3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(15):3028-3036. doi:10.7150/jca.20467 This issue Cite
Research Paper
MZF-1/Elk-1/PKCα is Associated with Poor Prognosis in Patients with Hepatocellular Carcinoma
1. Bachelor Program of Senior Services, Southern Taiwan University of Science and Technology, Tainan, Taiwan;
2. Institute of Biochemistry and Biotechnology, Medical College, Chung-Shan Medical University, Taichung, Taiwan;
3. Clinical Laboratory, Chung Shan Medical University Hospital, Taichung, Taiwan;
4. Department Physical Therapy, School of Medical and Health Sciences, Fooyin University, Kaohsiung, Taiwan;
5. Institute of Nutrition, College of Biopharmaceutical and Food Sciences, China Medical University, Taichung, Taiwan;
6. Department of Medical Applied Chemistry, College of Health Care and Management, Chung Shan Medical University, Taichung, Taiwan;
7. Department of Medical Research, Tungs' Taichung MetroHarbor Hospital, Taichung, Taiwan;
8. Center for Molecular Medicine, China Medical University Hospital, Taichung, Taiwan;
9. Graduate Institute of Biomedical Sciences, China Medical University, Taichung, Taiwan.
Received 2017-4-8; Accepted 2017-7-15; Published 2017-9-2
Abstract
Background: Protein kinase C alpha (PKCα) is a key signaling molecule in human cancer development. As a therapeutic strategy, targeting PKCα is difficult because the molecule is ubiquitously expressed in non-malignant cells. PKCα is regulated by the cooperative interaction of the transcription factors myeloid zinc finger 1 (MZF-1) and Ets-like protein-1 (Elk-1) in human cancer cells.
Methods: By conducting tissue array analysis, herein, we determined the protein expression of MZF-1/Elk-1/PKCα in various cancers.
Results: The data show that the expression of MZF-1/Elk-1 is correlated with that of PKCα in hepatocellular carcinoma (HCC), but not in bladder and lung cancers. In addition, the PKCα down-regulation by shRNA Elk-1 was only observed in the HCC SK-Hep-1 cells. Blocking the interaction between MZF-1 and Elk-1 through the transfection of their binding domain MZF-160-72 decreased PKCα expression. This step ultimately depressed the epithelial-mesenchymal transition potential of the HCC cells.
Conclusion: These findings could be used to develop an alternative therapeutic strategy against patients with the PKCα-derived HCC.
Keywords: MZF-1, Elk-1, PKCα, HCC.
Introduction
Protein kinase C alpha (PKCα) is a member of the protein kinase C family, consisting of at least ten isoforms (α, βI, βII, γ, δ, ε, η, θ, and ζ, and ι), that regulates multiple biological processes, including cell proliferation, apoptosis, differentiation, migration, and adhesion [1]. The enhanced expression of PKCα has been reported in the tumor tissues of various cancer types [2-8]. Such expression has been demonstrated to promote tumor growth and metastasis [9]. Thus, developing therapeutic agents that target PKCα has become the focus of many research laboratories [10]. However, the off-target effects of targeting PKCα and the limited understanding of the signaling mechanisms upstream of PKCα have hampered this effort.
Several mechanisms that contribute to PKCα overexpression have been investigated. These mechanisms include I) the shift in signaling from the epidermal growth factor receptor to the platelet-derived growth factor receptor during the progression from non-stem cells to cancer stem cells (CSCs) [11], II) the epithelial-mesenchymal transition (EMT), which results in the dominance of pro-invasive pathways downstream G-protein receptors [12], and III) the overexpression or activation of ErbB2 [13]. We recently discovered that the transcription factors Ets-like protein-1 (Elk-1) and myeloid zinc finger-1 (MZF-1) regulate PKCα expression in human hepatocellular carcinoma (HCC) cells [14-17]. These transcription factors are also direct upstream inducers of the expression of the PKCα protein and mRNA; the former molecules bind to the PKCα promoters. The Elk-1 transcription factor activates the c-fos promoter by associating with serum response factors; moreover, Elk-1 is a target of both extracellular-signal-regulated kinase and c-Jun N-terminal kinase cascades [18]. The transcription factor also controls the expression of genes involved in cell-cycle progression, differentiation, and apoptosis in response to extracellular signals [19-21]. MZF-1 belongs to the Kruppel family of zinc finger proteins and is preferentially expressed in myeloid progenitor cells [22-25]. MZF-1 plays an important role in cell growth, differentiation, and apoptosis.
We previously found that knocking down Elk-1 and/or MZF-1 results in significantly decreased PKCα mRNA and protein expression [14]. This finding suggests that the transcription factors regulate PKCα cooperatively. In the present study, we investigated the correlation between MZF-1/Elk-1 and PKCα expression in human HCC and explored a peptide-based strategy that inhibits EMT in malignant cells.
