3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(17):3585-3591. doi:10.7150/jca.21368 This issue Cite
Research Paper
DNA Methylation Analysis of the SHOX2 and RASSF1A Panel in Bronchoalveolar Lavage Fluid for Lung Cancer Diagnosis
Department of Laboratory Medicine, Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai 200030, China.
*These authors contributed equally to this work.
Received 2017-6-7; Accepted 2017-8-28; Published 2017-9-30
Abstract
Introduction: Currently the majority of lung cancer patients are diagnosed as advanced diseases for no sensitive and specific biomarkers exist, noninvasive biomarkers with high sensitivity and specificity are urgently needed in lung cancer diagnosis. Bronchoscopy is a standard procedure of the diagnostic work-up of patients with suspected lung cancer despite of the limited diagnostic accuracy. Besides, epigenetic changes through DNA methylation play an important role in tumorigenesis. Thus, we examined the aberrant methylation of the SHOX2 and RASSF1A in bronchoalveolar lavage fluid (BALF) in comparing with conventional cytology examination and serum CEA in order to evaluate the new diagnostic method.
Patients and Methods: BALF and serum samples were collected from 322 patients at the time of diagnosis, 284 of them were pathologically confirmed lung cancer, 35 were benign lung diseases and 3 were malignancies in other systems. For all of the 322 patients, the methylation status of the SHOX2 and RASSF1A gene were detected by a new RT-PCR platform and then confirmed by sanger sequencing. Serum CEA were detected using electrochemiluminescence immunoassay.
Results: Profiling data showed the consistency of RT-PCR and sanger sequencing in detecting the methylation of the SHOX2 and RASSF1A. Besides, the combination of SHOX2 and RASSF1A methylation in BALF yielded a diagnostic sensitivity of 81.0% and specificity of 97.4%. When compared with established cytology examination (sensitivity: 68.3%, specificity: 97.4%) and serum biomarker carcinoembryonic antigen (CEA) (sensitivity: 30.6%, specificity: 100.0%), the SHOX2 and RASSF1A methylation panel showed the highest diagnostic efficiency. Notably, the combination of cytology and the SHOX2 and RASSF1A methylation panel could significantly improve the diagnostic efficacy.
Conclusion: The methylation analysis of the SHOX2 and RASSF1A panel in BALF with RT-PCR achieved a satisfactory sensitivity and specificity in lung cancer diagnosis, especially in an early stage. It could be used as a promising noninvasive biomarker for auxiliary diagnosis of lung cancer.
Keywords: Lung cancer, diagnosis, DNA methylation, SHOX2, RASSF1A
Introduction
Lung cancer is a major public health problem and the leading cause of cancer-related deaths worldwide [1, 2]. Computed tomography (CT) has been used as an early detection tool with high sensitivity that reduces mortality of lung cancer, but the diagnostic accuracy is limited due to its poor specificity [3]. At present, fiberoptic bronchoscopy is a standard procedure of the diagnostic work-up of patients with suspected lung cancer to obtain specimen for either histologic or cytologic examination. Although the technique is less invasive than other tissue procurement methods, carries a small risk of complications, and has high specificity, the diagnostic accuracy of bronchoscopy is relatively poor, with sensitivities ranging from 30%-69% depending on the size of the primary tumor and the number of parallel tests performed per bronchoscopy[4, 5]. Comparing to traditional blood, bronchoalveolar lavage fluid (BALF) from bronchoscopy represents an alternative source of lung cancer biomarkers due to its vicinity to tumor cells. Besides, BALF could be obtained by minimally invasive method, which made it potential in the clinical use of lung cancer diagnosis.
