3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(19):3933-3938. doi:10.7150/jca.20342 This issue Cite
Short Research Communication
FAM13A as a Novel Hypoxia-Induced Gene in Non-Small Cell Lung Cancer
1. Institute of Human Genetics, Polish Academy of Sciences, Strzeszyńska 32, 60-479 Poznań, Poland;
2. Department of Thoracic Surgery, University of Medical Sciences, Szamarzewskiego 62, 60-569 Poznań;
3. Department of Biochemistry and Biotechnology, Poznan University of Life Sciences , Dojazd 11, 60-632 Poznan, Poland.
Received 2017-3-30; Accepted 2017-8-4; Published 2017-10-23
Abstract
Several genome-wide association studies (GWASs), have identified that FAM13A and IREB2 loci are associated with lung cancer, but the mechanisms by which these genes contribute to lung diseases susceptibility, especially in hypoxia context, are unknown. Hypoxia has been identified as a major negative factor for tumor progression in clinical observation. It has been suggested, that lower oxygen tension, may modulate the IREB2 and FAM13A activity. However, the role of these genes in hypoxia response has not been explained. To precise the role of these genes in hypoxia response, we analyzed the FAM13A and IREB2 expression, in lung cancer cells in vitro and lung cancer tissue fragments cultured ex vivo.
Three cell lines: non-small cell lung cancer (A549, CORL-105), human lung fibroblasts (HL) and 37 lung cancer tissue fragments were analyzed. The expression of IREB2, FAM13A and HIF1α after sustained 72 hours of hypoxia versus normal oxygen concentration were analyzed by TaqMan® Gene Expression Assays and Western Blot.
The expression of FAM13A was significantly up-regulated by hypoxia in two lung cancer cell lines (A549, CORL-105, P<0.001), both at the level of protein and mRNA, and in lung cancer tissue fragments (P=0.0004). The IREB2 was down-regulated after hypoxia in A549 cancer cells (P<0.001).
Conclusions: We found that FAM13A overexpression in human lung cancer cell lines overlapped with hypoxia effect on lung cancer tissues. FAM13A is strongly induced by hypoxia and may be identified as a novel hypoxia-induced gene in non-small cell lung cancer.
Keywords: non-small cell lung cancer, IREB2 gene, FAM13A gene, hypoxia, gene expression.
Introduction
Hypoxia in solid tumors has been identified as a major negative prognostic factor, because decreased availability of oxygen in the tumor increases treatment resistance and enhances tumor progression and metastasis. Low oxygen level in tumor cells is mainly due to rapid proliferation of cells and deficiency of blood distribution in the tumor mass [1]. Several genome-wide association studies (GWASs), also ours [2], have identified that IREB2 and FAM13A loci are associated with lung cancer, but the mechanisms by which these genes contribute to lung diseases susceptibility, are unknown. It has been suggested, that lower oxygen tension, may modulate the IREB2 and FAM13A activity, but its function in hypoxia response has not been elucidated.
The biological function of the FAM13A (family with sequence similarity 13, member A, MIM 147582) gene product is poorly understood. The highest expression of FAM13A gene was detected in the brain and ovaries, followed by the lungs and kidneys. It is suggested that the most important part of the FAM13A protein is its N-terminal extension, containing the Rho-GAP domain, which presents tumor suppressor activity through inhibition of the intracellular signal transduction molecule RhoA [3].
The IREB2 (iron-responsive element binding protein 2, MIM 613299) gene belongs to a group of genes which regulate mammalian iron homeostasis. DeMeo et al. have observed increased levels of IREB2 mRNA and protein in lung tissue samples from chronic obstructive pulmonary disease patients compared to healthy donors [4]. Dysfunctions of IREB2 gene, as a major regulator of iron homeostasis, may lead to oxidative failures, which may be relevant in pathogenesis of lung cancer.
The mechanisms by which these genes contribute to hypoxia response in lung cancer cells are unknown. It has been observed that IREB2 is posttranslationally regulated by hypoxia [4]. FAM13A gene expression analyses in several cells from various tissues, but no lung cancer cells, have shown a consistent increase of expression in response to hypoxia [5]. However, the precise role of these genes in hypoxia response has not been fully explained. In this study, we investigated the effect of long-term exposure to hypoxia on the IREB2 and FAM13A expression at mRNA and protein level, from 3 cell lines and fresh human lung cancer tissue fragments cultured ex vivo. Herein, we used complementary approach to provide evidence that FAM13A and IREB2 activity are modulated by hypoxia.
