3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(3):450-459. doi:10.7150/jca.23151 This issue Cite
Research Paper
The Unknown microRNA Expression of Male Breast Cancer. Similarities and Differences with Female Ductal Carcinoma. Their Role as Tumor Biomarker
1. Translational Surgical Pathology Section, Laboratory of Pathology. Center for Cancer Research, National Cancer Institute, National Institutes of Health.
2. Cancer Institute, Salamanca University, Spain.
3. University Clinic Hospital, Madrid.
Received 2017-10-3; Accepted 2017-10-27; Published 2018-1-1
Abstract
Mature microRNAs (miRNAs) are small non-protein coding RNAs that modulate gene expression after transcription. Few studies have shown that male breast cancer (MBC) shows distinctive miRNAs pattern, suggesting its relevance in this pathology. To study this, we performed a profile of 800 miRNAs in 9 MBC samples and in normal epithelial cells of 3 MBC cases.
Experimental Design: Of FFPE tissues, miRNA was extracted for profiles using the NanoString method. miRNAs were obtained by comparing tumor samples versus normal epithelium. Quantitative real-time PCR analyzes were performed by the TaqMan approach for specific miRNAs.
Results: The profile of 800 miRNAs showed a different microRNA expression pattern between MBC and its normal counterpart, suggesting a specific microRNA cancer expression profile for MBC. Forty-nine miRNAs showed greater expression, while 26 were found to be down-regulated in MBC, compared to normal tissue. The lower expression of miR-125b correlated significantly with tumors> 2 cm, suggesting that its down-regulation may be implicated in mechanisms to more aggressive tumors.
Conclusions: These results suggest that MBC has a unique expression profile compared to normal breast tissue and expression profile of female breast cancer. Differentially expressed miRNAs provide insights of this uncommon but highly aggressive pathology.
Keywords: microRNAs, male breast cancer
Introduction
Male breast cancer (MBC) represents 1% of all breast cancer cases in the United States, with 2,240 new cases and 410 deaths expected by 2013 [1, 2]. Analysis of the Surveillance, Epidemiology and End Results (SEER) data from 1973-2008 has shown the overall incidence of MBC is 1.5 cases per 100,000 males [3] with White males being more frequently affected than other ethnic groups. MBC can be hereditary or sporadic. Genetic risk factors for MBC include BRCA2 carriers [4] and patients with Cowden and Klinefelter syndromes. Recently, the MBC Pooling Project from the National Cancer Institute has shown that there may be a significant risk for MBC associated with increased body mass index and gynecomastia after combination of data from case-control and large cohort studies [5].
MBC presents at a later onset compared to breast cancer in women (65-69 vs. 60-64 years), aspect that contributes to its unfavorable overall prognosis, often being discovered in advanced stages with lymph node metastases [6, 7]. In the SEER analysis, 12% of men were diagnosed with breast carcinoma before the age of 50 whereas 40% were diagnosed after age 70 [3].
Clinically, MBC presents as a palpable mass with or without nipple involvement, manifested as retraction, fixation, bloody discharge or erythematous scaling. MBC shares some morphological characteristics with female breast cancer (FBC). The most common histological type is ductal carcinoma whereas lobular carcinoma is extremely rare. Papillary carcinomas are more common than in its female counterpart [8]. MBC is characteristically positive for estrogen and progesterone receptors and negative for Her2. Differential diagnosis includes gynecomastia and metastatic carcinoma or melanoma [8].
At the molecular level, MBC is a heterogeneous disease that differs from FBC. By gene expression profiling Johansson et al. [9] have identified two different subgroups of MBC -designated luminal M1 and luminal M2- which appear to occur only in males and have a unique biology. Although both subgroups show expression of estrogen receptor, luminal M1 tumors are associated with aggressive behavior and worse prognosis, whereas luminal M2 tumors may behave less aggressive and contain high levels of immune response related genes and genes associated with estrogen receptor signaling [9].
However, because of the rareness of MBC and the lack of large series studies, the current knowledge about its biology and treatment is still limited and based on what is known about its female presentation. The majority of patients diagnosed with loco-regional MBC undergo total mastectomy. However, breast conservative surgery seems to have equivalent outcomes and less secondary effects than radical surgery in this set of patients [3, 10].
Mature microRNAs (miRNAs) are small (approximately 18-25 nucleotides long) non-protein coding RNAs. miRNAs modulate gene expression by acting post-transcriptionally, through either messenger RNA degradation or translation inhibition [11]. Since the discovery of miRNAs [12], several studies have demonstrated their involvement in carcinogenesis by promoting expression of proto-oncogenes or by inhibiting the expression of tumor suppressor genes [13]. miRNA expression profile platforms have been especially used to characterize several types of cancers, helping to understand tumor development and progression [14, 15]. Numerous studies show that different solid tumors in fact display a cancer type-specific deregulation of miRNAs [16-18]. One of the first studies demonstrating miRNA deregulation through a comparison of cancerous and normal mammary tissues by miRNA microarray identified a panel of 29 miRNAs modulated specifically in breast cancer samples from female patients [16]. However, only few studies have been reported regarding miRNA expression profiling in MBC [13, 19, 20, 21], all showing different miRNA signatures associated with breast tumors in males; therefore, the current knowledge about the involvement of miRNAs in MBC development is scarce and not conclusive.
To further elucidate the possible differences between MBC and previously reported FBC miRNA profiles, we performed a comprehensive profiling of 800 miRNAs in 9 MBC samples and in normal epithelial cells from 3 of the same MBC cases.