Materials and Methods
Immunohistochemical (IHC) staining
Array slides (HCC BS03014, lung cancer LC10011, and bladder cancer BL482) were purchased from US Biomax, Inc. (Rockville, MD, USA). The BS03014 slides included 60 cases of carcinoma,but 1 case was lost during evaluation. The LC10011 slides included 40 cases of non-small cell carcinomas, but 5 cases were lost during evaluation. The BL482 slides included 48 cases of transitional cell carcinomas. Detailed information on this array can be viewed at http://www.biomax.us/tissue-arrays/.
The slides were deparaffinized in xylene and rehydrated in an alcohol series. The sections were then incubated with 3% H2O2 for 5 min. After washing with PBS, the sections were boiled in EDTA solution (1 mM EDTA, 0.1% NP-40; pH 8.0) for 5 min (for PKCα) or in citric acid solution (10 mM citric acid monohydrate; pH 6.0) for 15 min (for Elk-1 and MZF-1 detection) in a microwave oven. After cooling for 1 h, the sections were washed thrice in PBS for 5 min. Then,the sections were incubated in PBS with 5% normal bovine serum for 25 min. The sections were washed with PBS and incubated with antibodies against PKCα (10 ng/ml PBS plus 0.2% BSA) (Sigma-Aldrich, St Louis, MO), Elk-1 (1:400) (Santa Cruz, CA), and MZF-1 (1:400) (Santa Cruz) at 4 °C overnight. After washing thrice with PBS for 5 min, the sections were incubated with biotinylate-labeled goat anti-rabbit IgG or rabbit anti-mouse IgG (Sigma-Aldrich) at room temperature for 1 h. Then, the sections were washed with PBS and incubated with peroxidase-conjugated ABC reagent (Avidin/Biotin kit, Vector Laboratories, Inc., Burlingame, CA, USA) at room temperature for 30 min. The sections were visualized by adding 3,3′-diaminobenzidine substrate (Sigma-Aldrich). The reaction was terminated by rinsing the sections with distilled water. The sections were counterstained with Gill's hematoxylin V (Mute Pure Chemicals Ltd., Tokyo, Japan) and dehydrated in an alcohol series. Afterwards, the sections were cleared with xylene before mounting with Malinol (Muto Pure Chemicals Ltd., Tokyo, Japan) and examined under a BX40 system microscope (Olympus, Tokyo, Japan) with a CCD DPII camera (Olympus). Resulting images were analyzed using Image-Pro® Plus software (Media Cybernetics, Silver Spring, MD, USA). PKCα/Elk-1/MZF-1 expression was scored by staining intensity as follows: 1+, weak; 2+, moderate; and 3+, strong.
Plasmid construction
Plasmids containing different fragments of MZF-1-c-Myc (encoding amino acids 60-72) were amplified from pcDNA-MZF-1-c-Myc by PCR (25). Then, the PCR products were isolated and cloned into a pcDNA™ 3.1/myc-His vector (reverse-transcription PCR and cloned into the pcDNATM 3.1/myc-His vector [Invitrogen]).
Cell lines
Cancer cells from various human organs were obtained directly from the ATCC (Manassas, VA, USA). The liver cancer cells include HCC HA22T (BCRC no.60168), Hep3B (BCRC no.60434), HepG2 (BCRC no.RM60025), SK-Hep-1 (ATCC no. HTB-52), and Huh-7 (ATCC no. JCRB-0403) cells. Meanwhile, the lung cancer cells were A549 (ATCC no. CCL185), H322 (ATCC no. CRL-5806), H1299 (ATCC no. CRL-5803), and H928. All the cells were cultured in media specific to each cell line and supplemented with 10% fetal bovine serum, 100 units/mL penicillin G, and 100 µg/mL streptomycin (Gibico, Grand Island, NY, USA) in a humidified atmosphere containing 5% CO2 at 37°C.
Transfection and stable clone establishment
Lipofectin was used for transfection. Cells were cultured in 60 mm dishes containing minimum essential medium (MEM) supplemented with 10% fetal calf serum (FCS-MEM) at 37 °C for 24 h before rinsing with serum-free MEM. Then, the sample was transferred to 1 mL serum-free MEM containing 15 µg/mL Lipofectamine 2000 transfection reagent (Invitrogen) and various doses of the indicated plasmid. After incubating for a minimum of 6 h, 1 mL MEM supplemented with 20% FCS was added to the medium. After incubating for another 18 h, the medium was replaced with fresh FCS-DMEM. Then, the cells were incubated for at least 48 h before they were lysed for subsequent assays.
Stable clones were established by seeding low-passage cells at a density of 3 × 105 cells in 60mm tissue culture dishes. The cells were transfected with the MZF-160-72 plasmid (5 µg/6 mL) using Lipofectamine 2000. Then 5 h post-transfection, the cells were washed thrice in serum-free MEM and allowed to recover for 24 h in fresh medium. Stable clones were selected by growing the cells at 1:10 to 1:15 (vol/vol) in DMEM medium supplemented with geneticin (G418; 600 µg/ml) at 37 °C for 5 weeks. Individual clones were then transferred to 96-well plates and grown until confluence. After being transferred to flasks, the cells were cultured until confluence, harvested, and frozen in liquid nitrogen for further experimentation.
