3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(4):638-649. doi:10.7150/jca.21180 This issue Cite
Review
Human salivary microRNAs in Cancer
1. Department of Surgery and Medical Surgical Specialties, Medicine and Dentistry School, University of Santiago de Compostela, Spain. Health Research Institute of Santiago (IDIS); Santiago de Compostela, Spain.
2. Liquid Biopsy Analysis Unit, Translational Medical Oncology, Health Research Institute of Santiago (IDIS), CIBERONC, Complexo Hospitalario Universitario de Santiago de Compostela (SERGAS), Santiago de Compostela, Spain.
3. Cell Cycle and Cancer Lab, Biomedical Research Group in Gynecology, Vall Hebron Research Institute (VHIR), Universitat Autònoma de Barcelona (UAB), Barcelona, Spain.
4. Medical Digestive Service, Complexo Hospitalario Universitario de Santiago de Compostela (SERGAS); Santiago de Compostela, Spain.
5. Department of Gynecology Oncology, Vall d'Hebron University Hospital, Barcelona, Spain.
Received 2017-5-25; Accepted 2017-12-11; Published 2018-1-6
Abstract
Circulating microRNAs (miRNAs) have emerged as excellent candidates for cancer biomarkers. Several recent studies have highlighted the potential use of saliva for the identification of miRNAs as novel biomarkers, which represents a great opportunity to improve diagnosis and monitor general health and disease. This review summarises the mechanisms of miRNAs deregulation in cancer, the value of targeting them with a therapeutic intention and the evidence of the potential clinical use of miRNAs expressed in saliva for the detection of different cancer types. We also provide a comprehensive review of the different methods for normalising the levels of specific miRNAs present in saliva, as this is a critical step in their analysis, and the challenge to validate salivary miRNAs as a reality to manage cancer patients.
Keywords: miRNAs, saliva, cancer, normalization, biomarkers, liquid biopsy
Introduction
Detection of cancer in the early stages is paramount for determining the prognosis, therapeutic interventions, survival rates and recurrence of the disease [1]. Numerous researchers have focused on finding specific biomarkers of disease in body fluids such as blood, urine or cerebrospinal fluid [2-4]. Recently, several studies have highlighted the usefulness of saliva for identifying biomarkers, noting that it represents a great opportunity to improve diagnosis and monitoring of general health and disease [5-7].
MicroRNAs (miRNAs or miRs) constitute one of the most abundant classes of gene-regulatory molecules. Different mechanisms have been established as responsible of the miRNAs deregulation in cancer. Genetic alterations such as chromosomal rearrangements, genomic amplifications, deletions or point mutations have been established to play an important role in cancer initiation and progression through the aberrant expression of miRNAs located in these affected regions and, subsequently, by the deregulation of their downstream mRNAs targets [8]. Besides, a substantial number of miRNA genes are subjected to epigenetic alterations such as DNA hypermethylation of tumor suppressor miRNAs, extensive genomic DNA hypomethylation and alteration of histone modification patterns. Altogether, these events can be related with aberrant miRNA expression in cancer [9, 10]. miRNA biogenesis is a complex phenomenon where several transcriptional and post-transcriptional factors are involved. Despite their small size, miRNA production, maturation, and regulatory function require the action of a large number of proteins, including the two RNase III proteins, DROSHA and DICER, and Argonaute (AGO) proteins [11]. They are generally transcribed by the RNA polymerase II (Pol II), generating a long transcript called primary miRNA (pri-miRNA). The processing of pri-miRNA to pre-miRNA occurs within the nucleus by the microprocessor complex (including DROSHA) and, after exportation to the cytoplasm, pre-miRNA is processed to the mature miRNA by DICER RNase. The mature miRNA along with AGO proteins form miRISC complex, which drives the specific silencing of the targeted mRNAs [11]. In oncogenesis, several mutations have been found in components of the miRNA biogenesis machinery as in DROSHA and DICER1, which have been characterized to affect to the expression of several miRNAs in cancer [12]. In addition, at transcriptional level, several factors have been described to modulate miRNA levels. For example, the oncogene MYC regulates the transcriptional activation of miR-17-92 cluster, an oncogenic miRNA over-expressed in several malignances [13]. Another example of cancer-involved transcriptional regulation of miRNA genes is the tumor-suppressor protein p53 that promotes the transcription of all miR-34 family members [14]. Due to the important role of miRNAs in cancer biology, future work will uncover additional pathways that regulate the expression of individual miRNAs or miRNA clusters in tumourigenesis.