Material and Methods
Patient Samples
Forty-two cases of MBC were retrieved from the surgical archives of the Laboratory of Pathology, National Cancer Institute, Bethesda, MD, USA after IRB approval. Hematoxylin and eosin (H&E) stained slides were reviewed to confirm the diagnosis and their clinicopathological features were reviewed, including tumor size, lymph node metastasis and status of estrogen receptor (ER), progesterone receptor (PR), androgen receptor (AR), Her-2 and Ki-67. For the profiling study of 800 microRNAs, 9 tumor and 3 normal epithelium samples were available. For a detailed description of all MBC cases included in this study see Table 1.
RNA Isolation
Isolation of total RNA from formalin-fixed paraffin embedded (FFPE) specimens was performed as described previously Further support the hypothesis that MBC have a different microRNA expression profile in comparison with normal mammary tissue and with that reported for FBC[22]. Briefly, ten micron formalin-fixed paraffin-embedded (FFPE) tissue sections were deparaffinized in three consecutive Xylene baths, rehydrated in graded ethanol and manually microdissected under light microscope to obtain separate amounts of tumor and normal breast tissue. RNA isolation was performed using the RecoverAll™ Total Nucleic Acid Isolations Kit (Ambion® by Life Technoligies, Foster City, CA, USA). The concentration of all RNA samples was quantified using NanoDrop 2000 (Thermo Scientific, Wilmington, DE, USA). RNA concentration of samples used for profiling was normalized at 33 ng/µl following the recommendations from NanoString Technologies.
MicroRNA profiling using NanoString nCounter miRNA assay
Total RNA samples were analyzed according to manufacturer's instructions for the nCounter Human miRNA Expression Assay kit (NanoString Technologies®, Seattle, WA). Briefly, 100 ng of each total RNA sample was used as input into the nCounter Human miRNA sample preparation. Hybridization was conducted for 16 h at 65°C. Subsequently, probes were purified and counted on the nCounter Prep Station. Each sample was scanned for 600 FOV (fields of view) on the nCounter Digital Analyzer. Data was extracted using the nCounter RCC Collector.
NanoString nCounter miRNA data analysis
For platform validation using synthetic oligonucleotides, NanoString nCounter miRNA raw data was normalized for lane-to-lane variation with a dilution series of six spike-in positive controls. The sum of the six positive controls for a given lane was divided by the average sum across lanes to yield a normalization factor, which was then multiplied by the raw counts in each lane to give normalized values. For each sample, the mean plus 2 times the standard deviation of the 8 negative controls was subtracted from each miRNA count in that sample. Only miRNAs with non-negative counts across all samples were retained for downstream analysis. The relative miRNA levels were indicated as median fold changes and a cutoff of 2 fold-change (up or down) was used.
Quantitative real-time PCR
Detection and quantification of selected mature miRNAs was carried out using Reverse transcription quantitative real-time PCR (RT-qPCR) and the TaqMan miRNA Assay (Assay ID: 000387 Applied Biosystems, Foster City, CA) according to the manufacturer's protocol. RNA samples were measured in triplicates. Preparation of cDNA was performed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). Reverse transcription reactions were performed in a GeneAmp PCR system 9700 thermal cycler (Applied Biosystems) with samples incubated at 16°C for 30 minutes, 42°C for 30 minutes and 85°C for 5 minutes. An RT-negative control was included in each batch of reactions. Each reaction consisted of 15 μl final volume, with 10 ng of total RNA including miRNAs. The qPCR reaction mix consisted of 20 μl final volume with RT product, TaqMan 2X Universal PCR Master Mix II and the appropriate 20X MicroRNA Assay Mix containing the probe for the miRNA of interest. PCR reactions were initially incubated for 10 minutes at 95°C, followed by 40 cycles of 95°C for 15 seconds and 60°C for 60 seconds. Inter-assay control and calibrator were included in each 96-well plate. All TaqMan assays (See Table 2) were run in triplicate using an AB StepOnePlus RealTime PCR System. Ct values were calculated with the StepOne Software v2.2.2 using automatic baseline settings. RNU6B (assay ID: 001093) was used as endogenous control for normalizing the expression level of selected miRNAs. The mean of Ct values was subtracted from the corresponding Ct value for the selected miRNAs resulting in the ΔCt value which was used for relative quantification of miRNA expression (ΔΔCt method). Changes is expression levels in tumor samples are shown as relative (fold change) to normal tissue.
Statistical Analysis
Welch based t-tests were carried out using MultiExperiment Viewer software (MeV version 4.8, http://www.tm4.org) to compare the two groups (normal vs. tumor) and to evaluate differences between miRNA expression and patient characteristics. Only mature miRNAs that showed at least a 2-fold change in expression are reported. Regarding RT-qPCR data, results are presented as the mean values and standard error of the mean. P-values were calculated for each miRNA between the normal and tumor samples using the biological replicates and genes were considered statistically significant if their P value was less than 0.05.
Results
Characterization of the studied population
The clinicopathologic characteristics of the 42 cases included in the present study are shown in Table 1. Patient's age range was 36-85 (mean 63 ± 12.9). Majority of the tumors were moderate to high grade and ranged between 0.2 and 7 cm in size. Histologically, 76% of the tumors were invasive ductal carcinomas (four papillary type); eight cases were DCIS, 4 of which were papillary; and 1 was mucinous. Thirty-six percent were grade 2 and 26.2% grade 3. Lymph node status was known in 71% of the cases, of these, 70% had lymph node metastasis. Prognostic factors were known in 28 cases, 23 (82%) were ER+/PR+, 2 (7.1%) ER+/PR-, 1 (3.5%) ER-/PR+, Her2 amplification was seen in 6 cases (21%) and 2 cases were triple negative (7.1%). AR was heterogeneous and was strongly + in DCIS.
Human male breast cancer tissue presents a different MicroRNA expression profile compare to its normal counterpart and to female breast cancer
To determine if MBC possesses a tumor-specific pattern of miRNAs expression, a panel of 800 miRNAs was analyzed in 9 MBC samples and compared against the expression of the same miRNAs on 3 normal epithelial components obtained from the same group of patients.