DNA methylation is essential for the regulation of gene expression and maintenance of cellular identity, epigenetic changes through altered DNA methylation play an important role in tumorigenesis of a variety of cancer types [6, 7]. Some studies have revealed that DNA methylation occurs mainly at CpG islands, where clustered by CG dinucleotides and usually presented in the gene promoters [8, 9]. Relevant evidences show that the promotor methylation of the short stature homeobox gene two (SHOX2) and the RAS association domain family 1, isoform A (RASSF1A) have been identified as diagnostic and prognostic biomarkers for lung cancer[10, 11]. DNA methylation analysis of the SHOX2 gene in blood showed a sensitivity of 60% and specificity of 90% in the diagnosis of lung cancer in the study performed by Chrisoph et al [12]. Previous studies illustrated that the methylation analysis of the SHOX2 and SEPT9 in pleural fluid showed a promising diagnostic and prognostic ability in thoracoabdominal malignancies and a combined use of cytology examination and SHOX2 and SEPT9 methylation detection in pleural fluid resulted in an increase of positive detecting rate comparing to cytology examination alone[13, 14]. Similar results achieved from a combined detection of the SHOX2 and PTGER4 gene methylation in lung cancer diagnosis [15, 16]. Aberrant methylation of the SHOX2 and RASSF1A gene in plasma samples were separately reported to be highly correlated with lung cancer diagnosis and prognosis, but the combination detection of SHOX2 and RASSF1A in BALF has hardly been reported.
In our study, we detected the aberrant methylation of the SHOX2 and RASSF1A gene in BALF of 322 patients using a new RT-PCR platform and then confirmed by sanger sequencing. Cytology examination in BALF and serum CEA assay were performed at the same time. The diagnostic value and clinical use of both the three methods were evaluated.
Demographic and clinical features of the patients.
| Total | Lung Cancer | Control | ||||||
|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |||
| Age (years) | 322 | 284 | 38 | |||||
| ≤50 | 56 | 17.4% | 37 | 13.0% | 19 | 50.0% | ||
| 51-60 | 102 | 31.7% | 95 | 33.5% | 7 | 18.4% | ||
| 60-70 | 116 | 36.0% | 109 | 38.2% | 7 | 18.4% | ||
| >70 | 48 | 14.9% | 43 | 15.1% | 5 | 13.2% | ||
| Median age | 61 | 61 | 51 | |||||
| Age range | 29-85 | 31-85 | 29-75 | |||||
| Sex | ||||||||
| Male | 240 | 74.5% | 212 | 74.6% | 28 | 73.7% | ||
| Female | 82 | 25.5% | 72 | 25.4% | 10 | 26.3% | ||
| Histology subtype | ||||||||
| Squamous cell carcinoma | - | - | 107 | 37.7% | - | - | ||
| Adenocarcinoma | - | - | 92 | 32.4% | - | - | ||
| Small cell lung cancer | - | - | 42 | 14.8% | - | - | ||
| Large cell lung cancer | - | - | 5 | 1.8% | - | - | ||
| Unkown | - | - | 38 | 13.3% | - | - | ||
| Benign lung diseases | - | - | - | - | 35 | 92.1% | ||
| Malignancies in other systems | - | - | - | - | 3 | 7.9% | ||
| Tumor stage | ||||||||
| Stage I | 28 | 9.9% | ||||||
| Stage II | 30 | 10.6% | ||||||
| Stage III | 133 | 46.8% | ||||||
| Stage IV | 93 | 32.7% | ||||||
Benign lung diseases including pulmonary infection, tuberculosis and bronchiectasis etc.
Malignancies in other systems including thyroid carcinoma, renal carcinoma and esophageal cancer.