In cancer biology, miRNAs have been established as key molecular components in tumour growth, invasion, angiogenesis, and immune evasion [15]. miRNAs have the ability to act as oncogenes or tumour suppressor genes in different steps of the tumourigenic process, affecting several cell-signaling pathways essential to carcinogenesis. Since miRNAs regulate important cellular processes by regulating multiple targets, anticancer therapies based on miRNAs are now under investigation. For the treatment of cancer, there have been discovered different potential therapeutic strategies as the following: RNA interference (RNAi)-based therapy (sandwich and multiplex RNAi inhibition strategy), miRNA inhibition therapy and miRNA mimetic agents [16]. Sandwich RNAi inhibition strategy focuses on the use of multiples agents to target one specific molecular defect whereas multiplex RNAi inhibition strategy targets multiple molecular defects accumulated in the same pathway of a specific cancer [17]. In the second RNA therapeutic drugs, specific miRNAs (anti-miR oligonucleotides, locked nucleic acid anti-miRNAs, antagomiRs, miRNA sponges, miRNA masks and small molecule inhibitors of miRNAs) act upon the mature miRNA inhibiting the interaction of that miRNA with its target mRNA [18-21]. For a similar purpose, it has emerged the “SMIR-approach”, a novel inhibitory-miRNA therapy based on small molecule inhibitors of miRNAs (SMIRs) that act inhibiting miRNA biogenesis or impeding miRNA-target interaction [22]. Another therapeutic alternative is the “miRNA mimics”, which goal is to recover the function of tumour suppressive miRNAs using synthetic miRNA-like molecules [23]. Nevertheless, the study of miRNA-based therapies is still in its infancy and side effects of these therapies need a more exhaustive evaluation. Research efforts must be addressed to the research of miRNA-based targeted therapies for cancer since they could represent a great advance to personalized medicine for cancer treatment [16].
Saliva has a very complex composition including enzymes, antibodies, hormones, antimicrobial elements and cytokines [24]. As a liquid biopsy, saliva presents many advantages over blood as collection is easy, safe, non-invasive and cost-effective [25]. Salivary diagnostic studies have reported the existence of several molecular indicators of local and systemic disorders, including cancer [26]. In the recent years miRNAs have emerged as excellent candidates for cancer biomarkers due to their remarkable stability and resistance to degradation [27-29]. miRNAs are small, noncoding RNA molecules consisting of 18-22 nucleotides that act as regulators of many cellular processes [30]. They are one of the most abundant classes of gene-regulatory molecules involved in gene expression [15]. A single miRNA can regulate the expression of multiple genes involved in various cellular process associated with cancer development [31, 32]. Nowadays it is well known that miRNAs play an important role in the initiation and progression of cancer and depending on their transcript targets they act as “oncomiRs” or “tumour suppressive miRNAs” [31, 33]. In this sense, carcinogenesis is associated with aberrant expression of miRNA profiles. Alterations in miRNAs could arise from various genetic alterations such as those involving chromosomal abnormalities, genomic mutations, epigenetic changes and anomalies in miRNA biogenesis [34]. Biologically, miRNAs have been identified in twelve body fluids: amniotic fluid, breast milk, bronchial lavage, cerebrospinal fluid, colostrum, peritoneal fluid, plasma, pleural fluid, seminal fluid, tears, urine and saliva [35]. There are multiple sources of circulating miRNAs including tumour cells, immune cells and blood cells [36, 37], and several proposals have been made on how miRNAs are released into circulation (e.g. packaged into microvesicles or exosomes, or in vesicle-free form, that is in association with protein complexes) [38].
In the last few years it has been reported that circulating miRNAs in serum and plasma may serve as diagnostic biomarkers [39]. Regarding human cancer, several authors [27, 40, 41] have reported different levels of salivary expression of miRNAs. Hence, saliva has attracted increased attention as a novel non-invasive tool for cancer diagnosis. In this review we describe findings related to the potential clinical use of salivary miRNAs for cancer detection. We also provide a comprehensive review of the different methods of normalising the levels of specific miRNAs present in saliva, as this is a critical step in their analysis.
Circulating miRNAs in saliva
There are several hypotheses for the presence of cell-free nucleic acids and proteins into saliva. These molecules could enter into the saliva from the blood via various cellular mechanisms, such as transcellular (passive intracellular diffusion and active transport) or paracellular routes (extracellular ultrafiltration) [7]. miRNAs could also be produced locally by apoptosis or cell necrosis and they could also be released by normal epithelial or cancerous cells in exosomes or microvesicles [42-44]. It is important to note that circulating miRNAs in saliva could act as signalling molecules in the context of cancer biology [45].