Methods
Approval
All experiments were performed in accordance with relevant guidelines and regulations approved by the Ethics Committee of the Poznan University of Medical Sciences (decision no. 802/10).
Informed consent
All participants and/or their legal guardian/s gave their written informed consent to participate.
Materials
Human cell lines
Human non-small cell lung carcinoma cell line A549, Caucasian lung adenocarcinoma cell line COR-L105 and human lung fibroblast HL cell line (from normal human lung tissue) were purchased from The European Collection of Cell Cultures (ECACC), supplied by Sigma Aldrich. The early passages were used for the experiments. Cells were cultured in RPMI medium (CORL-105, HL) or DMEM medium (A549), supplemented with 2mM Glutamine and 10% fetal bovine serum. The cells were growth to the confluence of 60-80% at 37°C in ambient (21%) oxygen level. Then part of cells, where left in the same condition (as “normal” or “control cells”), whereas cells subjected to sustained hypoxia were transferred to the BINDER CB53 incubator with 1% oxygen in atmosphere, for 72 hours. Each experiment was performed in triplicate.
Lung cancer fragments
Tumor tissue samples from 9 consecutive patients with non-small cell lung cancer (NSCLC) who were referred for surgical resection to the Department of Thoracic Surgery, University of Medical Sciences in Poznań, from September to December 2015, were included in the study. Histological subtypes of NSCLC were: 4 carcinoma planoepitheliale, 3 adenocarcinoma and 2 carcinoma macrocellulare. Patients with pre-operative chemotherapy were excluded from the study. 37 tumor fragments were analyzed as described by K. Leithner et al. [6]. Surgical specimens were dissected into small fragments and were incubated in 6-well culture plates (up to ten fragments per well) in DMEM medium + 2mM Glutamine + 10% FBS and antibiotic, antimycotic solution (Sigma). Fragments were cultured and let attached for 16 hours, thereafter cells were cultured for three days at 37°C in ambient (21%) oxygen or 1% oxygen in the BINDER CB53 incubator.
RNA extraction and cDNA synthesis
Total RNA was extracted using the TRI Reagent® (Sigma-Aldrich) according to the manufacturer's protocol and then converted into cDNA using the QuantiTect Reverse Transcription Kit (Qiagen).
Quantitative analysis of mRNA expression
Real-Time PCR analysis was performed using TaqMan probes directed at FAM13A, IREB2, HIF1α and GUSB as reference gene. The reactions were performed with HOT FIREPol Probe qPCR Mix Plus (no ROX) according to the manufacturer's instructions Solis Biodyne (Tartu, Estonia) and under conditions specified in the ABI TaqMan Gene Quantification assay protocol. Thermal cycling was performed using a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, Hercules, California, U.S.). The fold change value was calculated using 2-ΔΔCt, described by K. Livak et al. [7].
Western Blot
Cell lysis and protein extraction from three human lung cell lines was made using RIPA buffer (Sigma). Protein concentration was determined using the Bicinchoninic Acid Kit, BCA (Sigma). Proteins were mixed with Laemmli Sample Buffer, separated by SDS-PAGE and transferred to PVDF membranes using the BioRAD MiniProtean3® system. Membranes were blocked with 5% milk and incubated overnight at 4°C with antibodies (Abcam): IREB2 (ab181153), FAM13A (ab122440), HIF-1-alpha (ab51608) and β-Actin (ab115777). After incubation with horseradish peroxidase-conjugated secondary antibody (ab97051), they were visualized by chemiluminescence using ChemiDoc™ Imaging Systems BioRad.
Statistical analysis
The one-way ANOVA followed by Newman-Keuls test was performed to determine the significance. All experiments were performed at least three separate times.
Results and Discussion
We have previously confirmed the association between IREB2 gene and lung cancer and between FAM13A gene and chronic obstructive pulmonary disease (COPD) in Polish patients [2]. The mechanisms by which these genes contributes to lung diseases susceptibility are still explored. To our best knowledge, this is the first report in which the regulation of FAM13A and IREB2 gene expression by hypoxia in lung cancer cells was examined. Changes in gene expression in hypoxic cancer cells studied in vitro may differ from the in vivo due to the microenvironment of the tumor [6]. Here we studied gene expression in cancer tissue ex vivo which contains both tumor and stroma cells and mimics the in vivo situation.