A T-test based analysis was performed comparing the normal epithelium group versus the tumor group within MBC samples. Unsupervised hierarchical clustering of the miRNA expression data showed clear separation between normal and tumor mammary tissue, indicating a clear cancer-specific expression patter of a number of miRNAs in our MBC samples (Figure 1). In total, 49 miRNAs were up-regulated ranging from 2.05 fold up 6.57 fold change while 25 miRNAs showed to be down-regulated ranging from -2.08 fold to -18.86 fold change (Table 2). Some of the miRNA genes showed no statistically significant difference, which not necessarily means that the difference is not biologically significant. It is possible that these results respond to the fact that a small sample was used or the data were quite variable.
Among the up-regulated miRNAs, hsa-miR-200a-3p, hsa-miR-30a-5p, hsa-miR-29b-3p, hsa-miR-222-3p, hsa-miR-141-3p, hsa-miR-342-3p, hsa-miR-135b-5p, hsa-miR-29c-3p and hsa-miR-183-5p were the most prominently up-regulated (p<0.05). In particular, hsa-miR-200a-3p and hsa-miR-183-5p have been reported as over-expressed in MBC[19] as well as in its female counterpart[23, 24] while hsa-miR-30a-5p, hsa-miR-29b-3p, hsa-miR-222-3p, hsa-miR-141-3p, hsa-miR-342-3p, hsa-miR-135b-5p and hsa-miR-29c-3p have no reports to the present in MBC. On the other hand, hsa-miR-125b-5p, hsa-miR-4516, hsa-miR-143-3p, hsa-miR-214-3p, hsa-miR-575, hsa-miR-548ai, hsa-miR-203, hsa-miR-630, hsa-miR-145-5p and hsa-miR-1253 were the most down-regulated. From these, hsa-miR-145-5p has been reported as significantly down-regulated in both male and female breast cancer [19, 25].
The miRNA profile found in our study showed some differences from previously published miRNA profiles in FBC (Table 2). From the 74 miRNAs found to be deregulated in MBC, 37 (50%) have shown the same expression pattern in FBC and another 30% (22 miRNAs), to our knowledge, have no reports in this regard in FMC. On the other hand, and important 20% (15 miRNAs) showed an opposite expression pattern between male and female breast tumors. For instance, expression of hsa-miR-296-5p have shown to be down-regulated in FBC[26], while in the present study showed to be up-regulated more than two fold in the male tumors. Similarly, hsa-miR-135b-5p, hsa-miR-1180, hsa-miR-503, hsa-miR-9-5p, hsa-miR-34a-5p, hsa-miR-362-5p, hsa-181c-5p, hsa-miR-25-3p, hsa-miR-125a, hsa-miR-424-5p, hsa-miR-425-5p, and hsa-miR-194-5p showed to be significantly up-regulated in our cohort of male tumors while in FBC these miRNAs have shown to be down-regulated[23, 27-29]. Among the most down-regulated miRNAs found in the present study (more than 2 fold decrease in expression), hsa-miR-155-5p and hsa-miR-203 have shown to be up-regulated in female breast tumors [25, 30].
Clinicopathological features of male breast cancer patients.
| Characteristics | Number (%) |
|---|---|
| Age in years | |
| Median | 63 |
| Range | 36-89 |
| Histology | |
| IDC | 29 (69%) |
| DCIS | 4 (9.5%) |
| IPC | 3 (7.1%) |
| PCIS | 4 (9.5%) |
| MuC | 1 (2.4%) |
| Grade | |
| Low | 5 (11.9%) |
| Moderate | 15 (35.7%) |
| High | 11 (26.2%) |
| Unknown | 11 (26.2%) |
| Tumor size | |
| <2 cm | 16 (38%) |
| ≥2 cm | 18 (43%) |
| Unknown | 8 (19%) |
| Lymph node metastasis | |
| Positive | 21 (50%) |
| Negative | 9 (21.4%) |
| Unknown | 12 (28.6%) |
| Hormone receptors status | |
| ER-positive | 25 (59.5%) |
| ER-negative | 3 (7.1%) |
| Unknown | 14 (33.3%) |
| PR-positive | 24 (57.1%) |
| PR-negative | 4 (9.5%) |
| Unknown | 14 (33.3%) |
| Her-2/neu-positive | 6 (21%) |
| Her-2/neu-negative | 16 (38.1%) |
| Unknown | 18 (42.8%) |
IDC: Infiltrating Ductal Carcinoma; DCIS: Ductal Carcinoma In Situ; IPC: Infiltrating Papillary Carcinoma; PCIS: Papillary Carcinoma In Situ; MuC: Mucinous Carcinoma
Validation of differentially expressed microRNAs using Quantitative real-time PCR
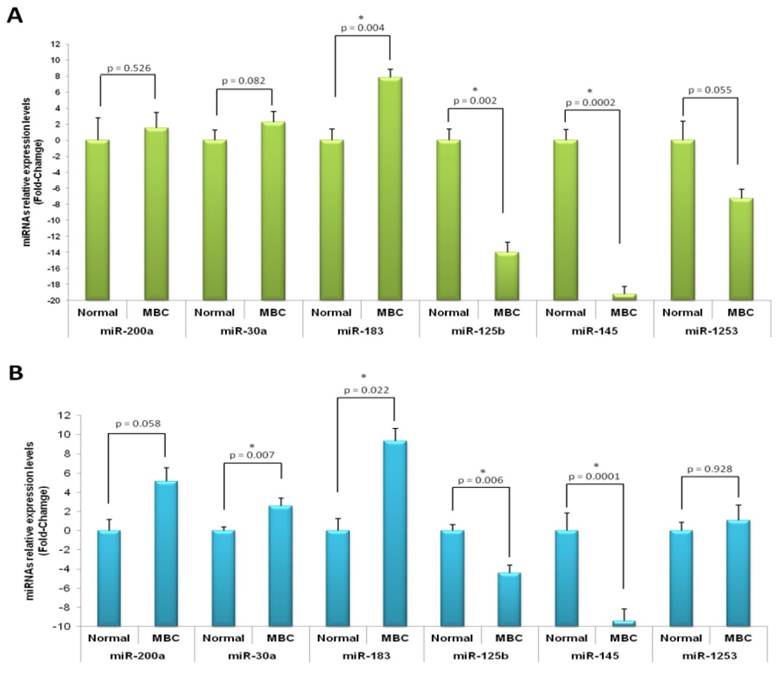
In order to validate the most deregulated miRNA we performed RT-qPCR. We tested the differential expression of several miRNAs including hsa-miR-200a, hsa-miR-30a, hsa-miR-183, as they were the most up-regulated, and hsa-miR-125b, hsa-miR-145 and hsa-miR-1253, as they behaved as the most down-regulated. For this, 16 MBC cases (profiling cohort, ntumor=8 and nnormal=4; independent cohort, ntumor=8 and nnormal=3) were used. The quantitative real-time PCR data confirmed the results obtained by miRNA profiling analysis (Figures 2). As shown in Figure 2, miR-200a, miR-30a and miR-183 were over-expressed in cancer samples, whereas miR-125b, miR-145 and miR-1253 were under-expressed. The differential expression among tumor and normal samples for hsa-miR-183, hsa-miR-125b and hsa-miR-145 were statistically significant in both studied cohorts. The rest of the miRNAs either showed statistically significant difference only in one group (miR-30a) or only a trend in both cohorts (miR-200a and miR-1253).