Immunoblotting analysis
Cancer cell lysates were lysed in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 5 mM EDTA, 50 mM HEPES [pH 7.5], 0.5% [w/v] sodium deoxycholate, 1% Nonidet P-40 [NP-40], 10 mM 2-mercaptoethanol) containing 2 mM phenylmethylsulfonyl fluoride (PMSF), 50 mg/mL aprotinin A, 25 mg/mL leupeptin, and 25 mg/mL pepstatin. The cell lysates were then centrifuged at 12000 × g for 5 min and maintained on ice. The cell lysates were resolved by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, after which the proteins were transferred to polyvinylidine fluoride membranes (Immobilon-P; Millipore, Bedford, MA, USA). The membranes were then incubated in blocking buffer (5% [w/v] non-fat dry milk, 0.1% [v/v] Tween 20) in Tris-buffered saline (TBST) at room temperature for 30 min. Next, the membraneswere probed with the following specific antibodies: anti-PKCα (BD Biosciences, San Jose, CA, USA); anti-E-cadherin (CDH1); anti-VIM and anti-SNAI2l (Cell Signaling, Beverly, MA, USA); anti-p38 MAPK (p38), anti-phospho-p38 MAPK (P-p38), and anti-urokinase-type plasminogen activator (uPA) (GeneTex, Inc., Irvine, CA, USA); anti-Elk-1, anti-MZF-1; and anti-β-actin polyclonal antibodies (Santa Cruz). Then, the blots were incubated in blocking buffer at 4 °C overnight, after which they were incubated with horseradish peroxidaselabeled anti-mouse or anti-rabbit secondary antibodies (Promega) at room temperature for 2 h. After three washes with TBST buffer, antibody-reactive proteins were detected using a chemiluminescent substrate (Pierce, Rockford, IL, USA).
Antisense knockout assays
A shRNA Elk-1-expressing plasmid vector was constructed using the pcDNA-HU6 vector (a gift from Dr. J. Tsai Chang, Institute of Toxicology, College of Medicine, Chung Shan Medical University, Taichung, Taiwan) as the vector backbone. The shRNA Elk-1 duplex sequence, which was obtained from human Elk-1 genes (GenBank, NCBI), was designed using the BLOCK-iT™ RNAi Designer software available at http://www.invitrogen.com. The sequence corresponded to the coding regions relative to the first nucleotide of the start codon. The sequences designed to produce hairpin RNAs identical to the oligonucleotide shRNA duplex sequences were as follows: sense, 5′-GATCCGCAAGAACAAGACCAACATTTCAAGAGAT-3′ and antisense, 5′-AGCTTAAAAAGCAAGAACAAGACCAACATTCTCTTGAA-3′. To generate the shRNA duplex, 40 µM each of sense and antisense oligonucleotides were annealed in the thermocycler using the following profile: 37°C for 30 min and 65°C for 15 min. The resulting shRNA duplex was then cloned into the pcDNA-HU6 vector in frame with the BamHI and HindIII sites. The insert was screened by PCR, with the HU6 primer and confirmed by sequencing with the HU6 primer. After transfection for 6 h, the cells were washed thrice in serum-free MEM and allowed to recover for 24 h in fresh medium. Stable clones were selected with geneticin (G418; 600 µg/ml) at 37°C for 4 weeks.
Statistical analysis
Data were expressed as mean ± standard deviation and were analyzed by ANOVA. Pearson's chi-squared test [26] and Student's t-test were used in two-group comparisons. Here, p< 0.05 was considered as statistically significant.
Results
PKCα expression is correlated with MZF-1/Elk-1 expression in HCC
To determine the clinical relevance of the correlation between PKCα and Elk-1 and/or MZF-1, we examined the expression of these proteins in tissue microarrays of liver, lung, and bladder cancers by IHC staining. We observed a positive correlation between moderate-to-strong PKCα staining and either Elk-1 and/or MZF-1 staining in HCC (Figure 1) but not in lung or bladder cancers (Figures 2 and 3, respectively). Moreover, the moderate-to-strong staining of PKCα/Elk-1/MZF-1 was most common in the grades 2 and 3 HCCs (Figure 1). We also validated these results in the HCC cells (Figure 4), and found that high PRKCA expression levels were significantly correlated with those of Elk-1 and MZF-1 in poorly differentiated HCC (r = 0.86, p < 0.05 and r = 0.92, p< 0.05, respectively). Reducing Elk-1 expression by siRNA gene silencing decreased the PKCα protein expression in the poorly differentiated HCC SK-Hep-1 cells (Figure 5). Collectively, these results suggest that PKCα, along with Elk-1 and MZF-1, function as important mediators of tumor progression in HCC.
