3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(1):198-204. doi:10.7150/jca.22145 This issue Cite
Research Paper
KDM5B overexpression predicts a poor prognosis in patients with squamous cell carcinoma of the head and neck
1. Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, Xiangya Road 87, Changsha 410008, Hunan, China;
2. Otolaryngology Major Disease Research Key Laboratory of Hunan Province, Xiangya Road 87, Changsha 410008, Hunan, China.
Received 2017-7-29; Accepted 2017-10-18; Published 2018-1-1
Abstract
Purpose: Lysine demethylase (KDM) 5B, as a member of the histone lysine demethylase family, is overexpressed and functions abnormally in various human cancers. However, its expression in the squamous cell carcinoma of the head and neck (SCCHN) remains unclear.
Methods: KDM5B expression was analyzed by immunohistochemistry and correlated with clinicopathological parameters in 103 archival SCCHN tissue samples and 24 adjacent noncancerous epithelial tissues.
Results: We found that KDM5B expression was higher in SCCHN than that in adjacent noncancerous tissues. This was closely associated with lymph node metastasis and tumor recurrence. In addition, Kaplan-Meier analysis revealed that patients with high KDM5B expression had shorter disease-free and overall survival times than those with low KDM5B expression. Importantly, both univariate and multivariate analysis demonstrated that KDM5B level was an independent prognostic factor in SCCHN patients.
Conclusions: These results indicate that KDM5B is a valuable biomarker that can be used to predict SCCHN patient outcome.
Keywords: Head and neck cancer, Squamous cell carcinoma, KDM5B, Metastasis, Prognosis.
Introduction
Squamous cell carcinoma of the head and neck (SCCHN) is one of the most common malignancies worldwide with more than 600,000 cases diagnosed annually, representing a major cause of cancer-associated illness and death. Despite progress in early diagnosis and multimodal interventions, including surgery, chemotherapy, radiotherapy and immunotherapy, SCCHN patients still exhibit unsatisfactory clinical response and have poor prognosis, especially at advanced stages of the disease [1]. Besides tobacco use, alcohol consumption, and human papillomavirus (HPV) infection [2, 3], local and distant lymph node metastases also contribute to the development and progression of SCCHN patients. In particular, patients with ipsilateral lymph node metastasis and/or distant metastasis have relatively low cure rates. Therefore, it is important to clarify the molecular changes associated with metastasis in order to develop more effective treatments and reduce mortality in SCCHN patients.
Epigenetic alterations such as DNA methylation, histone modification and chromatin remodeling have been implicated in the initiation and progression of cancers including SCCHN [4, 5]. Histone methylation, as one kind of histone modification, affects gene expression by directly altering chromatin structure and/or by promoting the recruitment of effector proteins [6]. Trimethylated lysine 4 on histone H3 (H3K4me3), an epigenetic marker, shows abnormal expression and function in diverse solid tumors [7]. H3K4me3 level is dynamically modulated by several histone methyltransferases and histone demethylases [8].
Lysine demethylase (KDM) 5B, also known as PLU-1 and JARID1B, functions as a transcriptional repressor by removing H3K4 di- and tri-methylation marks from target genes [9]. H3K4me3 removal by KDM5B is required for normal zygotic genome activation and early embryo development [10]. Additionally, KDM5B has diverse, context-specific roles in the epigenetic regulation of cell phenotype during normal development and carcinogenesis [11]. Increasing reports have demonstrated that KDM5B is overexpressed in multiple human cancers, which is associated with malignant behaviors such as unlimited proliferation, invasion, metastasis, and treatment resistance, suggesting that KDM5B can serve as a potential therapeutic target in human malignancies [9]. However, the expression and function of KDM5B in SCCHN remains unclear, particularly with respect to disease progression and patient outcome. To address this issue, the present study investigated KDM5B expression and its relationship to clinical parameters by analyzing a large set of archival SCCHN tissue specimens.