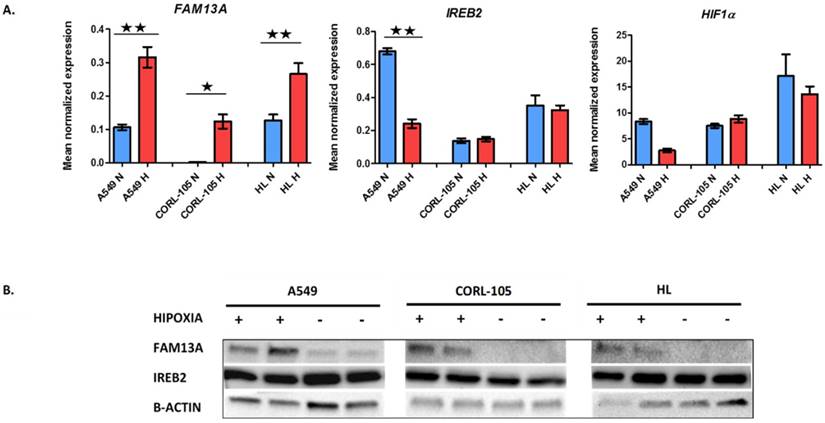
The FAM13A mRNA was significantly up-regulated by chronic hypoxia in cancer cell lines (A549, CORL-105; P<0.001). The expression fold change values, corrected for GUSB (glucuronidase-β gene), relative to normoxia, were 3.1 and 54.8 for A549 and CORL-105 cells, respectively. The overexpression of FAM13A was confirmed at the protein level by Western blot analysis in all lung cancer cell lines (Figure 1). We observed also the marginally increase FAM13A expression in normal human lung fibroblasts (HL; P<0.001), with fold change value 2.3, but the induction by hypoxia was weaker, compared to lung cancer cells, measured on the level of protein and mRNA.
Hypoxia-induced changes in expression of FAM13A, IREB2 and HIF1α in lung cancer cell lines and human lung fibroblasts. A. Quantitative analysis of FAM13A, IREB2 and HIF1α mRNA expression in 3 cell lines. Cells were cultured in hypoxia or ambient oxygen (normoxia) for 72 hours. Gene expression was normalized to the endogenous control, GUSB. Mean normalized expression levels in hypoxia and normoxia are shown. Data are expressed as the mean ± SEM of triplicate values. Results are mean from three independent experiments. A549, human non-small cell lung carcinoma cell line; CORL-105, Caucasian lung adenocarcinoma cell line; HL, human lung fibroblast; N, normoxia (blue); H, hypoxia (red). *P < 0.001, **P < 0.0001. B. Western blot analysis of FAM13A, IREB2 and β-actin proteins expression in 3 cell lines, from two separate experiments. Cells were cultured in hypoxia or ambient oxygen (normoxia) for 72 hours and total proteins were extracted. A549, human non-small cell lung carcinoma cell line; CORL-105, Caucasian lung adenocarcinoma cell line; HL, human lung fibroblast; Hypoxia exposition +; Normoxia, -.
Quantitative analysis of FAM13A, IREB2 and HIF1α mRNA expression in cultured lung cancer fragments. Non-small cell lung cancer (NSCLS) samples from 9 donors (different colors) were cultured in hypoxia or ambient oxygen (normoxia) for 72 hours. Data are presented as fold changes (FCh) in gene expression. FCh were calculated using the 2-ΔΔCT method as described by Livak et al. Briefly, mean Ct values for the gene of interest and a reference gene in hypoxia (CtT,GOI and CtT,Ref, respectively) and normoxia (CtN,GOI and CtN,Ref, respectively) were used to calculate ΔCt (CtGOI - CtRef) for each sample. Then to derive the fold change (FCh) of the gene in the hypoxia compared with the normoxia FCh = 2-ΔΔCt, where ΔΔCt = ΔCtH - ΔCtN. An FCh of >2 was defined as overexpression, and an FCh of <0.5 was defined as under-expression of the gene.