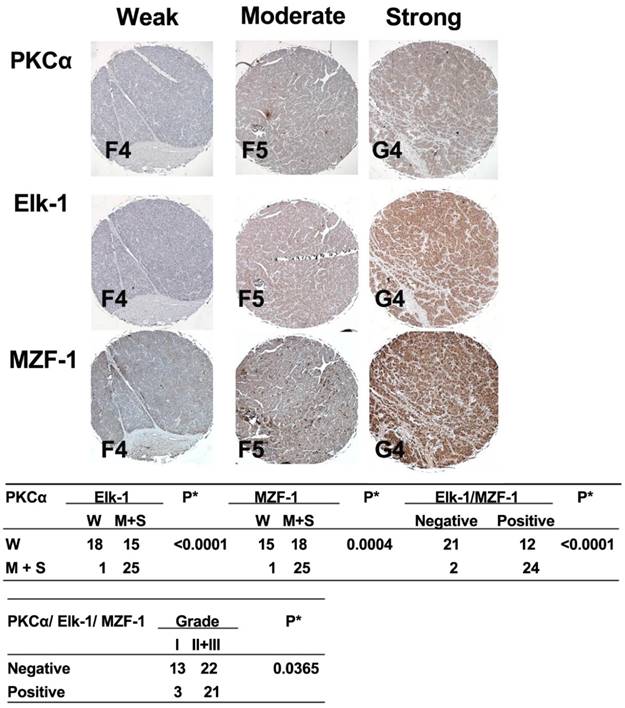
Correlations between the expression of PKCα and Elk-1/MZF-1 in human HCC.IHC analyses and correlations of PKCα and Elk-1/MZF-1 expression in human HCC. The up-panel shows the representative staining results for the samples scored by visual assessment as “weak,” “moderate,” or “strong” on the basis of staining intensity. The label (i.e. F4) at the bottom-left corner of each sample is the serial number of the patients indicated by US Biomax, Inc. (Rockville, MD, USA), and can be viewed at http://www.biomax.us/tissue-arrays/. The numbers of each group classified on the basis of PKCα, Elk-1, or MZF-1 staining intensity or grade are depicted in the down-panel. A positive rating was given to the moderate or strong expression of the genes of interest; otherwise, a negative rating was given. The clinical characteristic grades of I, II, and III were obtained from US Biomax, Inc. *p< 0.05, Pearson's chi-squared test.
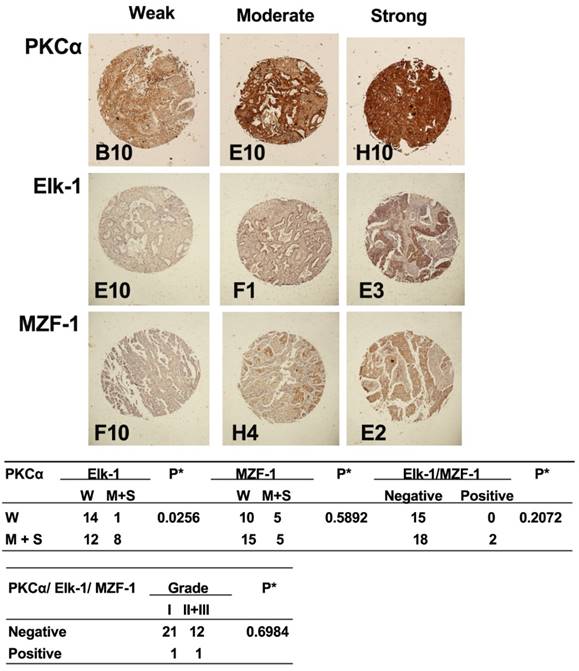
Correlations between the expression of PKCα and Elk-1/MZF-1 in human lung cancer. IHC analyses and correlations of PKCα and Elk-1/MZF-1 expression in human lung cancer. The representative staining results, labels, and clinical characteristic grades for the samples were described as in Figure 1. *p< 0.05, Pearson's chi-squared test.
Inhibition of MZF-1 and Elk-1heterodimer formation attenuates the malignant phenotypes of HCC cells by reducing PKCα expression
In our previous work, we reported that the expression of MZF-1∆DBD (contains MZF-160-72 but lacks the DNA-binding domain) or Elk-1∆DBD (contains Elk-1145-157 but lacks the DNA-binding domain) decreased PKCα expression, cell migration and invasion, and cell proliferation in HCC cells [27]. Compared with the tumors that developed in the mice injected with empty-vector-treated cancer cells, the tumors in the mice with MZF-1∆DBD-treated cancer cells were smaller throughout almost the entire experimental time interval. Furthermore, the MB-231 and Hs578T breast cancer cells stably expressing MZF-160-72(MB-231-M(v3), MB-231-M(v4), Hs578T-M(s2), and Hs578T-M(s3)) were more rounded than the elongated parental and vector control cells [28]. These observations suggest that MZF-160-72hindered the endogenous Elk-1 and MZF-1 interaction, subsequently moderatedthe transcription factors' binding to the PRKCA promoter, and ultimately inhibited PKCα and EMTcore genes.