Differentially expressed miRNAs between normal epithelium and tumor tissue from male breast cancer patients. Unsupervised hierarchical clustering of the miRNA expression data showed clear separation between normal and tumor mammary tissue, indicating a cancer-specific expression pattern of a number of miRNAs in our MBC samples.
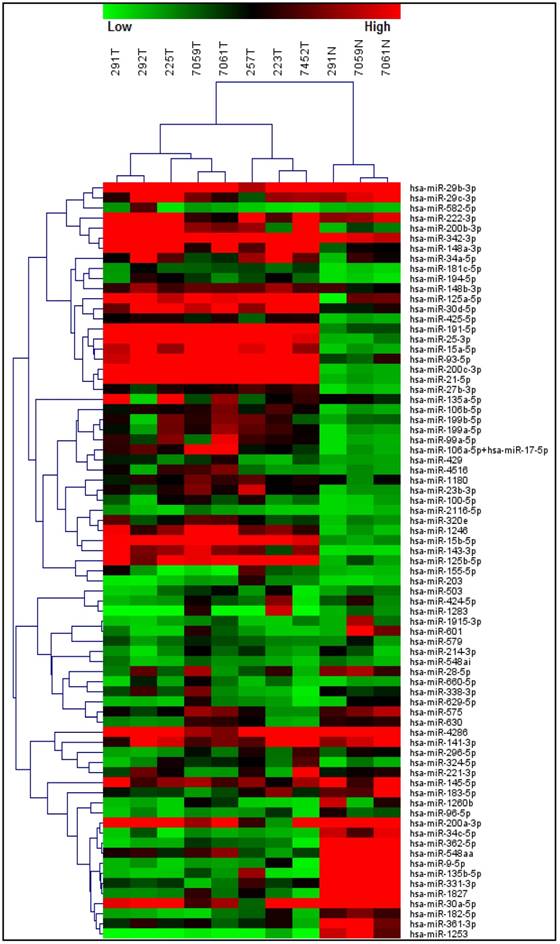
MicroRNAs differentially expressed in male breast cancer samples versus normal epithelium
| MicroRNA | Tumor (normalized) | Normal (normalized) | Fold change | p-value | Reported expression in FMC1 |
|---|---|---|---|---|---|
| hsa-miR-200a-3p | 511 | 78 | 6.57 | 0.0052 | Up-regulated |
| hsa-miR-30a-5p | 767 | 118 | 6.53 | 0.0160 | Up-regulated |
| hsa-miR-29b-3p | 895 | 143 | 6.25 | 0.0101 | Up-regulated |
| hsa-miR-222-3p | 1471 | 307 | 4.79 | 0.0585 | Down-regulated |
| hsa-miR-141-3p | 385 | 84 | 4.60 | 0.0059 | Up-regulated |
| hsa-miR-342-3p | 7185 | 1586 | 4.53 | 0.0339 | Up-regulated |
| hsa-miR-135b-5p | 64 | 14 | 4.50 | 0.2240 | Down-regulated |
| hsa-miR-29c-3p | 338 | 80 | 4.21 | 0.0281 | Up-regulated |
| hsa-miR-183-5p | 105 | 25 | 4.13 | 0.0273 | Up-regulated |
| hsa-miR-135a-5p | 691 | 186 | 3.71 | 0.2496 | Up-regulated |
| hsa-miR-191-5p | 1829 | 555 | 3.30 | 0.0005 | Up-regulated |
| hsa-miR-182-5p | 45 | 14 | 3.25 | 0.0463 | Up-regulated |
| hsa-miR-30d-5p | 580 | 181 | 3.20 | 0.0056 | Up-regulated |
| hsa-miR-1180 | 152 | 48 | 3.15 | 0.0024 | Down-regulated |
| hsa-miR-503 | 53 | 17 | 3.12 | 0.0431 | Down-regulated |
| hsa-miR-200b-3p | 430 | 138 | 3.11 | 0.0185 | Up-regulated |
| hsa-miR-15a-5p | 441 | 144 | 3.06 | 0.0002 | Up-regulated |
| hsa-miR-582-5p | 46 | 15 | 2.96 | 0.2454 | No Reports found |
| hsa-miR-200c-3p | 1129 | 398 | 2.84 | 0.0005 | No Reports found |
| hsa-miR-9-5p | 38 | 13 | 2.83 | 0.0510 | Down-regulated |
| hsa-miR-34a-5p | 258 | 92 | 2.80 | 0.0465 | Down-regulated |
| hsa-miR-106a-5p+hsa-miR-17-5p | 269 | 97 | 2.76 | 0.1426 | Up-regulated |
| hsa-miR-28-5p | 122 | 45 | 2.70 | 0.0994 | No Reports found |
| hsa-miR-4286 | 3509 | 1344 | 2.61 | 0.2325 | No Reports found |
| hsa-miR-361-3p | 93 | 36 | 2.58 | 0.0148 | No Reports found |
| hsa-miR-362-5p | 40 | 15 | 2.58 | 0.0237 | Down-regulated |
| hsa-miR-181c-5p | 64 | 25 | 2.56 | 0.0060 | Down-regulated |
| hsa-miR-324-5p | 90 | 36 | 2.52 | 0.0463 | No Reports found |
| hsa-miR-25-3p | 1409 | 561 | 2.51 | 0.0081 | Down-regulated |
| hsa-miR-125a-5p | 668 | 267 | 2.50 | 0.0238 | Down-regulated |
| hsa-miR-93-5p | 668 | 272 | 2.46 | 0.0063 | Up-regulated |
| hsa-miR-106b-5p | 146 | 59 | 2.46 | 0.0063 | Up-regulated |
| hsa-miR-148a-3p | 835 | 339 | 2.46 | 0.0642 | No Reports found |
| hsa-miR-660-5p | 48 | 20 | 2.41 | 0.0472 | No Reports found |
| hsa-miR-424-5p | 79 | 33 | 2.40 | 0.2037 | Down-regulated |
| hsa-miR-425-5p | 117 | 50 | 2.36 | 0.0056 | Up-regulated |