Materials and Methods
Patient information and tissue preparation
Archival paraffin-embedded SCCHN tissue samples from 103 patients were analyzed in this retrospective study, in which 24 adjacent noncancerous epithelial tissues were included [12, 13]. Samples were collected from January 2002 to October 2004 from patients who had no history of previous malignancies, or radio- or chemotherapy. Recurrence and metastasis were established based on physical examinations, imaging results, operation, and post-operative pathological examination. Informed consent was obtained from all patients before surgery. Pathological tumor-node-metastasis (TNM) stage was determined according to the 7th American Joint Committee on Cancer staging system. Patient characteristics such as sex, age, smoking history, alcohol consumption, tumor site, histological grade, tumor size, clinical stage, and lymph node metastasis were recorded (Table 1). The study was approved by the Research Ethics Committee of Central South University, Changsha, China.
Correlations between KDM5B expression and clinicopathological parameters
| KDM5B expression | |||||
|---|---|---|---|---|---|
| Parameters | No. of patients | Low | High | χ2 value | P-value |
| Age | |||||
| <58 | 47 | 29 | 18 | 0.220 | 0.690 |
| ≥58 | 56 | 32 | 24 | ||
| Sex | |||||
| Female | 4 | 2 | 2 | 0.147 | 1.000 |
| Male | 99 | 59 | 40 | ||
| Smoking | |||||
| Yes | 54 | 29 | 25 | 1.432 | 0.316 |
| No | 49 | 32 | 17 | ||
| Alcohol consumption | |||||
| Yes | 55 | 33 | 22 | 0.029 | 1.000 |
| No | 48 | 28 | 20 | ||
| Histological grade& | |||||
| G1 + G2 | 37 | 19 | 18 | 1.482 | 0.296 |
| G3 | 66 | 42 | 24 | ||
| Primary tumor sites | |||||
| Glottic | 63 | 41 | 22 | 2.304 | 0.153 |
| Others | 40 | 20 | 20 | ||
| T classifications | |||||
| T1 + T2 | 52 | 33 | 19 | 0.781 | 0.426 |
| T3 + T4 | 51 | 28 | 23 | ||
| Clinical stages | |||||
| I + II | 41 | 29 | 12 | 3.736 | 0.066 |
| III + IV | 62 | 32 | 30 | ||
| Lymph node metastasis | |||||
| N0 | 64 | 45 | 19 | 8.607 | 0.003 |
| N+ | 39 | 16 | 23 | ||
| Recurrence† | |||||
| Yes | 59 | 30 | 29 | 7.160 | 0.011 |
| No | 40 | 31 | 9 | ||
NOTE: & G1, G2 and G3 were defined as poorly-differentiated, moderately-differentiated and well-differentiated, respectively.
†. Four patients were lost to follow-up due to changes in address or telephone number.
Immunohistochemistry
Immunohistochemistry was performed as reported in our previous studies [12-14], using the PV-6001 Two-Step IHC Detection Reagent (ZhongShan Goldenbridge Bio, Beijing, China). Briefly, antigen retrieval was carried out for slides harboring tissue sections in 10 mmol/l citrate buffer (pH 6.0) for 15 min at 100°C in a microwave oven. Endogenous peroxidase activity was blocked by incubation in 3% hydrogen peroxide for 10 min at room temperature. The slides were then incubated overnight at 4℃ with mouse monoclonal anti- KDM5B antibody at 1:600 dilution (MA5-24626; Invitrogen, Carlsbad, CA, USA), followed by horseradish peroxidase-labeled goat anti-rabbit IgG. Immunoreactivity was visualized with 3′, 3′-diaminobenzidine and samples were counterstained with Mayer's hematoxylin. Negative control slides were prepared by applying normal goat serum instead of the primary antibody under the same experimental conditions.
Evaluation of staining
Sections were evaluated and scored by two independent investigators, who were blinded to the clinicopathological parameters. Nuclear staining was considered as positive for KDM5B expression and the entire tissue section was examined to determine the final score. Staining intensity was scored as 0 (negative), 1 (weak), 2 (medium) and 3 (strong). Degree of staining was stratified as 0 (0%), 1 (1%-25%), 2 (26%-50%), 3 (51%- 75%) and 4 (76%-100%), and was defined as the percentage of the positively stained area relative to the total area of tumor infiltration. The final staining score (ranges: 0-7) for KDM5B was determined as the sum of staining intensity and degree. Tumors were categorized into two groups: high KDM5B expression (scored as 5-7) and low KDM5B expression (scored as 0-4) based on the final staining score [12].