Patients and Methods
Study Subjects and Samples
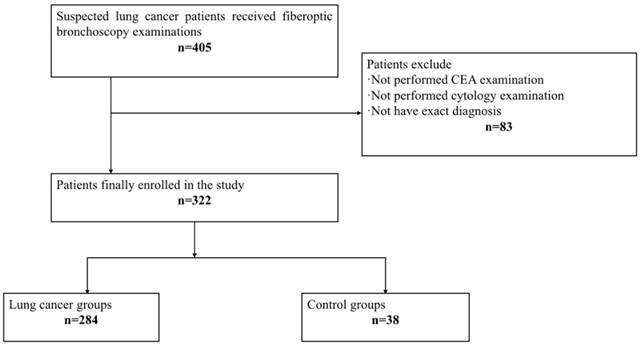
All samples were collected from consenting individuals according to protocols approved by the ethics committee of Shanghai Chest Hospital. The registration number of this clinical study is LS1418. In this study, we recruited 405 patients who received fiberoptic bronchoscopy examination in Shanghai Chest Hospital from December 2014 to June 2015, and performed a 2-year follow-up. After a series of exclusion criteria including absence of CEA detection and cytology examination or without exact diagnosis, 322 patients finally enrolled in our research (Figure 1). Among 322 patients, 284 were diagnosed as lung cancer, including107 squamous cell carcinomas, 92 adenocarcinomas, 42 small cell lung cancer, 5 large cell lung cancer, and 38 unclassified non-small cell lung cancer patients. The other 38 cases were controls, of which 35 were benign lung diseases including pulmonary infection, tuberculosis and bronchiectasis etc., and 3 patients were malignancies in other systems, including 1 thyroid carcinoma, 1 renal carcinoma, and 1 esophageal cancer. The final diagnosis of all patients were made according to histologically results, and the tumor stages were defined following the American Joint Committee on Cancer (AJCC) staging system as revised in 2010 (7th edition). Table 1 summarizes the demographic and clinical features of the patients.
CEA analysis
A total of 3 mL of venous blood was collected and serum was isolated by centrifugation at 3000 rpm for 10 min for CEA quantitative detection. Using electrochemiluminescence immunoassay kit (Tellgen Co., Ltd., Shanghai China) according to the manufacture's instruction. The critical value was: CEA>5.0ng/mL [17]. Serum samples were detected immediately.
Flow chart.
Cytological and pathological analysis
BALF samples were gathered by washing the affected lung segment with 20-40 mL of normal saline through fiberoptic bronchoscopy examinations. 10 mL BALF sample was centrifuged at 2000 rpm for 10 min, then pour off the supernatant and re-suspended the cellular button with 10-15 mL of Scott's tap water substitute. Next, the specimen was re-centrifuged at 2000rpm for 10 min and the cellular precipitant was used to make alcohol-fixed slides and stained with the Papanicolaou stain. Bronchial biopsy samples or surgical samples were processed and stained with haematoxylin and eosin. All the slides were reviewed by experienced pathologists in our hospital.
DNA extraction and processing
Precipitation of 10 mL BALF centrifuged at 10000 rpm for 5 min was used for DNA extraction. Genomic DNA was extracted using a standard kit-based method (TIANamp Genomic DNA Kit, Tiangen Biotech Co., Ltd., Beijing China), and unmethylated cytosine was modified to uracil with sodium bisulfite using the EZ DNA methylation-Directtm kit (Zymo Research, USA). DNA samples were detected immediately or stored at -80℃ [4].
Sanger sequencing
The primers were designed using Primer Premier 5.0. Here followed the primer sequences: SHOX2 forward primer: 5'-GGTGTTGTGTCGTATAGGGAGT-3', reverse primer: 5'- TCCGCCTCCTACCTTCTAAC-3'; RASSF1A forward primer: 5'- GAGGGAAGGAAGGGTAAGG-3', reverse primer:5'- GAGGGAAGGAAGGGTAAGG-3'. Each reaction was performed in 40 μL volume containing 5 μL cDNA, 0.8 μL (10 μM) forward primer, 0.8 μL (10 μM) reverse primer, 20 μL 2 × Taq buffer (including dNTPs and Taq polymerase), and 13.4 μL ddH20. The thermal cycling profile for PCR was set up as follows: pre-denaturation at 95℃ for 10 min, 45 cycles of denaturation for 30 s at 95℃, annealing for 35 s at 58℃, and extension for 30 s at 72℃, followed by a final extension at 72℃ for 8 min. PCR products were sequenced by Sangon Biotech Co., Shanghai, Ltd.