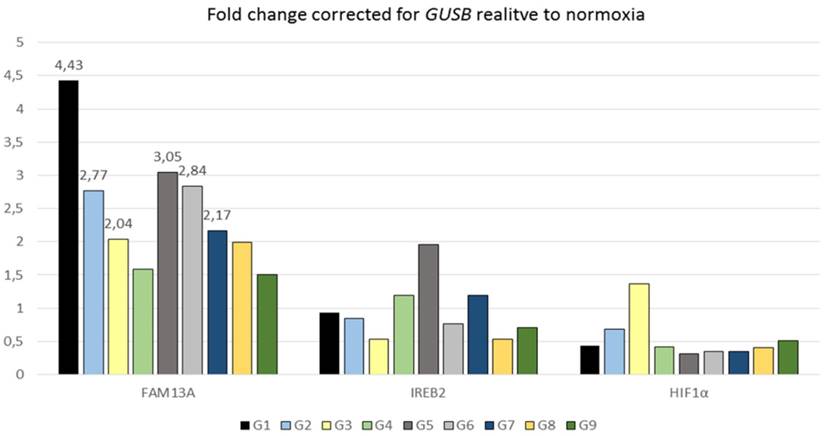
The FAM13A gene was also significantly regulated in the same direction under hypoxia in NSCLC cancer fragments (P=0.0004) (Figure 2). The average of fold change in FAM13A expression was >2 in case of 6 out of 9 tumor samples. In conclusion, we found that FAM13A is strongly induced by hypoxia in lung cancer cells both at the level of protein and mRNA.
This is the first report in which overexpression of FAM13A in lung cancer cells, was indicated both in vitro and ex vivo model in short-term cultures after prolonged hypoxia. Although little is known of the FAM13A regulation, gene expression analyses in cell lines from several tissues (renal epithelial cells, breast epithelial cells, smooth muscle cells and endothelial cells), not comprising the lung, have shown a consistent increase in its expression in response to hypoxia [5]. Global gene expression profiling on three tumor cell lines (melanoma, prostate and ovarian cancer), have indicated the statistically increase in FAM13A mRNA expression under chronic hypoxia conditions in prostate and ovarian cells. Opposite, in melanoma cells, the authors have observed weak decrease of FAM13A expression inducted by experimental cycling hypoxia and no changes after prolonged hypoxia [8]. The differences in response to hypoxia conditions, among various cell types confirm that gene expression profiles induced by hypoxia strongly depend on the cell type. It seems that each cell type has a specific panel of hypoxia-response genes and it is very difficult to find the “universal” hypoxia genes [8]. Additional work will be required to determine the mechanism of FAM13A induction by hypoxia and its contribution in features of lung cancer cells. It will worth to check, how much is altered FAM13A expression specific for lung cancer cells and if it may contribute to enhanced aggressiveness of cancer. Recently, FAM13A was identified as key regulator of NSCLC growth and progression. It was confirmed that FAM13A is involved in tumor cell proliferation, downstream of TGFβ and HIF1α, association with Th1 gene expression and lung tumor cell migration [9].
Differences in the expression of FAM13A gene have also been noted, in normal oxygen tension, in respiratory epithelial cells during differentiation into pulmonary type II alveolar cells in vitro model of cystic fibrosis [10]. Eisenhut F. et al. found increased numbers of FAM13A protein expressing cells in the tumoral region of lung tissues from a cohort of patients with NSCLC [9]. However, according to Human Protein Atlas database [11], the FAM13A transcript and protein is detected at medium level in normal lung tissue. In two lung cancer cell lines (A549 and SCLC-21H - small cell lung carcinoma), transcripts were detected at low level. FAM13A protein is also expressed by lung cancer tissue in 11 patients with squamous cell carcinoma and adenocarcinoma. Most cancer cells showed moderate cytoplasmic immunoreactivity. Only in 2 out of 11 patients with lung cancer (squamous cell carcinoma) showed low protein expression level, compare with high level detected in 3 patients with lung adenocarcinomas. These observations have showed that in normal oxygen tension, in the vast majority of cells, FAM13A is expressed at low or moderate level. Our findings suggested that hypoxia strongly induces the FAM13A expression, especially in lung cancer cells.
The potential role of FAM13A gene in development or/and occurrence of cancer and lung diseases is still not well established. Known findings about FAM13A functional role are limited and mainly concerns COPD and emphysema. Recently, Z. Jin et al. showed that depletion of FAM13A in human lung cancer cells (A549 cell line) causes a reduction in Wnt signaling activity which provides evidence that FAM13A may contribute to human lung diseases [12]. Another study by Z. Jiang et al. indicated that FAM13A influences COPD susceptibility by promoting β-catenin degradation [13]. With the use of two mouse models, authors have observed that knockdown of FAM13A leads to increased activation of the Wnt pathway, stimulating epithelial cell proliferation and thus lung repair [13]. This FAM13A interaction with Wnt pathway needs to be clarify and connected to the effect of carrying the FAM13A risk allele. This translation from risk allele to biological function in COPD, was described in detail by C.A. Brandsma [14].