Follow-up
Follow-up materials after surgery were obtained from 99 patients (96.1%); four patients were lost to follow-up due to changes in address or telephone number. Recurrence and metastasis were determined by clinical and pathologic examination and imaging. Overall and disease-free survival were calculated from the day of surgery to the date of death or tumor relapse. Death from other causes was treated as censored cases. The follow-up time ranged from 2 to 60 months, with a median follow-up time of 45.97 months (SD = 19.435).
The Cancer Genome Atlas (TCGA) data analysis
Publicly available human SCCHN datasets TCGA were analyzed. The mRNA expression profiles (Illumina Hiseq RNAseq V2, RSEM normalized) downloaded from the TCGA portal (https://cancergenome.nih.gov/) included 44 normal and 512 SCCHN tissue samples.
Statistical analysis
Continuous variables are expressed as mean ± SD. Positive rates of KDM5B expression in SCCHN tissues and adjacent noncancerous tissues were compared by χ2 test, which was also used to analyze the difference between KDM5B protein levels and clinicopathological parameters. Survival analysis was performed with the Kaplan-Meier method and survival curves were compared using the log-rank test. Cox's proportional hazards model was applied in univariate and multivariate analyses to identify factors that were independent indicators of prognosis. Statistical analyses were performed using SPSS v.17.0 software (SPSS Inc., Chicago, IL, USA). P < 0.05 was considered statistically significant.
Results
KDM5B expression is elevated in SCCHN patients
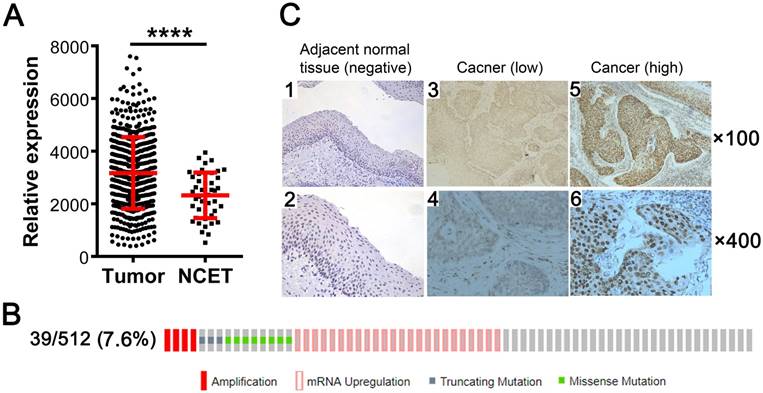
We investigated the expression status of KDM5B in SCCHN by analyzing TCGA RNA-Seq data and found that KDM5B mRNA level was higher in the 512 SCCHN tissue samples than in the 44 adjacent non-cancerous epithelial tissue samples (Fig.1A). Genetic alterations in KDM5B including gene amplification, mRNA upregulation, and truncating mutation and missense mutations, were detected in 39/512 (7.6%) of SCCHN patients (Fig.1B). To confirm the expression of KDM5B protein, immunohistochemistry was performed for 103 paraffin-embedded, archival primary SCCHN tumor samples. KDM5B immunoreactivity was mostly distributed in the nuclei of carcinoma and non-cancerous epithelial cells (Fig.1C). A total of 80 (77.7%) primary tumor samples were positive for KDM5B expression, including 42 patients with high KDM5B expression and 61 patients with low expression, respectively. However, KDM5B expression was only positive in 9 (9/24 = 37.5%) cases of non-cancerous adjacent tissues (P < 0.01). These results indicate that KDM5B is overexpressed in SCCHN.
KDM5B expression is positively associated with SCCHN progression
We investigated the associations between KDM5B expression and diverse clinicopathological parameters. As summarized in Table 1, KDM5B overexpression was significantly associated with cervical lymph node metastasis (P = 0.003) and recurrence (P = 0.011), respectively. However, KDM5B expression did not differ between groups classified by age (P = 0.690), sex (P = 1.000), alcohol history (P = 1.000), smoking (P = 0.316), tumor grade (P = 0.296), tumor site (P = 0.153), tumor T classification (P = 0.426), and advanced clinical stage (P = 0.066). These results indicate that KDM5B expression is closely correlated with SCCHN progression.