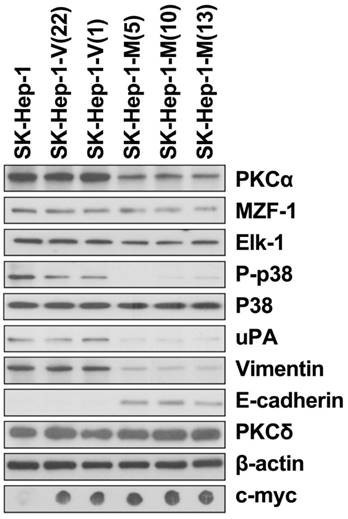
Next, we examined the effects of MZF-160-72on HCC cells. The changes in the protein expressions of these EMTcore genes were also similar to those in the SK-Hep-1 HCC cells stably expressing MZF-160-72(Figure 6). The changes in the HCC SK-Hep-1 cellswere consistent with those observed in the cells with PKCα knockdown by transfection with shRNA [28]. The expressions of PKCα, phosphorylated-mitogen-activated protein kinase (MAPK) p38, and urokinase-type plasminogen activator (uPA) were reduced. The expression of the EMTcore genes was also altered.
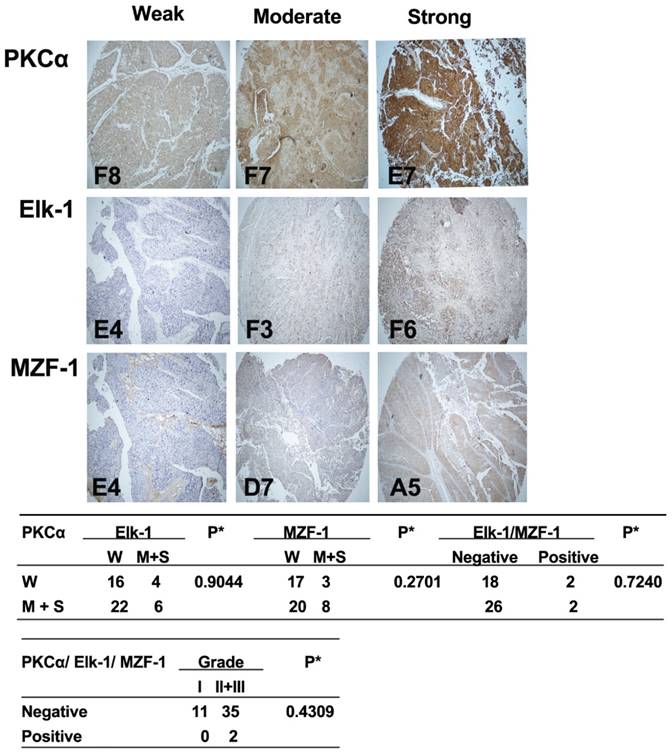
Correlations between the expression of PKCα and Elk-1/MZF-1 in human bladder cancer.IHC analyses and correlations of PKCα and Elk-1/MZF-1 expression in human bladder cancer. The representative staining results, labels, and clinical characteristic grades for the samples were described as in Figure 1. *p< 0.05, Pearson's chi-squared test.
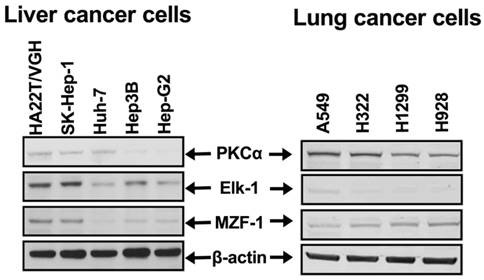
PKCα expression correlated with MZF-1/Elk-1 expression in HCC cell lines. Immunoblotting analysis of the protein levels of PKCα, Elk-1, and MZF-1 in HCC and lung cancer cells.
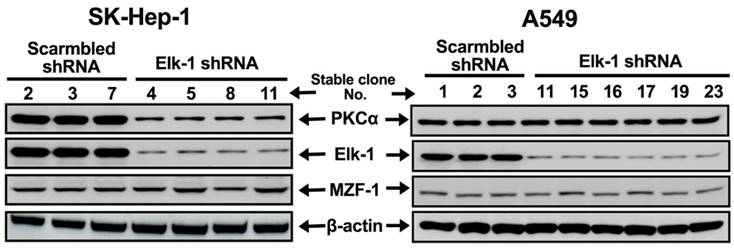
PKCα expression regulated by Elk-1 expression in HCC cell lines. Immunoblotting analysis of the expressions of PKCα, Elk-1, and MZF-1 in HCC SK-Hep-1 and lung A549 cancer cells transfected with Elk-1 shRNA. β-Actin was used as internal control.
Discussion
We previously demonstrated that MZF-1 and Elk-1 interact cooperatively to regulate PKCα expression [14, 26, 29], and a decrease in either endogenous MZF-1 or Elk-1 level affects PKCα expression, cell migration activity, and tumorigenesis [14-17]. We further identified specific domains within MZF-1 and Elk-1, which are responsible for the disruption of their interactions. Such disruption leads to decreased DNA binding activity, followed by reduced PKCα expression, and the eventual attenuation of cell migration and EMT potential. Present findings indicate a positive correlation between moderate-to-strong PKCα expression and either Elk-1 and/or MZF-1 staining in HCC.The moderate-to-strong staining of PKCα/Elk-1/MZF-1 was most commonly observed in the grades 2 and 3 HCCs. Thus, introducing peptides that saturate the binding surfaces of the molecules of interest is a potential strategy for developing alternative anti-cancer therapies.