RT-PCR
The unmodified DNA was analyzed by the Methylated Human SHOX2 and RASSF1A Gene Detection Kit (Tellgen Co., Ltd., Shanghai China). The unmethylated C bases in genomic DNA were modified to U bases by sulfite, and then to T bases in the process of PCR amplification, while the methylated C bases remain unchanged, so the methylated and unmethylated C bases can be distinguished. Products of PCR amplification using the specific primers targeting different sequences before and after modification were detected by TaqMan probes, using methylated SHOX2 and RASSF1A DNA plasmids as controls.
Statistical analysis
Statistical analyses were performed using SPSS 19.0 software package (SPSS Inc., Chicago, IL). The methodological consistency between RT-PCR and Sanger sequencing in detecting the aberrant methylation of the SHOX2 and RASSF1A panel were analyzed using Kappa-test. The frequency of methylation in SHOX2 and RASSF1A gene, CEA detection, and cytology examination was analyzed using chi-square test. For each diagnostic method, we established a receiver operating characteristic (ROC) curve to calculate the area under the ROC curve (AUC) to evaluating the diagnostic efficacy. A P-value<0.05 was considered statistically significant.
Results
A new DNA methylation detection assay based on RT-PCR
We first developed a new DNA methylation detection assay based on RT-PCR and confirmed it with sanger sequencing. The results shown in Table 2 revealed an almost perfect consistency (Kappa=0.9698, 95%CI: 0.9388-0.9980) between RT-PCR and sanger sequencing in detecting the methylation of the SHOX2 and RASSF1A gene. Four cases got positive results detected by RT-PCR, while negative by sanger sequencing. Three of these four cases were finally diagnosed lung cancer, while the other one turned out to be pulmonary infection. Results detected by RT-PCR were chosen to represent the aberrant methylation of SHOX2 and RASSF1A.
The consistency of RT-PCR and Sanger sequencing in detecting aberrant methylation of the SHOX2 and RASSF1A gene.
| PCR | Sequencing | Total | Kappa | 95%CI | ||
|---|---|---|---|---|---|---|
| Positive | Negative | |||||
| SHOX2+RASSF1A | Positive | 227 | 4 | 231 | 0.9698 | 0.9388-0.9980 |
| Negative | 0 | 91 | 91 | |||
| Total | 227 | 95 | 322 | |||
Diagnostic sensitivity of SHOX2 and RASSF1A methylation in BALF in different groups
We analyzed the diagnostic sensitivity of SHOX2 and RASSF1A methylation in BALF in different histological subtypes and pathological stages of lung cancer (Table 3), our data showed that the positive detection rate of the SHOX2 and RASSF1A methylation panel in BALF was 81.0% in lung cancer group and 2.6% in control group. The positive rate of SHOX2 and RASSF1A methylation in BALF was higher in the lung cancer group than the control group (p<0.001, 95%CI: 0.714-0.853). Besides, the sensitivity of cytology and serum CEA was 68.3% and 30.6% respectively, which is dominantly lower than that of the SHOX2 and RASSF1A methylation in BALF (P<0.001, P<0.001, respectively). In a more detailed analysis, we noticed that the SHOX2 and RASSF1A methylation panel showed a 100% detection rate in large cell carcinoma, when CEA and cytology missed more than half of them. Besides, results from other three histological types also showed the same performance, which means the SHOX2 and RASSF1A methylation panel in BALF is a noninvasive biomarker with wide application in almost all histological subtypes of lung cancer. However, the cases size for large cell lung cancer is not enough to represent for this histological subtype (n=5), further studies should perform with more large cell lung cancer patients included.