KDM5B expression is negatively associated with disease-free and overall survival
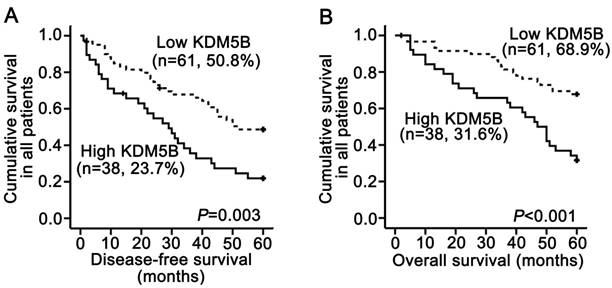
A survival analysis was performed using data from 99 patients for whom follow-up information was available. The Kaplan-Meier analysis revealed that KDM5B overexpression in SCCHN patients was significantly correlated with shorter disease-free survival time (P = 0.003; Fig.2A) and shorter overall survival time (P < 0.001; Fig.2B). The log-rank test confirmed that survival time differed between patients with high and low KDM5B expression. Univariate Cox regression analyses determined that cancer site, clinical stages, T classifications, metastasis and KDM5B expression were significantly associated with both disease-free survival and overall survival (All P < 0.05). Additionally, smoking history was also correlated with disease-free survival (P = 0.028, Table 2). However, metastasis was only determined to be an independent prognostic factor for overall survival (P = 0.049), and KDM5B expression was an independent prognostic factor for post-surgery outcome (All P < 0.05; Table 2).
Stratified survival analysis
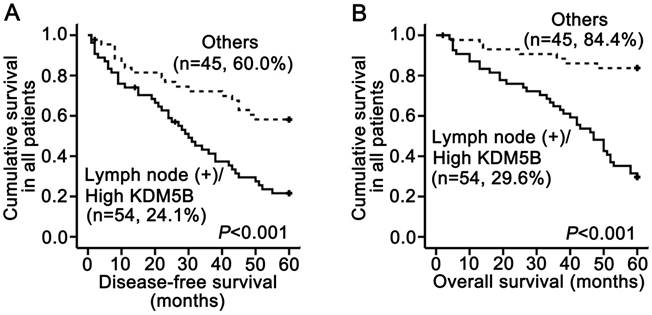
In our previous study using the same SCCHN tissue samples, we demonstrated that lymph node metastasis was significantly associated with shorter disease-free and overall survival and was an independent prognostic factor for reduced overall survival by multivariate analysis [12, 14]. We therefore carried out a subset analysis that combined KDM5B expression and lymph node status. The results demonstrated that patients with a high KDM5B expression/lymph node (+) phenotype had shorter disease-free and overall survival than other patients (P < 0.001 and P < 0.001, respectively; Fig.3A-B). These results demonstrate that the combination of KDM5B expression and lymph node metastasis can more effectively predict SCCHN patient prognosis.
KDM5B is overexpressed in SCCHN. (A) KDM5B mRNA levels in SCCHN and corresponding non-cancerous adjacent tissue samples from TCGA dataset were compared. NCET, noncarcinoma epithelial tissues. (B) Frequencies of KDM5B alterations—including amplification, mRNA upregulation, truncation and missense mutations—from SCCHN TCGA database were analyzed in cBioportal. (C) Representative images of KDM5B protein expression in SCCHN and corresponding adjacent non-cancerous epithelial tissues, as determined by immunohistochemistry. Original magnification: 100× (upper) and 400× (lower).
KDM5B expression is correlated with poor clinical outcome in SCCHN patients. (A, B) Kaplan-Meier analysis and log-rank test for disease-free survival (A) and overall survival (B) in SCCHN patients with differential expression of KDM5B.