Several GWASs, beside of FAM13A, have identified IREB2 gene is associated with lung cancer, but the mechanisms of contribution to lung diseases susceptibility, are still being explored. Known suggestions, about modulation of IREB2 expression by hypoxia, are limited. In the current study, we observed that the IREB2 mRNA was significantly down-regulated after hypoxia only in A549 cancer cells, with expression fold change 0.36. The decrease in the IREB2 expression was also observed at the protein level by Western blot analysis in A549 cell line (Figure 1). The IREB2 gene was regulated in the same direction under hypoxia in NSCLC cancer fragments, with the average fold change <0.7 in case of 3 out of 9 samples (Figure 2). Our results indicated that the IREB2 gene, in contest of hypoxia, may have not an impact on lung tumors progression. However, studies on mouse models showed that overexpression of IREB2 promoted the growth of tumor xenografts in nude mice [15]. Opposite, quantitative analysis of IREB2 mRNA levels in normal lung and lung adenocarcinoma demonstrated the increased expression of CHRNA5, but no change in IREB2 [16]. In another study, the authors have failed to reveal that IREB2 mediates effects on lung cancer cell growth in vitro [17].
In a panel of analyzed cell lines and cancer fragments, HIF1α showed no increase in expression, as measured on mRNA and protein levels (data not shown), which confirmed previous observations that HIF1α is not proper marker of sustained chronic hypoxia [18], in lung cancer epithelial cells. The mechanism of HIF1-α mRNA stabilization is not fully explained. HIF-1α antisense transcript, has been shown to down-regulate HIF-1α mRNA in lung epithelial cells, which may be a part of adaptation process to chronic hypoxia. The expression of HIF-1α mRNA is suppressed in prolonged hypoxia, suggesting that the control of HIF1A gene transcription is regulated negatively. Recently, the Repressor Element 1-Silencing Transcription factor (REST) was proposed as HIF-1α transcription repressor in prolonged hypoxia [19], potentially to prevent an over activated HIF response, that could be detrimental to cells. Further supporting this protective role of reduced HIF-1α mRNA expression in prolonged hypoxia, high levels of HIF-1α mRNA have been observed in some cancers (hepatocellular carcinoma, gastric and prostate cancer) and often associated to poor prognosis [19]. In addition, recently proposed model, in which HIF-1α isoform was described to drive the initial response to hypoxia (<12 hours). Shift from HIF-1α to HIF-2α expression was reported as being evident in some solid tumors. It appears that expression of each isoform, depends on the cell type, the tissue, the level of O2 tension and the time of exposure to hypoxia [20].
Conclusions
FAM13A overexpression in human lung cancer cell lines overlapped with hypoxia effect on lung cancer tissues. It allowed to described the FAM13A gene as a novel hypoxia-induced gene in non-small cell lung cancer.