Detection sensitivity of CEA, cytology, and the SHOX2 and RASSF1A methylation panel in different histological subtype groups.
| Tumor classification | CEA | Cytology | SHOX2+RASSF1A | |||
|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |
| Lung cancer | ||||||
| Squamous cell carcinoma (n=107) | 22 | 20.6% | 80 | 74.8% | 95 | 88.8% |
| Adenocarcinoma (n=92) | 46 | 50.0% | 54 | 58.4% | 64 | 69.6% |
| Small cell lung cancer (n=42) | 6 | 9.5% | 34 | 81.0% | 38 | 90.5% |
| Large cell lung cancer (n=5) | 1 | 20.0% | 1 | 20.0% | 5 | 100.0% |
| Unkown (n=38) | 12 | 31.6% | 25 | 65.8% | 28 | 73.7% |
| Total (n=284) | 87 | 30.6% | 194 | 68.3% | 230 | 81.0% |
| Control | ||||||
| Benign lung diseases (n=35) | 0 | 0.0% | 1 | 2.9% | 1 | 2.9% |
| Malignancies in other systems (n=3) | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% |
| Total (n=38) | 0 | 0.0% | 1 | 2.6% | 1 | 2.6% |
Next, we evaluated the detection rates of the SHOX2 and RASSF1A methylation panel in BALF in different tumor stages (Table 4). Our results showed that the methylation analysis of SHOX2 and RASSF1A in BALF showed the highest diagnostic ability with a total positive detection rate of 81.0%, while CEA and cytology were 30.6% and 68.3%, respectively. Interestingly, the SHOX2 and RASSF1A methylation panel in BALF got an extremely high detection rate of 85.7% in stage I lung cancer patients, while CEA and cytology were 10.7% and 46.4%, respectively. The above data suggested that the SHOX2 and RASSF1A methylation panel in BALF was a potential tool for lung cancer diagnosis, especially in an early stage.
Detection sensitivity of CEA, cytology, and the SHOX2 and RASSF1A methylation panel in different tumor stage groups.
| Tumor stage | CEA | Cytology | SHOX2+RASSF1A | |||
|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |
| Stage I (n=28) | 3 | 10.7% | 13 | 46.4% | 24 | 85.7% |
| Stage II (n=30) | 7 | 23.3% | 17 | 56.7% | 24 | 80.0% |
| Stage III (n=133) | 33 | 24.8% | 92 | 69.2% | 104 | 78.2% |
| Stage IV (n=93) | 44 | 47.3% | 72 | 77.4% | 78 | 83.9% |
| Total (n=284) | 87 | 30.6% | 194 | 68.3% | 230 | 81.0% |
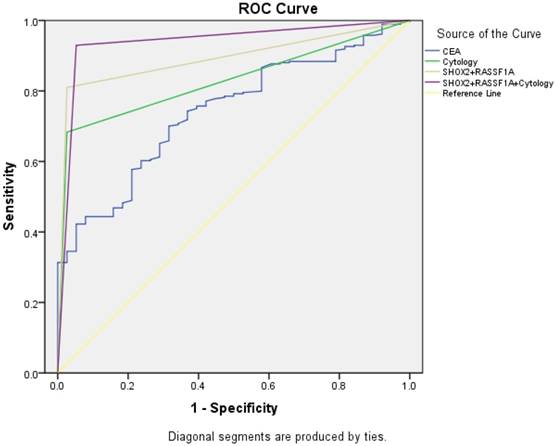
ROC curve analysis of the SHOX2 and RASSF1A methylation panel in BALF
We performed a ROC curve analysis to compare the diagnostic efficacy of the SHOX2 and RASSF1A methylation panel in BALF with cytology and serum CEA (Figure 2). As shown in Figure 2 and Table 5, the SHOX2 and RASSF1A methylation panel in BALF showed the highest AUC value of 0.892 (95%CI: 0.849-0.943), comparing to CEA (AUC value: 0.741, 95%CI :0.670-0.812) and cytology (AUC value: 0.828, 95%CI: 0.777-0.880). Besides, methylation analysis of SHOX2 and RASSF1A panel showed the highest diagnostic sensitivity of 81.0%, comparing to CEA (30.6%) and cytology (68.3%). Notably, when combining the SHOX2 and RASSF1A methylation panel with cytology, the AUC was 0.938 (95% CI: 0.894-0.983), and the sensitivity and specificity were improved to 93.0% and 94.7%, respectively, which suggested that the SHOX2 and RASSF1A methylation detection of BALF could be an effective complementary tool of cytology in lung cancer diagnosis.