Cox model analysis of disease-free survival and overall survival
| Disease-free survival | Overall survival | |||
|---|---|---|---|---|
| Parameters | Relative risk (95%CI) | P-value | Relative risk (95%CI) | P-value |
| Univariate | ||||
| Age | 0.917 (0.550-1.530) | 0.741 | 0.879 (0.490-1.577) | 0.665 |
| Gender | 1.262 (0.395-4.038) | 0.695 | 1.791 (0.554-5.791) | 0.331 |
| Smoking | 1.810 (1.067-3.071) | 0.028 | 1.388 (0.764-2.521) | 0.281 |
| Alcohol consumption | 0.878 (0.526-1.463) | 0.617 | 1.285 (0.708-2.334) | 0.409 |
| Histological grades | 1.007 (0.591-1.718) | 0.978 | 1.093 (0.594-2.014) | 0.775 |
| Primary tumor sites | 0.337 (0.225-0.632) | <0.001 | 0.270 (0.147-0.496) | <0.001 |
| T classifications | 2.021 (1.193-3.423) | 0.009 | 3.175 (1.661-6.068) | <0.001 |
| Clinical stages | 3.295 (1.797-6.041) | <0.001 | 7.661 (3.011-19.492) | <0.001 |
| Lymph node metastasis | 3.175 (1.875-5.375) | <0.001 | 7.184 (3.652-14.133) | <0.001 |
| KDM5B expression | 2.155 (1.298-3.604) | 0.003 | 2.787 (1.539-5.049) | 0.001 |
| Multivariate | ||||
| KDM5B expression | 1.956 (1.117-3.424) | 0.019 | 2.089 (1.079-4.045) | 0.029 |
| Lymph node metastasis | 1.111 (0.477-2.589) | 0.807 | 2.798 (1.004-7.794) | 0.049 |
NOTE: All the clinicopathological variables listed in the table were included in the univariate and multivariate analyses.
Abbreviation: 95% CI, 95% confidence interval.
KDM5B expression is inversely related to SCCHN patient survival. (A, B) Stratified analysis of disease-free survival (A) and overall survival (B) according to the combination of lymph node metastasis and KDM5B expression.
Discussion
The results of our study demonstrate that KDM5B expression was increased in SCCHN tissues compared to that in adjacent noncancerous tissues, and was associated with progression parameters including lymph node metastasis and recurrence in patients with SCCHN. Importantly, high levels of KDM5B protein were predictive of poor prognosis, and could be used as an independent prognostic parameter, especially when combined with lymph node metastasis status.
We found here that KDM5B overexpression was indicative of SCCHN progression. KDM5B functions as a demethylase to maintain the balance of H3K4me3 [15, 16]; its aberrant expression has been reported in a variety of human malignancies including breast [17], bladder [18], liver [19, 20], lung [21], prostate [22], ovarian [23], oral [24] and cervical [25] cancers. Our data are consistent with previous reports that high levels of KDM5B are linked to cancer aggressive parameters including metastasis and recurrence [17, 20, 24]. Thus, KDM5B may have critical roles in cancer initiation and progression.
Loss-of-function experiments have validated that KDM5B inhibition suppresses the growth of breast cancer cells in vitro and colony formation in soft agar, as well as mammary tumor formation in vivo [26, 27]. KDM5B overexpression is required for the stem cell-like phenotype in oral squamous cancer cells [24] and neuroblastoma [28], and confers resistance to chemo- and radiotherapy. These findings suggest a pro-oncogenic role for KDM5B in human cancers.
The results of our study indicate that KDM5B is associated with metastasis in patients with SCCHN. Epithelial-mesenchymal transition (EMT) has been extensively reported to initiate and enhance cancer metastasis [29]. KDM5B can inhibit the expression of microRNAs in the miR-200 family by altering their histone H3 methylation status, and then increase the expression of transcription factor zinc finger E-box-binding homeobox (ZEB)1 and ZEB2 to induce EMT and metastasis in lung cancer [30]. In hepatocellular carcinoma, KDM5B promotes metastasis and EMT via phosphatase and tensin homolog (PTEN)/AKT signaling [19]. On the contrary, KDM5B repressed the expression of chemokine (C-C motif) ligand 14 (CCL14), an epithelium-derived chemokine, to reduce the angiogenic and metastatic potential of breast cancer cells in vivo [31]. These contradictory functions suggest that KDM5B has complex roles in metastasis and that the final phenotype is determined by downstream genes that are epigenetically regulated by KDM5B. Therefore, the exact molecular mechanisms regarding metastasis regulation are urgent for detailed investigations in future.