| hsa-miR-429 | 70 | 30 | 2.34 | 0.0251 | Up-regulated |
| hsa-miR-1260b | 38 | 17 | 2.29 | 0.1265 | No Reports found |
| hsa-miR-194-5p | 78 | 35 | 2.25 | 0.0205 | Down-regulated |
| hsa-miR-96-5p | 37 | 17 | 2.23 | 0.0290 | Up-regulated |
| hsa-miR-296-5p | 78 | 35 | 2.23 | 0.0749 | Down-regulated |
| hsa-miR-338-3p | 87 | 39 | 2.23 | 0.2052 | No Reports found |
| hsa-miR-331-3p | 82 | 38 | 2.17 | 0.0396 | Up-regulated |
| hsa-miR-629-5p | 43 | 20 | 2.15 | 0.1488 | Up-regulated |
| hsa-miR-1827 | 65 | 31 | 2.14 | 0.0860 | No Reports found |
| hsa-miR-148b-3p | 237 | 111 | 2.13 | 0.0013 | Up-regulated |
| hsa-miR-15b-5p | 801 | 386 | 2.08 | 0.0032 | Up-regulated |
| hsa-miR-34c-5p | 32 | 16 | 2.08 | 0.0817 | Up-regulated |
| hsa-miR-221-3p | 337 | 164 | 2.05 | 0.4990 | Up-regulated |
| hsa-miR-199b-5p | 164 | 342 | -2.08 | 0.2868 | Down-regulated |
| hsa-miR-1915-3p | 34 | 72 | -2.10 | 0.0649 | Down-regulated |
| hsa-miR-199a-5p | 169 | 367 | -2.17 | 0.3680 | Down-regulated |
| hsa-miR-1283 | 69 | 165 | -2.40 | 0.3898 | No Reports found |
| hsa-miR-320e | 133 | 343 | -2.59 | 0.4138 | Down-regulated |
| hsa-miR-99a-5p | 182 | 508 | -2.79 | 0.2010 | Down-regulated |
| hsa-miR-27b-3p | 139 | 392 | -2.82 | 0.1810 | No Reports found |
| hsa-miR-601 | 46 | 141 | -3.06 | 0.4475 | Down-regulated |
| hsa-miR-579 | 58 | 178 | -3.08 | 0.1265 | No Reports found |
| hsa-miR-2116-5p | 30 | 95 | -3.20 | 0.3402 | No Reports found |
| hsa-miR-155-5p | 64 | 209 | -3.27 | 0.0925 | Up-regulated |
| hsa-miR-548aa | 130 | 431 | -3.32 | 0.1340 | No Reports found |
| hsa-miR-23b-3p | 167 | 566 | -3.39 | 0.0834 | No Reports found |
| hsa-miR-1246 | 374 | 1401 | -3.75 | 0.1105 | Down-regulated |
| hsa-miR-100-5p | 79 | 301 | -3.81 | 0.1118 | Down-regulated |
| hsa-miR-125b-5p | 1359 | 5610 | -4.13 | 0.1087 | Down-regulated |
| hsa-miR-4516 | 114 | 497 | -4.38 | 0.3296 | No Reports found |
| hsa-miR-143-3p | 296 | 1502 | -5.07 | 0.1961 | Down-regulated |
| hsa-miR-214-3p | 51 | 296 | -5.85 | 0.1025 | Down-regulated |
| hsa-miR-575 | 135 | 844 | -6.25 | 0.4096 | Down-regulated |
| hsa-miR-548ai | 37 | 243 | -6.49 | 0.2672 | No Reports found |
| hsa-miR-203 | 43 | 366 | -8.47 | 0.4531 | Up-regulated |
| hsa-miR-630 | 77 | 704 | -9.13 | 0.3829 | Down-regulated |
| hsa-miR-145-5p | 326 | 4174 | -12.80 | 0.2319 | Down-regulated |
| hsa-miR-1253 | 6 | 106 | -18.86 | 0.1520 | No Reports found |
Quantitative real-time PCR validation of microRNA expression profile results on male breast cancer. Relative expression of microRNAs in male breast cancer compared with normal tissue by real-time PCR in profiling (A) and independent (B) cohorts. miR-200a, miR-30a and miR-183 were over-expressed in cancer samples, whereas miR-125b, miR-145 and miR-1253 were under-expressed. Asterisk, p values less than 0.05; error bars, standard deviation.