Acknowledgements
This work was supported by the National Science Centre, Poland: grant No. 2011/01/D/NZ5/02841 and, in part, No. 2016/21/D/NZ5/00072.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Yu L, Hales CA. Long-term exposure to hypoxia inhibits tumor progression of lung cancer in rats and mice. BMC Cancer. 2011;11:331
2. Ziolkowska-Suchanek I, Mosor M, Gabryel P, Grabicki M, Zurawek M, Fichna M. et al. Susceptibility loci in lung cancer and COPD: association of IREB2 and FAM13A with pulmonary diseases. Sci Rep. 2015;5:13502
3. Young RP, Hopkins RJ, Hay BA, Whittington CF, Epton MJ, Gamble GD. FAM13A locus in COPD is independently associated with lung cancer - evidence of a molecular genetic link between COPD and lung cancer. Appl Clin Genet. 2011;4:1-10
4. DeMeo DL, Mariani T, Bhattacharya S, Srisuma S, Lange C, Litonjua A. et al. Integration of genomic and genetic approaches implicates IREB2 as a COPD susceptibility gene. Am J Hum Genet. 2009;85:493-502
5. Chi JT, Wang Z, Nuyten DS, Rodriguez EH, Schaner ME, Salim A. et al. Gene expression programs in response to hypoxia: cell type specificity and prognostic significance in human cancers. PLoS Med. 2006;3:e47
6. Leithner K, Wohlkoenig C, Stacher E, Lindenmann J, Hofmann NA, Galle B. et al. Hypoxia increases membrane metallo-endopeptidase expression in a novel lung cancer ex vivo model - role of tumor stroma cells. BMC Cancer. 2014;14:40
7. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-8
8. Olbryt M, Habryka A, Student S, Jarzab M, Tyszkiewicz T, Lisowska KM. Global gene expression profiling in three tumor cell lines subjected to experimental cycling and chronic hypoxia. PLoS One. 2014;9:e105104
9. Eisenhut F, Heim L, Trump S, Mittler S, Sopel N, Andreev K. et al. FAM13A is associated with non-small cell lung cancer (NSCLC) progression and controls tumor cell proliferation and survival. Oncoimmunology. 2017;6:e1256526
10. Wright JM, Merlo CA, Reynolds JB, Zeitlin PL, Garcia JG, Guggino WB. et al. Respiratory epithelial gene expression in patients with mild and severe cystic fibrosis lung disease. Am J Respir Cell Mol Biol. 2006;35:327-36
11. Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A. et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419
12. Jin Z, Chung JW, Mei W, Strack S, He C, Lau GW. et al. Regulation of nuclear-cytoplasmic shuttling and function of Family with sequence similarity 13, member A (Fam13a) by B56-containing PP2As and Akt. Mol Biol Cell. 2015
13. Jiang Z, Lao T, Qiu W, Polverino F, Gupta K, Guo F. et al. A Chronic Obstructive Pulmonary Disease Susceptibility Gene, FAM13A, Regulates Protein Stability of beta-catenin. Am J Respir Crit Care Med. 2016
14. Brandsma CA, Timens W. The Translation from Risk Allele to Biological Function in Chronic Obstructive Pulmonary Disease. What's in It for FAM13A? Am J Respir Crit Care Med. 2016;194:130-2
15. Maffettone C, Chen G, Drozdov I, Ouzounis C, Pantopoulos K. Tumorigenic properties of iron regulatory protein 2 (IRP2) mediated by its specific 73-amino acids insert. PLoS One. 2010;5:e10163
16. Falvella FS, Galvan A, Frullanti E, Spinola M, Calabro E, Carbone A. et al. Transcription deregulation at the 15q25 locus in association with lung adenocarcinoma risk. Clin Cancer Res. 2009;15:1837-42
17. Liu Y, Liu P, Wen W, James MA, Wang Y, Bailey-Wilson JE. et al. Haplotype and cell proliferation analyses of candidate lung cancer susceptibility genes on chromosome 15q24-25.1. Cancer Res. 2009;69:7844-50
18. Li QF, Wang XR, Yang YW, Lin H. Hypoxia upregulates hypoxia inducible factor (HIF)-3alpha expression in lung epithelial cells: characterization and comparison with HIF-1alpha. Cell Res. 2006;16:548-58
19. Cavadas MA, Mesnieres M, Crifo B, Manresa MC, Selfridge AC, Scholz CC. et al. REST mediates resolution of HIF-dependent gene expression in prolonged hypoxia. Sci Rep. 2015;5:17851
20. Bordji K, Grandval A, Cuhna-Alves L, Lechapt-Zalcman E, Bernaudin M. Hypoxia-inducible factor-2alpha (HIF-2alpha), but not HIF-1alpha, is essential for hypoxic induction of class III beta-tubulin expression in human glioblastoma cells. FEBS J. 2014;281:5220-36
Author contact
![]() Corresponding author: Iwona Ziółkowska-Suchanek, PhD, Institute of Human Genetics, Polish Academy of Sciences, Department of Molecular Pathology, Strzeszyńska 32, 60-479 Poznań, Poland e-mail: iwona.ziolkowskapoznan.pl Tel: +48 61 6579281; Fax: +48 61 823323
Corresponding author: Iwona Ziółkowska-Suchanek, PhD, Institute of Human Genetics, Polish Academy of Sciences, Department of Molecular Pathology, Strzeszyńska 32, 60-479 Poznań, Poland e-mail: iwona.ziolkowskapoznan.pl Tel: +48 61 6579281; Fax: +48 61 823323

 Global reach, higher impact
Global reach, higher impact