The major finding of this study is the validation of KDM5B as a prognostic marker in SCCHN patients. Currently, the standard management of SCCHN patients is based on anatomic considerations, TNM stage, and histopathologic grade [1]. However, recent studies have suggested that other factors such as molecular and cellular characteristics of primary tumors can improve our ability to predict patient outcome [12, 32]. In the present study, KDM5B expression was inversely correlated with overall survival and disease-free survival, which is consistent with findings in breast [17] and hepatocellular carcinoma [20]. In particular, patients with a high KDM5B expression/lymph node metastasis (+) phenotype had shorter disease-free and overall survival times. Therefore, evaluating KDM5B protein expression in combination with other clinical variables such as lymph node metastasis may provide a more accurate measure of SCCHN patient prognosis.
In conclusion, our study indicates that KDM5B is increased in human SCCHN and that KDM5B overexpression is closely correlated with tumor malignant progression and poor survival in SCCHN patients, suggesting KDM5B can serve as a specific and novel prognostic protein in SCCHN.
Acknowledgements
This study was funded by grants from the National Natural Science Foundation of China (grant numbers 81772903, 81773243, 81202128, 81602684, 81372426, 81602389 and 81472696), and the Natural Science Foundation of Hunan Province (grant number 2015JJ3137).
Abbreviations
EMT, epithelial-mesenchymal transition; H3K4me3, trimethylated lysine 4 on histone H3; KDM5B, lysine demethylase; SCCHN, squamous cell carcinoma of the head and neck; TCGA, The Cancer Genome Atlas; TNM, tumor-node-metastasis
Competing Interests
The authors have declared that no competing interest exists.
References
1. Cohen EE, LaMonte SJ, Erb NL, Beckman KL, Sadeghi N, Hutcheson KA. et al. American Cancer Society Head and Neck Cancer Survivorship Care Guideline. CA Cancer J Clin. 2016;66:203-39
2. Blot WJ, McLaughlin JK, Winn DM, Austin DF, Greenberg RS, Preston-Martin S. et al. Smoking and drinking in relation to oral and pharyngeal cancer. Cancer Res. 1988;48:3282-7
3. Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E. et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294-301
4. Ning B, Li W, Zhao W, Wang R. Targeting epigenetic regulations in cancer. Acta Biochim Biophys Sin (Shanghai). 2016;48:97-109
5. D'Souza W, Saranath D. Clinical implications of epigenetic regulation in oral cancer. Oral Oncol. 2015;51:1061-8
6. Tran TQ, Lowman XH, Kong M. Molecular Pathways: Metabolic control of histone methylation and gene expression in cancer. Clin Cancer Res. 2017
7. Spangle JM, Dreijerink KM, Groner AC, Cheng H, Ohlson CE, Reyes J. et al. PI3K/AKT Signaling Regulates H3K4 Methylation in Breast Cancer. Cell Rep. 2016;15:2692-704
8. Vallianatos CN, Iwase S. Disrupted intricacy of histone H3K4 methylation in neurodevelopmental disorders. Epigenomics. 2015;7:503-19
9. Han M, Xu W, Cheng P, Jin H, Wang X. Histone demethylase lysine demethylase 5B in development and cancer. Oncotarget. 2017;8:8980-91
10. Dahl JA, Jung I, Aanes H, Greggains GD, Manaf A, Lerdrup M. et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature. 2016;537:548-52
11. Facompre ND, Harmeyer KH, Basu D. Regulation of oncogenic PI3-kinase signaling by JARID1B. Oncotarget. 2017;8:7218-9
12. Liu Y, Xie C, Zhang X, Huang D, Zhou X, Tan P. et al. Elevated expression of HMGB1 in squamous-cell carcinoma of the head and neck and its clinical significance. Eur J Cancer. 2010;46:3007-15
13. Liu Y, Zhang X, Qiu Y, Huang D, Zhang S, Xie L. et al. Clinical significance of EphA2 expression in squamous-cell carcinoma of the head and neck. J Cancer Res Clin Oncol. 2011;137:761-9
14. Liu Y, Su Z, Li G, Yu C, Ren S, Huang D. et al. Increased expression of metadherin protein predicts worse disease-free and overall survival in laryngeal squamous cell carcinoma. Int J Cancer. 2013;133:671-9