Correlation between microRNA expression and clinicopathological features in patients with male breast cancer
Using the expression profiling data, we also evaluated whether a correlation exists between various clinicopathological features associated with tumor specimens and the expression of the most deregulated miRNAs. We analyzed the group of MBC cases selected for expression profiling regarding tumor grade (moderate to high grade), degree of lymph nodes metastasis, tumor size and Ki-67 expression level. Whereas no statistically significant association were found between miRNA expression and degree of lymph nodes metastasis and Ki-67 expression levels (data not shown), a statistically significant association between tumor size and hsa-miR-125b and tumor grade and hsa-miR-200a and hsa-miR-30a, were noticed (P < 0.05) (Table 3).
Correlation between clinicopathological features and miRNA expression
| Median Expression | |||
|---|---|---|---|
| Tumor size | |||
| <2 cm | ≥2 cm | P-value | |
| hsa-miR-125b | 2212.9 | 949.0 | 0.03 |
| Tumor grade | |||
| High | Moderate | ||
| hsa-miR-200a | 176.91 | 537.952 | 0.04 |
| hsa-miR-30a | 57.37 | 943.958 | 0.02 |
P-values of <0.05 were considered significant
Discussion
In the present study we present a comprehensive profiling of 800 miRNA in MBC, which showed differences between miRNA expression of the breast cancer and their normal counterpart, suggesting a cancer-specific miRNA expression profile for MBC. Due to the low incidence of MBC and lack of large series studies, the current knowledge about its biology is limited. Most knowledge about genetic and molecular alterations has been extrapolated from FBC. Only few studies have analyzed the miRNA profiling in both female and male breast cancer [13, 19, 20, 21]. In fact, several of the most recent analyses on miRNAs and MBC analyzed cases of familiar MBC. When compared with previously reported profiling studies in FBC [23, 25, 28], some of the altered miRNAs found in this study had been also identified to be deregulated in the female group. Forty-nine miRNAs showed increased expressions while 25 miRNAs were found to be down-regulated in MBC tissue when compared with normal tissue. Particularly, hsa-miR-200a-3p, hsa-miR-30a-5p, hsa-miR-29b-3p, hsa-miR-222-3p, hsa-miR-141-3p, hsa-miR-342-3p, hsa-miR-135b-5p, hsa-miR-29c-3p, hsa-miR-183-5p were the most prominently up-regulated and hsa-miR-125b-5p, hsa-miR-4516, hsa-miR-143-3p, hsa-miR-214-3p, hsa-miR-575, hsa-miR-548ai, hsa-miR-203, hsa-miR-630, hsa-miR-145-5p and hsa-miR-1253 were the most down-regulated in MBC.
From this group of 19 most deregulated miRNAs, 12 have been reported to be differentially expressed in other tumors, including FBC (hsa-miR-200a-3p, hsa-miR-30a-5p, hsa-miR-29b-3p, hsa-miR-222-3p, hsa-miR-141-3p, hsa-miR-342-3p, hsa-miR-183-5p, hsa-miR-125b-5p, hsa-miR-214-3p, hsa-miR-575, hsa-miR-203 and hsa-miR-145-5p)[24, 25, 31-34] whereas the remaining 7 miRNAs had no previous reports in this regard.
Among the seventy-four miRNA that were found to be deregulated in MBC, 37 of them had been reported to show the same expression pattern in FBC (Table 2). However, an important fraction (20%) showed opposite expression pattern between the two populations. Among these group, hsa-miR-222, hsa-miR-34a, hsa-miR-181c has been shown to be differentially expressed in tamoxifen-resistant and tamoxifen-sensitive breast cancer cell lines. The authors suggest that these miRNAs with opposite expression between the two cell lines may be involved in endocrine resistance [35]. The cell lines used in the mentioned study are well-known models of FBC, which emphasize the significance of our findings regarding differentially expressed miRNAs possibly implicated in resistance development. These findings highlight the relevance of considering the differences in miRNA expression patterns between male and female breast tumors when considering treatment options and overall disease management.
Among the several miRNAs we found to be deregulated in MBC and that had been suggested to be involved in the development and progression of cancer, hsa-miR-200a-3p and hsa-miR-183-5p have been reported as over-expressed and hsa-miR-145-5p as significantly down-regulated in both male and female breast cancer. It was recently reported that the hsa-miR-200a suppresses the epithelial-mesenchymal transition (EMT), the initiating step of metastasis, which has being supported by data suggesting that hsa-miR-200a, targeting of YAP1 (Yes-associated protein 1), suppresses the pro-apoptotic protein expression and allows the cells to evade anoikis [24]. Other studies have demonstrated that some miRNAs, including hsa-miR-141, hsa-miR-183, hsa-miR-200b/c and miR-21 may be implicated in the progression of the disease. We found that hsa-miR-183, hsa-miR-141 and hsa-miR-200b/c, that have been noted to increase during the Normal-atypical ductal hyperplasia (ADH) transition, and maintained their high expression profiles during later stages, were over-expressed in our cases suggesting tumor progression [23].
