men will have an insignificant cancer and would be more clinically useful to be monitored using this test in watchful waiting program.
Since our model incorporated patients with prior history of PCa Gleason 3+3, this test has the potential to be used for monitoring patients on active surveillance. However, additional study with large number of patients is needed to explore the value of this test in active surveillance. Patients who are negative for specific cut-off point at the initiation of the active surveillance can be monitored using this test.
While the testing predicts biopsy results, it does not provide information on the prostatectomy results. Long follow up and correlation with prostatectomy results is needed in future studies for selecting more precise cut-off points. We expect underestimation of the aggressiveness or the grade of the cancer as well as missing cancers on biopsy16. Therefore, correlation with prostatectomy or fusion biopsy is necessary for better evaluation of the reliability of such testing. However, because of the expected underestimation of cancer, we expect that the specificity of our test might improve when correlated with fusion biopsy or prostatectomy data. Furthermore, recent study suggested that not all Gleason 3+4 will have aggressive disease17. Some patients with Gleason 3+4 detected in <10% of the tumor may have indolent disease. This is another limitation of this study that should be addressed in future studies.
Conclusions
In summary, the results of this multi-center, community-based trial for the validation of a urine/plasma test for the prediction of prostate biopsy results in the purpose of reducing unnecessary biopsy prove that the selected biomarkers are reliable and provide reproducible results, but the addition of clinical information in the test algorithm improves the sensitivity and NPV of the test significantly. The test is practical and less dependent on PSA or its isoforms and can be used for early detection of prostate cancer as well as for monitoring patients on active surveillance as well as watchful waiting.
Competing Interests
Ma W and Albitar M are employed by NeoGenomics Laboratories, which offer testing for prostate cancer.
Lund L, Shore N, Feddersen S, Wojno K, Uchio E, Moylan D: No financial conflict to disclose.
References
1. Potosky AL, Miller BA, Albertsen PC, Kramer BS. The role of increasing detection in the rising incidence of prostate cancer. JAMA. 1995;273:548
2. Stanford JL, Stephenson RA, Coyle LM, Correa R Eley JW, Gilliland F et al. Prostate Cancer Trends 1973-1995, Publication no. 99-4543, SEER Program, National Cancer Institute, Bethesda, MD. 1999
3. Brett AS, Ablin RJ. Prostate-cancer screening-what the U.S. Preventive Services Task Force left out. N Engl J Med. 2011;365:1949
4. Schröder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V. et al. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320
5. Wilt TJ, MacDonald R, Rutks I, Shamliyan TA, Taylor BC, Kane RL. Systematic review: comparative effectiveness and harms of treatments for clinically localized prostate cancer. Ann Intern Med. 2008;148:435
6. Andriole GL, Crawford ED, Grubb RL 3rd, Buys SS, Chia D, Church TR. et al. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360:1310
7. Whittemore AS, Cirillo PM, Feldman D, Cohn BA. Prostate specific antigen levels in young adulthood predict prostate cancer risk: results from a cohort of Black and White Americans. J Urol. 2005;174:872
8. Stamey TA, Yang N, Hay AR, McNeal JE, Freiha FS, Redwine E. Prostate-specific antigen as a serum marker for adenocarcinoma of the prostate. N Engl J Med. 1987;317:909
9. Filella X, Giménez N. Evaluation of [-2] proPSA and Prostate Health Index (phi) for the detection of prostate cancer: a systematic review and meta-analysis. Clin Chem Lab Med. 2013;51:729
10. Parekh DJ, Punnen S, Sjoberg DD, Asroff SW, Bailen JL, Cochran JS. et al. A multi-institutional prospective trial in the USA confirms that the 4Kscore accurately identifies men with high-grade prostate cancer. Eur Urol. 2015;68:464
11. Carroll PR, Parsons JK, Andriole G, Bahnson RR, Castle EP, Catalona WJ. et al. NCCN Guidelines Insights: Prostate Cancer Early Detection, Version 2.2016. J Natl Compr Canc Netw. 2016;14:509
12. McKiernan J, Donovan MJ, O'Neill V, Bentink S, Noerholm M, Belzer S. et al. A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA Oncol. 2016Jul1;2(7):882-9
13. Ma W, Diep K, Fritsche HA, Shore N, Albitar M. Diagnostic and prognostic scoring system for prostate cancer using urine and plasma biomarkers. Genet Test Mol Biomarkers. 2014Mar;18(3):156-63
14. Albitar M, Ma W, Lund L, Albitar F, Diep K, Fritsche HA, Shore N. Predicting Prostate Biopsy Results Using a Panel of Plasma and Urine Biomarkers Combined in a Scoring System. J Cancer. 2016Feb2;7(3):297-303
15. Thompson IM, Ankerst DP, Chi C, Goodman PJ, Tangen CM, Lucia MS. et al. Assessing prostate cancer risk: results from the Prostate Cancer Prevention Trial. J Natl Cancer Inst. 2006;98:529
16. Colleselli D, Pelzer A E, Steiner E, Ongarello S, Schaefer G, Bartsch G, Schwentner C. et al. Upgrading of Gleason score 6 prostate cancers on biopsy after prostatectomy in the low and intermediate tPSA range. Prostate Cancer and Prostatic Diseases. 2010;13:182-185
17. Schiavina R, Borghesi M, Brunocilla E, Romagnoli D, Diazzi D, Giunchi F. et al. The biopsy Gleason score 3+4 in a single core does not necessarily reflect an unfavourable pathological disease after radical prostatectomy in comparison with biopsy Gleason score 3+3: looking for larger selection criteria for active surveillance candidates. Prostate Cancer Prostatic Dis. 2015;18:270-275
Author contact
![]() Corresponding author: Maher Albitar, MD, NeoGenomics Laboratories, 31 Columbia, Aliso Viejo, CA 92656 Phone: (949) 275-7564 Fax: (949) 206-1865t e-mail: maheralbitarcom
Corresponding author: Maher Albitar, MD, NeoGenomics Laboratories, 31 Columbia, Aliso Viejo, CA 92656 Phone: (949) 275-7564 Fax: (949) 206-1865t e-mail: maheralbitarcom

 Global reach, higher impact
Global reach, higher impact