Discussion
The relationship between cancer diagnosis and the detection of aberrant methylation changes of the SHOX2 and RASSF1A gene has been revealed by previous studies [12, 18, 19]. Methylation analysis of these two genes in plasma samples has been performed for several times, separately. However, since the detection sensitivity of SHOX2 (60%) and RASSF1A (52.4%) in plasma alone is not satisfactory, besides, detection of somatic gene expression in plasma can be very challenging due to its complex component, we wonder whether the combination of the SHOX2 and RASSF1A gene methylation detection in BALF can improve the detection sensitivity in lung cancer diagnosis[12, 20]. Bronchoalveolar lavage fluid is a kind of non-invasive specimen that is easily obtained through fiberoptic bronchoscopy examination, a routine inspection item for suspected lung cancer patients. Due to its vicinity to tumor cells, BALF represents an alternative source of biomarkers for cancer diagnosis.
ROC curve for CEA, cytology, and the SHOX2 and RASSF1A methylation panel.
The diagnostic efficacy of CEA, cytology and the SHOX2 and RASSF1A methylation panel
| AUC | Sensitivity | Specificity | PPV | NPV | ||
|---|---|---|---|---|---|---|
| Value | 95%CI | |||||
| CEA | 0.741 | 0.670-0.812 | 30.6% | 100.0% | 100.0% | 16.2% |
| Cytology | 0.828 | 0.777-0.880 | 68.3% | 97.4% | 99.5% | 29.1% |
| SHOX2+RASSF1A | 0.892 | 0.849-0.934 | 81.0% | 97.4% | 99.6% | 40.7% |
| SHOX2+RASSF1A+Cytology | 0.938 | 0.894-0.983 | 93.0% | 94.7% | 99.3% | 64.3% |
The cut-off value for CEA was 5ng/mL.
Methylation signatures are proving to be significant markers for tumor diagnosis, aberrant methylation of some genes including SHOX2 and SEPT9 have already been translated into commercial clinical assays [21]. We detected the methylation changes in our 322 patients with both sanger sequencing and the Methylated Human SHOX2 and RASSF1A Gene Detection Kit (Tellgen Co. Ltd., Shanghai China). The consistency between the two platforms suggested that the method based on RT-PCR is credible in aberrant methylation analysis. It may be potential to be a substitution of sanger sequencing on methylation detection for its efficiency on operation and low cost.
Our data showed that the SHOX2 and RASSF1A methylation panel achieved a sensitivity of 81.0% and a specificity of 97.4% with an AUC value of 0.892 in lung cancer diagnosis. This suggests that the SHOX2 and RASSF1A methylation panel has an efficient diagnostic ability in lung cancer diagnosis. The performance of the methylation panel in all histological subtype groups were satisfying. The positive detection rates of the panel in large cell lung cancer, small cell lung cancer, squamous cell carcinoma and adenocarcinoma were 100%, 90.5%, 88.8% and 69.6%, respectively. However, results from large cell lung cancer detection may provide less reference because there only included 5 large cell lung cancer patients in our study. When compared in squamous cell carcinoma, adenocarcinoma, and small cell lung cancer, the SHOX2 and RASSF1A methylation panel showed better performance in small cell lung cancer and squamous cell carcinoma than that in adenocarcinoma, which was similar to previous studies [4, 11]. A probable explanation is that central type lung cancers have more chances to release tumor cells and biomarkers into BALF.