We found down-regulation of hsa-miR-145, a possible tumor suppressor miRNA which has been demonstrated in other cancers including FBC. Iorio et al. (2005) demonstrated that hsa-miR-145 is progressively down-regulated from normal breast to cancer with high proliferation index suggesting that deregulation of this miRNA may affect critical molecular events involved in tumor progression. Recent studies show that hsa-miR-145 is down-regulated in human cancer cell line MCF-7 and that when over-expressed by plasmid inhibits MCF-7 cell growth and induces apoptosis [27]. More recently, other study found that hsa-miR-145 exhibited a pro-apoptotic effect, which is dependent on TP53 activation, and that TP53 activation can, in turn, stimulate hsa-miR-145 expression, thus establishing a death promoting loop between miR-145 and TP53. It was also found that hsa-miR-145 can down-regulate ER-α protein expression. These findings support a view that hsa-miR-145 re-expression therapy could be mainly envisioned in the specific group of patients with ER-α-positive and/or TP53 wild-type tumors [36].
We also found hsa-miR-125b to be down-regulated in tumor samples. Previous studies demonstrate the involvement of miR-125b in the progression of breast cancer. Iorio et al. (2005) showed that breast cancer primary tumors and cell lines have decreased levels of miR-125b expression, suggesting that lack of this miRNA may impair differentiation capabilities of cancer cells [25]. Baffa et al. (2009), seeking a specific miRNA expression signature characterizing the metastatic phenotype of solid tumors, performed a miRNA microarray analysis on 43 paired primary tumors (ten colon, ten bladder, 13 breast, and ten lung cancers) and one of their related metastatic lymph nodes[37]. miR-125b was found to be among the specific miRNAs that may be directly involved in cancer metastasis and that it may represent a novel diagnostic tool in the characterization of metastatic cancer gene. A recent study demonstrates the implication of miR-125b in predicting clinical outcome in breast cancer patients [38]. The authors found that miR-125b was significantly associated with therapeutic response, exhibiting higher expression level in non-responsive patients. In addition, breast cancers with high miR-125b expression prior to chemotherapy had higher percentage of proliferating cells and lower percentage of apoptotic cells in the corresponding surgical specimens obtained after neo-adjuvant chemotherapy, suggesting that miR-125b expression in breast cancers was reversely correlated with apoptosis and proliferation inhibition induced by chemotherapy. This finding has important implications in the development of targeted therapeutics for overcoming chemotherapeutic resistance in novel anti-cancer strategies. Although the evidence supporting the important implication of miR-125 in breast cancer biology has been generated using FBC, the miR-125b behavior observed in the present study has also been found in an earlier study on MBC [13], suggesting a similar role for hsa-miR-125b in this pathology, but validation studies are in need for further corroboration.
The differential expression of one miRNA was determined to be correlated with some clinicopathological characteristics of the male breast tumors included in the present study. The expression of miR-125b was significantly correlated with tumor size, feature that has been recognized as a poor prognosis indicator [39]. Expression of miR-125b was significantly lower in tumors equal or larger than 2 cm when compare with tumors smaller than 2 cm, suggesting that its down-regulation may be implicated in mechanisms leading to more aggressive tumors. The behavior (up or down-regulation) of each of these miRNAs was consistent in all the samples, which posse several characteristics previously describe in other MBC cohorts. Among these, our cohort presented a high rate of ER and PR positivity and a low rate of Her-2 amplification, three features that distinguish very well between female and male breast carcinomas [39].
Overall, our report further supports previous studies about the existence of a tumor-specific miRNA expression signature and contributes to the understanding of miRNA expression patterns and their relationship to carcinogenesis in MBC. Also, our results are consistent with previous data from similar studies regarding miRNA profiling in MBC demonstrating the existence of a miRNA expression signature for this pathology when compare with normal epithelia and with FBC miRNA profiles. Moving forward, large series studies would need to be conducted to address and evaluate the specific role the miRNAs found in the present study potentially have in the biology of MBC, particularly its relationship with prognostic tumor characteristics.
Conclusions
In the present study, we present a comprehensive profiling of 800 miRNA in MBC. Our results further support the hypothesis that MBC have a different miRNA expression profile in comparison with normal mammary tissue and with that reported for FBC. In agreement with previous reports, high grade MBC tumors in our group were mostly ER and PR positive, one of the major differences between male and female breast cancer.
The identification of miRNAs as potential markers of poor prognosis may provide insights into the understanding of these aggressive malignant tumors and may prove to be useful as diagnostic and prognostic tools as well as have a potential roll in the development of new therapeutic approaches. Larger sample studies are required to confirm the results presented in this report and to correlate them with clinical outcome in patients with MBC.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Weiss JR, Moysich KB, Swede H. Epidemiology of male breast cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:20-6
2. Society AC. Cancer Facts & Figures 2013-2014. Atlanta: American Cancer Society, Inc. 2013
3. Fields EC, DeWitt P, Fisher CM, Rabinovitch R. Management of male breast cancer in the United States: a surveillance, epidemiology and end results analysis. Int J Radiat Oncol Biol Phys. 2013;87:747-52