15. He R, Kidder BL. H3K4 demethylase KDM5B regulates global dynamics of transcription elongation and alternative splicing in embryonic stem cells. Nucleic Acids Res. 2017;45:6427-41
16. Li X, Liu L, Yang S, Song N, Zhou X, Gao J. et al. Histone demethylase KDM5B is a key regulator of genome stability. Proc Natl Acad Sci U S A. 2014;111:7096-101
17. Bamodu OA, Huang WC, Lee WH, Wu A, Wang LS, Hsiao M. et al. Aberrant KDM5B expression promotes aggressive breast cancer through MALAT1 overexpression and downregulation of hsa-miR-448. BMC Cancer. 2016;16:160
18. Li X, Su Y, Pan J, Zhou Z, Song B, Xiong E. et al. Connexin 26 is down-regulated by KDM5B in the progression of bladder cancer. Int J Mol Sci. 2013;14:7866-79
19. Tang B, Qi G, Tang F, Yuan S, Wang Z, Liang X. et al. JARID1B promotes metastasis and epithelial-mesenchymal transition via PTEN/AKT signaling in hepatocellular carcinoma cells. Oncotarget. 2015;6:12723-39
20. Wang D, Han S, Peng R, Jiao C, Wang X, Yang X. et al. Depletion of histone demethylase KDM5B inhibits cell proliferation of hepatocellular carcinoma by regulation of cell cycle checkpoint proteins p15 and p27. J Exp Clin Cancer Res. 2016;35:37
21. Shen X, Zhuang Z, Zhang Y, Chen Z, Shen L, Pu W. et al. JARID1B modulates lung cancer cell proliferation and invasion by regulating p53 expression. Tumour Biol. 2015;36:7133-42
22. Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z. et al. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc Natl Acad Sci U S A. 2007;104:19226-31
23. Wang L, Mao Y, Du G, He C, Han S. Overexpression of JARID1B is associated with poor prognosis and chemotherapy resistance in epithelial ovarian cancer. Tumour Biol. 2015;36:2465-72
24. Lin CS, Lin YC, Adebayo BO, Wu A, Chen JH, Peng YJ. et al. Silencing JARID1B suppresses oncogenicity, stemness and increases radiation sensitivity in human oral carcinoma. Cancer Lett. 2015;368:36-45
25. Zhou Y, An Q, Guo RX, Qiao YH, Li LX, Zhang XY. et al. miR424-5p functions as an anti-oncogene in cervical cancer cell growth by targeting KDM5B via the Notch signaling pathway. Life Sci. 2017;171:9-15
26. Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H. et al. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25:801-12
27. Catchpole S, Spencer-Dene B, Hall D, Santangelo S, Rosewell I, Guenatri M. et al. PLU-1/JARID1B/KDM5B is required for embryonic survival and contributes to cell proliferation in the mammary gland and in ER+ breast cancer cells. Int J Oncol. 2011;38:1267-77
28. Kuo YT, Liu YL, Adebayo BO, Shih PH, Lee WH, Wang LS. et al. JARID1B Expression Plays a Critical Role in Chemoresistance and Stem Cell-Like Phenotype of Neuroblastoma Cells. PLoS One. 2015;10:e0125343
29. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871-90
30. Enkhbaatar Z, Terashima M, Oktyabri D, Tange S, Ishimura A, Yano S. et al. KDM5B histone demethylase controls epithelial-mesenchymal transition of cancer cells by regulating the expression of the microRNA-200 family. Cell Cycle. 2013;12:2100-12
31. Li Q, Shi L, Gui B, Yu W, Wang J, Zhang D. et al. Binding of the JmjC demethylase JARID1B to LSD1/NuRD suppresses angiogenesis and metastasis in breast cancer cells by repressing chemokine CCL14. Cancer Res. 2011;71:6899-908
32. Akbani R, Ng PK, Werner HM, Shahmoradgoli M, Zhang F, Ju Z. et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat Commun. 2014;5:3887
Author contact
![]() Corresponding author: Yong Liu, Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, Xiangya Road 87, Changsha 410008, Hunan, China. Tel: +86 731 84327469; fax: +86 731 88710883; e-mail: liuyongentedu.cn
Corresponding author: Yong Liu, Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, Xiangya Road 87, Changsha 410008, Hunan, China. Tel: +86 731 84327469; fax: +86 731 88710883; e-mail: liuyongentedu.cn

 Global reach, higher impact
Global reach, higher impact