Early diagnostic ability is essential for lung cancer patients because inadequate early diagnosis may lead to poor prognosis [22]. We compared the three diagnostic methods in lung cancer patients from stage I to stage IV. CEA showed an extremely low diagnostic ability in early stage patients of lung cancer. Among 28 patients in stage I, there were only 3 turned out to be positive in CEA examinations, while cytology examination and methylation detection of the SHOX2 and RASSF1A methylation panel distinguished 13 and 24 from the 28 patients, respectively. The SHOX2 and RASSF1A methylation panel achieved a relatively high diagnostic rate of 85.7% in stage I patients, which demonstrates that the panel may have the potential to become a promising biomarker for early diagnosis of lung cancer. A relevant study revealed that biomarkers in BALF had features such as earlier appearance and higher concentrations in comparing with serum samples [23]. Thus detection of biomarkers in BALF may provide more significant information for lung cancer diagnosis, especially in an early stage.
In our study, the methylation analysis of the SHOX2 and RASSF1A panel in BALF (AUC =0.892) showed a best diagnostic ability in lung cancer diagnosis compared with serum CEA detection (AUC=0.741) and cytology examination (AUC=0.828). A combined use with cytology further improved the diagnostic sensitivity and specificity to 93.0% and 94.7%, respectively. This suggests that the SHOX2 and RASSF1A methylation detection of BALF could be an effective complementary tool of cytology in lung cancer diagnosis.
However, there also exists some limitations in our research. Though 322 patients were included in our research, the exact number of patients in some histological subtype groups such as large cell lung cancer was insufficient that the diagnostic value may be over-evaluated, so more patients in this group needed to be enrolled in further studies. Besides, the control group should better consist of a group of healthy cases to evaluate the specificity and sensitivity, but it is difficult to obtain BALF samples from healthy individuals. What's more, whether the methylation changes of the two genes correlated to prognosis of lung cancer patients is not clear. Further studies could focus on the possibility of a better prognosis in lung cancer patients with unmethylated SHOX2 and RASSF1A gene.
In conclusion, the methylation analysis of the SHOX2 and RASSF1A panel in BALF showed an efficient diagnostic ability in lung cancer diagnosis, especially in an early stage. It may have the potential to become a promising biomarker in lung cancer diagnosis, and be a complementary tool of cytology in lung cancer diagnosis.
Acknowledgements
The study was supported by The National Natural Science Foundation of China (81672833), Research Project of Shanghai Health and Family Planning Commission (20154Y0185) and The Municipal hospital clinical auxiliary department capacity building project of Shanghai Shen Kang Hospital Development Center (SHDC22014011).