PKCα is an important signaling molecule in the progression of many carcinomas and plays a key role in EMT [11, 30-32]. The fluctuating intensities of stress factors (e.g., hypoxia, inflammation, and the either cooperative or hostile interactions of tumor intercellular networks), which are known to induce EMT, all increase the adaptation potential of cancer cells; such adaptation mechanisms include bypassing cellular senescence and the subsequent development of CSCs [33]. CSCs appear to be responsible for driving tumor growth, recurrence, and metastasis [34, 35]. These stem cells also possess mesenchymal phenotypes associated with highly aggressive cancer traits [36, 37]. Our data support previous findings, which indicate that EMT-inducing agents enriched CSCs and increased PKCα expression. The increase in PKCα expression correlated with increased Elk-1 and MZF-1 expression in the clinical tissue array in the present study. This observation demonstrated that the induction of PKCα overexpression by Elk-1 and MZF-1 expression may have induced the development of high-grade malignancy. However, further studies on the mechanism underlying the increase in Elk-1 and MZF-1 expression are still underway.
EMT reduction by the disruption of MZF-1/Elk-1 heterodimer formation in HCC cell lines. Changes in protein levels in the parental and MZF-160-72-transfected stable HCC SK-Hep-1 cells as detected by immunoblotting analysis.SK-Hep-1-V indicates the control vector-transfected stable cells; SK-Hep-1-M indicates the MZF-160-72 vector-transfected stable cells. The number in the () indicates the designation of the stable-clone cell. β-Actin was used as internal control.
In HCC research, where PKCα expression is higher in human HCC [5], PKCα overexpression is known to increase the molecular potential for activating the mitogen-activated protein kinase (MAPK) p38 signaling pathway [29]. PKCα overexpression is also known to be associated with cell malignancy. Thus, PKCα inhibition decreases the malignancy of HCC cells and suppresses metastasis [38]. Herein, we showed that inhibiting MZF-1/Elk-1 heterodimer formation decreased PKCα expression and MAPK p38 activation. Therefore, considering the high correlation between PKCα and Elk-1/MZF-1 expression in HCC patients, inhibiting the Elk-1/MZF-1 interaction represents a novel and feasible strategy to specifically inhibit PKCα expression, and eventually, tumor development. This approach may also increase the effectiveness of HCC treatment.
Conclusions
PKCα inhibitors are currently being investigated in human clinical trials, both alone and in combination with other modalities [10, 39]. However, formulating a viable treatment strategy, which specifically targets PKCα in cancer cells, is challenging due to the ubiquity of PKCα and potential off-target effects [10, 40]. Peptide-based therapy is currently both under clinical and preclinical stages of development, and this approach is available in the market for treating human diseases [41]. An example of such therapyis TAT-beclin 1 [42] derived from beclin1. The peptidehas been shown to induce autophagy, decrease the replication of several pathogens (including HIV-1), and increase the survivability of mice from viruses such as chikungunya and the West Nile virus. Another example is the PKCα antagonistic peptide [9], which is derived from a highly variable region V5 of the enzyme. The peptide inhibits intravasation, cell migration, and metastasis; protects against liver damage; and normalizes blood cell count in animal tests. The direct targeting of the Elk-1/MZF-1 interaction by PKCα treatment methods could allow the targeting of PKCα-, Elk-1-, and MZF-1-expressing malignant cells without any adverse effects on non-cancerous cells that show non-prominent Elk-1 and MZF-1 expression. Moreover, the discovery of the interactive sequences of Elk-1 and MZF-1 could later lead to a new strategy of treating PKCα-derived HCC involving the identification of small molecules that increase drug delivery potential and inhibitory effectiveness in metastasis.
Abbreviations
PKCα: protein kinase C alpha; CSCs: cancer stem cells; EMT: epithelial-mesenchymal transition; Elk-1: Ets-like protein-1; MZF-1: myeloid zinc finger-1; HCC: hepatocellular carcinoma; IHC: immunohistochemical; MAPK: mitogen-activated protein kinase; uPA: urokinase-type plasminogen activator.