4. Ding Y, Steele L, Kuan C-J, Greilac S, Neuhausen S. Mutations in BRCA2 and PALB2 in male breast cancer cases from the United States. Breast Cancer Research and Treatment. 2011;126:771-8
5. Brinton LA, Cook MB, McCormack V, Johnson KC, Olsson H, Casagrande JT. et al. Anthropometric and hormonal risk factors for male breast cancer: male breast cancer pooling project results. J Natl Cancer Inst. 2014;106:djt465
6. Ravandi-Kashani F, Hayes TG. Male breast cancer: a review of the literature. Eur J Cancer. 1998;34:1341-7
7. Joshi MG, Lee AK, Loda M, Camus MG, Pedersen C, Heatley GJ. et al. Male breast carcinoma: an evaluation of prognostic factors contributing to a poorer outcome. Cancer. 1996;77:490-8
8. Burga AM, Fadare O, Lininger RA, Tavassoli FA. Invasive carcinomas of the male breast: a morphologic study of the distribution of histologic subtypes and metastatic patterns in 778 cases. Virchows Arch. 2006;449:507-12
9. Johansson I, Nilsson C, Berglund P, Lauss M, Ringner M, Olsson H. et al. Gene expression profiling of primary male breast cancers reveals two unique subgroups and identifies N-acetyltransferase-1 (NAT1) as a novel prognostic biomarker. Breast Cancer Res. 2012;14:R31
10. Rushton M, Kwong A, Visram H, Graham N, Petrcich W, Dent S. Treatment outcomes for male breast cancer: a single-centre retrospective case-control study. Curr Oncol. 2014;21:e400-407
11. Corcoran C, Friel AM, Duffy MJ, Crown J, O'Driscoll L. Intracellular and extracellular microRNAs in breast cancer. Clin Chem. 2011;57:18-32
12. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-54
13. Fassan M, Baffa R, Palazzo JP, Lloyd J, Crosariol M, Liu CG. et al. MicroRNA expression profiling of male breast cancer. Breast Cancer Res. 2009;11:R58
14. Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD. et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A. 2004;101:11755-60
15. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297
16. Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259-69
17. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D. et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-8
18. Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ. et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214
19. Lehmann U, Streichert T, Otto B, Albat C, Hasemeier B, Christgen H. et al. Identification of differentially expressed microRNAs in human male breast cancer. BMC Cancer. 2010;10:109
20. Pinto R, De Summa S, Danza K, Popescu O, Paradiso A, Micale L. et al. MicroRNA expression profiling in male and female familial breast cancer. Br J Cancer. 2014;111:2361-8
21. Danza K, De Summa S, Pilato B, Carella M, Palumbo O, Popescu O. et al. Combined microRNA and ER expression: a new classifier for familial and sporadic breast cancer patients. J Transl Med. 2014;12:319
22. Walter BA, Gomez-Macias G, Valera VA, Sobel M, Merino MJ. miR-21 Expression in Pregnancy-Associated Breast Cancer: A Possible Marker of Poor Prognosis. J Cancer. 2011;2:67-75
23. Chen L, Li Y, Fu Y, Peng J, Mo MH, Stamatakos M. et al. Role of deregulated microRNAs in breast cancer progression using FFPE tissue. PLoS One. 2013;8:e54213
24. Yu SJ, Hu JY, Kuang XY, Luo JM, Hou YF, Di GH. et al. MicroRNA-200a promotes anoikis resistance and metastasis by targeting YAP1 in human breast cancer. Clin Cancer Res. 2013;19:1389-1399
25. Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S. et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065-70
26. Vaira V, Faversani A, Dohi T, Montorsi M, Augello C, Gatti S. et al. miR-296 regulation of a cell polarity-cell plasticity module controls tumor progression. Oncogene. 2012;31:27-38
27. Wang S, Bian C, Yang Z, Bo Y, Li J, Zeng L. et al. miR-145 inhibits breast cancer cell growth through RTKN. Int J Oncol. 2009;34:1461-6
28. Li S, Meng H, Zhou F, Zhai L, Zhang L, Gu F. et al. MicroRNA-132 is frequently down-regulated in ductal carcinoma in situ (DCIS) of breast and acts as a tumor suppressor by inhibiting cell proliferation. Pathol Res Pract. 2013;209:179-83
29. Arigoni M, Barutello G, Riccardo F, Ercole E, Cantarella D, Orso F. et al. miR-135b coordinates progression of ErbB2-driven mammary carcinomas through suppression of MID1 and MTCH2. Am J Pathol. 2013;182:2058-70
30. Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL. et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348-60
31. Chen WX, Hu Q, Qiu MT, Zhong SL, Xu JJ, Tang JH. et al. miR-221/222: promising biomarkers for breast cancer. Tumour Biol. 2013;34:1361-70
32. Wang Y, Zhang X, Li H, Yu J, Ren X. The role of miRNA-29 family in cancer. Eur J Cell Biol. 2013;92:123-8
33. Vosa U, Vooder T, Kolde R, Vilo J, Metspalu A, Annilo T. Meta-analysis of microRNA expression in lung cancer. Int J Cancer. 2013;132:2884-93
34. Radojicic J, Zaravinos A, Vrekoussis T, Kafousi M, Spandidos DA, Stathopoulos EN. MicroRNA expression analysis in triple-negative (ER, PR and Her2/neu) breast cancer. Cell Cycle. 2011;10:507-17
35. Manavalan TT, Teng Y, Appana SN, Datta S, Kalbfleisch TS, Li Y. et al. Differential expression of microRNA expression in tamoxifen-sensitive MCF-7 versus tamoxifen-resistant LY2 human breast cancer cells. Cancer Lett. 2011;313:26-43
36. Spizzo R, Nicoloso MS, Lupini L, Lu Y, Fogarty J, Rossi S. et al. miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells. Cell Death Differ. 2010;17:246-54
37. Baffa R, Fassan M, Volinia S, O'Hara B, Liu CG, Palazzo JP. et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219:214-21
38. Wang H, Tan G, Dong L, Cheng L, Li K, Wang Z. et al. Circulating MiR-125b as a marker predicting chemoresistance in breast cancer. PLoS One. 2012;7:e34210
39. Liu D, Xie G, Chen M. Clinicopathologic characteristics and survival of male breast cancer. Int J Clin Oncol. 2014;19:280-7
Author contact
![]() Corresponding author: Maria J. Merino M.D. Address: 10 Center Drive, Building 10 MSC Room 2B44, Bethesda MD 20892. Phone: (301) 496-2441 Fax: (301) 480-1458. E-mail: mjmerinonih.gov
Corresponding author: Maria J. Merino M.D. Address: 10 Center Drive, Building 10 MSC Room 2B44, Bethesda MD 20892. Phone: (301) 496-2441 Fax: (301) 480-1458. E-mail: mjmerinonih.gov

 Global reach, higher impact
Global reach, higher impact