Ethics
The study was approved by the Ethics Committee of Shanghai Chest Hospital, Shanghai Jiao Tong University, China.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10-29
2. Jemal A, Bray F, Center MM. et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69-90
3. Aberle DR, Adams AM, Berg CD. et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365(5):395-409
4. Ren M, Wang C, Sheng D. et al. Methylation analysis of SHOX2 and RASSF1A in bronchoalveolar lavage fluid for early lung cancer diagnosis. Ann Diagn Pathol. 2017;27:57-61
5. Grunnet M, Sorensen JB. Carcinoembryonic antigen (CEA) as tumor marker in lung cancer. Lung Cancer. 2012;76(2):138-143
6. Vizoso M, Puig M, Carmona FJ. et al. Aberrant DNA methylation in non-small cell lung cancer-associated fibroblasts. Carcinogenesis. 2015;36(12):1453-1463
7. Stelzer Y, Shivalila CS, Soldner F. et al. Tracing dynamic changes of DNA methylation at single-cell resolution. Cell. 2015;163(1):218-229
8. Sandoval J, Peiro-Chova L, Pallardo FV. et al. Epigenetic biomarkers in laboratory diagnostics: emerging approaches and opportunities. Expert Rev Mol Diagn. 2013;13(5):457-471
9. Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Curr Opin Genet Dev. 2012;22(1):50-55
10. Wei H, Fang N, Guo L. et al. [Meta-analysis of the Association between RASSF1A Gene Promoter Methylation and Non-small Cell Lung Cancer]. Zhongguo Fei Ai Za Zhi. 2015;18(7):443-450
11. Darwiche K, Zarogoulidis P, Baehner K. et al. Assessment of SHOX2 methylation in EBUS-TBNA specimen improves accuracy in lung cancer staging. Ann Oncol. 2013;24(11):2866-2870
12. Kneip C, Schmidt B, Seegebarth A. et al. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer in plasma. J Thorac Oncol. 2011;6(10):1632-1638
13. Dietrich D, Jung M, Puetzer S. et al. Diagnostic and prognostic value of SHOX2 and SEPT9 DNA methylation and cytology in benign, paramalignant and malignant pleural effusions. PLoS One. 2013;8(12):e84225
14. Jung M, Putzer S, Gevensleben H. et al. Diagnostic and prognostic value of SHOX2 and SEPT9 DNA methylation and cytology in benign, paramalignant, and malignant ascites. Clin Epigenetics. 2016;8:24
15. Weiss G, Hasinger O, Esche S. et al. 31PD DNA methylation of SHOX2 and PTGER4 as a plasma-based tool to differentiate between patients with malignant and benign lung disease. Journal of Thoracic Oncology. 2016;11(4):S68
16. Weiss G, Schlegel A, Kottwitz D. et al. Validation of the SHOX2/PTGER4 DNA Methylation Marker Panel for Plasma-Based Discrimination between Patients with Malignant and Nonmalignant Lung Disease. J Thorac Oncol. 2017;12(1):77-84
17. Zhao W, Yu H, Han Z. et al. Clinical significance of joint detection of serum CEA, SCCA, and bFGF in the diagnosis of lung cancer. Int J Clin Exp Pathol. 2015;8(8):9506-9511
18. Song L, Yu H, Li Y. Diagnosis of Lung Cancer by SHOX2 Gene Methylation Assay. Mol Diagn Ther. 2015;19(3):159-167
19. Shou F, Xu F, Li G. et al. RASSF1A promoter methylation is associated with increased risk of thyroid cancer: a meta-analysis. Onco Targets Ther. 2017;10:247-257
20. Zhai X, Li SJ. Methylation of RASSF1A and CDH13 genes in individualized chemotherapy for patients with non-small cell lung cancer. Asian Pac J Cancer Prev. 2014;15(12):4925-4928
21. Paska AV, Hudler P. Aberrant methylation patterns in cancer: a clinical view. Biochem Med (Zagreb). 2015;25(2):161-176
22. Hu J, Qian GS, Bai CX. Chinese consensus on early diagnosis of primary lung cancer (2014 version). Cancer. 2015;121(Suppl 17):3157-3164
23. Wang H, Zhang X, Liu X. et al. Diagnostic value of bronchoalveolar lavage fluid and serum tumor markers for lung cancer. J Cancer Res Ther. 2016;12(1):355-358
Author contact
![]() Corresponding author: Jiatao Lou, MD, Department of Laboratory Medicine, Shanghai Chest Hospital, 241 West Huaihai Road, Shanghai, 200030, China. Tel.: +86-21-2220000-1503, Fax: +86-21-62808279, E-mail: loujiataocom
Corresponding author: Jiatao Lou, MD, Department of Laboratory Medicine, Shanghai Chest Hospital, 241 West Huaihai Road, Shanghai, 200030, China. Tel.: +86-21-2220000-1503, Fax: +86-21-62808279, E-mail: loujiataocom

 Global reach, higher impact
Global reach, higher impact