Acknowledgements
We thank Shao-Hsuan Kao for insightful discussions and Andy Chang and Jennifer Hsu for reviewing the manuscript. This work was supported by grants from the Ministry of Science and Technology, Republic of China (MOST 104-2320-B-039-032), the China Medical University Hospital, Taichung, Taiwan (DMR-106-030), and the China Medical University, Taichung, Taiwan (CMU104-S-41), as well as in part by Taiwan Ministry of Health and Welfare Clinical Trial and Research Center of Excellence (MOHW106-TDU-B-212-113004).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Nakashima S. Protein kinase Cα (PKCα): regulation and biological function. J Biochem. 2002;132:669-675
2. Lahn M, Su C, Li S, Chedid M, Hanna KR, Graff JR. et al. Expression levels of protein kinase C-alpha in non-small-cell lung cancer. Clin Lung Cancer. 2004;6:184-189
3. Koren R, Ben Meir D, Langzam L, Dekel Y, Konichezky M, Baniel J. et al. Expression of protein kinase C isoenzymes in benign hyperplasia and carcinoma of prostate. Oncol Rep. 2014;11:321-326
4. Lahn M, Sundell K, Köhler G. The role of protein kinase C-alpha in hematologic malignancies. Acta Haematol. 2006;115:1-8
5. Wu TT, Hsieh YH, Wu CC, Hsieh YS, Huang CY, Liu JY. Overexpression of protein kinase C alpha mRNA in human hepatocellular carcinoma: a potential marker of disease prognosis. Clin Chim Acta. 2007;382:54-58
6. Chuize K, Yuyan Z, Zhe Z, Tao L, Meng Y, Qi Y. Protein kinase C-alpha is expressed and activated during the development of renal cell carcinoma. Urology. 2010;76:514.e1-514.e5
7. Lønne GK, Cornmark L, Zahirovic IO, Landberg G, Jirström K, Larsson C. PKCalpha expression is a marker for breast cancer aggressiveness. Mol Cancer. 2010;9:76
8. Lin SC, Chen WY, Lin KY, Chen SH, Chang CC, Lin SE. et al. Clinicopathological correlation and prognostic significance of protein kinase Cα overexpression in human gastric carcinoma. PLoS One. 2013;8:e56675
9. Kim J, Thorne SH, Sun L, Huang B, Mochly-Rosen D. Sustained inhibition of PKCα reduces intravasation and lung seeding during mammary tumor metastasis in an in vivo mouse model. Oncogene. 2011;30:323-333
10. Mochly-Rosen D, Das K, Grimes KV. Protein kinase C, an elusive therapeutic target? Nat Rev Drug Discov. 2012;11:937-957
11. Tam WL, Lu H, Buikhuisen J, Soh BS, Lim E, Reinhardt F. et al. Protein kinase C α is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell. 2013;24:347-364
12. Ouelaa-Benslama R, De Wever O, Hendrix A, Sabbah M, Lambein K, Land D. et al. Identification of a GαGβγ, AKT and PKCα signalome associated with invasive growth in two genetic models of human breast cancer cell epithelial-to-mesenchymal transition. Int J Oncol. 2012;41:189-200
13. Tan M, Li P, Sun M, Yin G, Yu D. Upregulation and activation of PKC alpha by ErbB2 through Src promotes breast cancer cell invasion that can be blocked by combined treatment with PKC alpha and Src inhibitors. Oncogene. 2006;25:3286-3295
14. Hsieh YH, Wu TT, Tsai JH, Huang CY, Hsieh YS, Liu JY. PKCα expression regulated by Elk-1 and MZF-1 in human HCC cells. Biochem Biophys Res Commun. 2006;339:217-225
15. Hsieh YH, Wu TT, Huang CY, Hsieh YS, Liu JY. Suppression of tumorigenicity of human hepatocellular carcinoma cells by antisense oligonucleotide MZF-1. Chin J Physiol. 2007;50:9-15
16. Ying TH, Hsieh YH, Hsieh YS, Liu JY. Antisense oligonucleotide Elk-1 suppresses the tumorigenicity of human hepatocellular carcinoma cells. Cell Biol Int. 2008;32:210-216
17. Wu TT, Hsieh YH, Hsieh YS, Liu JY. Reduction of PKC alpha decreases cell proliferation, migration, and invasion of human malignant hepatocellular carcinoma. J Cell Biochem. 2008;103:9-20
18. Wiren KM, Toombs AR, Zhang XW. Androgen inhibition of MAP kinase pathway and Elk-1 activation in proliferating osteoblasts. J Mol Endocrinol. 2004;32:209-226
19. Shao N, Chai Y, Cui JQ, Wang N, Aysola K, Reddy ES. et al. Induction of apoptosis by Elk-1 and deltaElk-1 proteins. Oncogene. 1998;17:527-532
20. Vanhoutte P, Nissen JL, Brugg B, Gaspera BD, Besson MJ, Hipskind RA. et al. Opposing roles of Elk-1 and its brain-specific isoform, short Elk-1, in nerve growth factor-induced PC12 differentiation. J Biol Chem. 2001;276:5189-5196
21. Khurana A, Dey CS. Involvement of Elk-1 in L6E9 skeletal muscle differentiation. FEBS Lett. 2002;527:119-124
22. Hromas R, Collins SJ, Hickstein D, Raskind W, Deaven LL, O'Hara P. et al. A retinoic acid-responsive human zinc finger gene, MZF-1, preferentially expressed in myeloid cells. J Biol Chem. 1991;266:14183-14187
23. Bavisotto L, Kaushansky K, Lin N, Hromas R. Antisense oligonucleotides from the stage-specific myeloid zinc finger gene MZF-1 inhibit granulopoiesis in vitro. J Exp Med. 1991;174:1097-1101
24. Hromas R, Morris J, Cornetta K, Berebitsky D, Davidson A, Sha M. et al. Aberrant expression of the myeloid zinc finger gene, MZF-1, is oncogenic. Cancer Res. 1995;55:3610-3614
25. Robertson KA, Hill DP, Kelley MR, Tritt R, Crum B, Van Epps S. et al. The myeloid zinc finger gene (MZF-1) delays retinoic acid-induced apoptosis and differentiation in myeloid leukemia cells. Leukemia. 1998;12:690-698
26. Naik PS, Deshmukh S, Khandeparkar SG, Joshi A, Babanagare S, Potdar J, Risbud NS. Epithelial ovarian tumors: Clinicopathological correlation and immunohistochemical study. Midlife Health. 2015;6:178-183
27. Yue CH, Huang CY, Tsai JH, Hsu CW, Hsieh YH, Lin H. et al. MZF-1/Elk-1 complex binds to protein kinase Cα promoter and is involved in hepatocellular carcinoma. PLoS One. 2015;10:e0127420
28. Lee CJ, Hsu LS, Yue CH, Lin H, Chiu YW, Lin YY. et al. MZF-1/Elk-1 interaction domain as therapeutic target for protein kinase Cα-based triple-negative breast cancer cells. Oncotarget. 2016;7:59845-59859
29. Hsieh YH, Wu TT, Huang CY, Hsieh YS, Hwang JM, Liu JY. p38 mitogen-activated protein kinase pathway is involved in protein kinase Calpha-regulated invasion in human hepatocellular carcinoma cells. Cancer Res. 2007;67:4320-4327
30. Wu WS, Tsai RK, Chang CH, Wang S, Wu JR, Chang YX. Reactive oxygen species mediated sustained activation of protein kinase C alpha and extracellular signal-regulated kinase for migration of human hepatoma cell Hepg2. Mol Cancer Res. 2006;4:747-758
31. Tang R, Yang C, Tao JL, You YK, An N, Li SM. et al. Epithelial-mesenchymal transdifferentiation of renal tubular epithelial cells induced by urinary proteins requires the activation of PKC-α and βI isozymes. Cell Biol Int. 2011;35:953-959
32. Kinehara M, Kawamura S, Mimura S, Suga M, Hamada A, Wakabayashi M. et al. Protein Kinase C-Induced Early Growth Response Protein-1 Binding to SNAIL Promoter in Epithelial-Mesenchymal Transition of Human Embryonic Stem Cells. Stem Cells Dev. 2014;23:2180-2189
33. Csermely P, Hódsági J, Korcsmáros T, Modos D, Perez-Lopez AR, Szalay K. et al. Cancer stem cells display extremely large evolvability: alternating plastic and rigid networks as a potential Mechanism: Network models, novel therapeutic target strategies, and the contributions of hypoxia, inflammation and cellular senescence. Semin Cancer Biol. 2014 Doi: 10.1016
34. Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983-3988
35. Dalerba P, Cho RW, Clarke MF. Cancer stem cells: models and concepts. Annu Rev Med. 2007;58:267-284
36. Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol. 2011;27:347-376
37. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871-890
38. Yuan X, Chen H, Li X, Dai M, Zeng H, Shan L, Sun Q, Zhang W. Inhibition of protein kinase C by isojacareubin suppresses hepatocellular carcinoma metastasis and induces apoptosis in vitro and in vivo. Sci Rep. 2015;5:12889
39. Li Z, Wang N, Fang J, Huang J, Tian F, Li C. et al. Role of PKC-ERK signaling in tamoxifen-induced apoptosis and tamoxifen resistance in human breast cancer cells. Oncol. Rep. 2012;27:1879-1886
40. Marengo B, De Ciucis C, Ricciarelli R, Pronzato MA, Marinari UM, Domenicotti C. Protein kinase C: an attractive target for cancer therapy. Cancers (Basel). 2011;3:531-567
41. Thundimadathil J. Cancer treatment using peptides: current therapies and future prospects. J Amino Acids. 2012;2012:967347
42. Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L. et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494(7436):201-206
Author contact
![]() Corresponding authors: Jer-Yuh Liu PhD., Graduate Institute of Cancer Biology, College of Medical, China Medical University, No 6, Hsueh-Shih Road, Taichung 404, Taiwan. Tel: +886-4-22052121 ext. 7932; Fax: +886-4-22333496; E-mail: jylcmu.edu.tw Chia-Jen Lee PhD., Department of Medical Research, Tungs' Taichung MetroHarbor Hospital, No.699, Sec. 8, Taiwan Blvd., Taichung City 435, Taiwan. Tel: +886-4-26581919 ext. 79004; E-mail: chiajenlee54com
Corresponding authors: Jer-Yuh Liu PhD., Graduate Institute of Cancer Biology, College of Medical, China Medical University, No 6, Hsueh-Shih Road, Taichung 404, Taiwan. Tel: +886-4-22052121 ext. 7932; Fax: +886-4-22333496; E-mail: jylcmu.edu.tw Chia-Jen Lee PhD., Department of Medical Research, Tungs' Taichung MetroHarbor Hospital, No.699, Sec. 8, Taiwan Blvd., Taichung City 435, Taiwan. Tel: +886-4-26581919 ext. 79004; E-mail: chiajenlee54com

 Global reach, higher impact
Global reach, higher impact