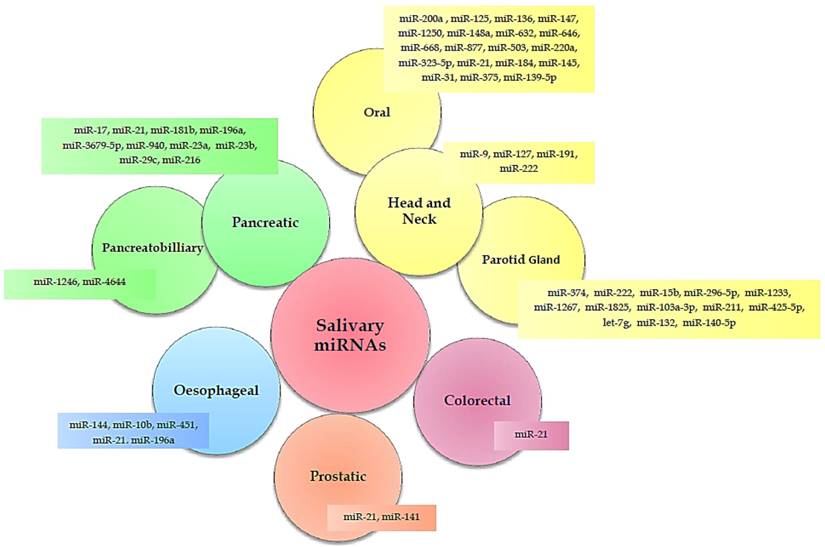
Numerous studies (Table 1) have identified a variety of salivary miRNAs (Figure 1) as promising non-invasive diagnostic biomarkers of cancer. Below, we summarised these studies according to the tumour type, moving from tumours localized close to the oral cavity to more distant tumours.
Summary of studies with salivary miRNAs identified in different types of cancer.
| Cancer Location | Patients (No) | Tumor stage | miRNAs | Status | Normalization method | Authors |
|---|---|---|---|---|---|---|
| Head and neck | DP: 12 OSCC, 12 HC VP: 50 OSCC, 50 HC | I-IV | miR-200a, miR-125a | Downregulated | U6-snRNA | [41] |
| 9 OSCC, 8 OSCC-R, 8 OLP, 9 HC | NA | miR-136, miR-147, miR-1250, miR-148a, miR-632, miR-646, miR-668, miR-877, miR-503, miR-220a, miR-323-5p, miR-24, miR-27b | Downregulated (Upregulated miR-24 and miR-27b) | miR-191 | [51] | |
| 20 OSCC, 20 HC, 40 OPMDs, 20 RAS | NA | miR-21, miR-184, miR-145 | Upregulated (Downregulated miR-145) | SNORD68 | [53] | |
| 45 OSCC, 10 OVL, 24 HC | I-IV | miR-31 | Upregulated | miR-16 | [48] | |
| 15 OSCC, 7 HC | NA | miR-375 | Downregulated | miR-191 | [50] | |
| DP: 3 TSCC, 4 HC | I-III | miR-139-5p | Downregulated | U6-snRNA | [54] | |
| VP: 25 TSCC, 25 HC | ||||||
| DP: 5 HNSCC, 5 HC | II-IV | miR-9, miR-127, miR-191, miR-222 | Upregulated | SNORD96A | [46] | |
| VP: 35 HNSCC, 35 HC | ||||||
| DP: 10 C, 10 BT VP: 28 C, 19 BT | NA | miR-374, miR-222, miR-15b, let-7g, miR-132, miR-140-5p | Upregulated | U6-snRNA | [58] | |
| DP: 20 PGNs, 10 HC | NA | miR-296-5p, miR-1233, miR-1267, miR-1825, miR-103a-3p, miR-211, miR-425-5p | Upregulated | U6-snRNA | [59] | |
| VP (whole saliva): 46 PGNs, 14 HC | ||||||
| Oesophageal | DP: 7 C, 3 HC | I-IV | miR-144, miR-10b, miR-451, miR-21 | Upregulated | miR-16 | [27] |
| VP: 39 C, 19 HC | ||||||
| DP: 8C, 4 HC | II-IV | miR-21 | Upregulated | miR-16 | [64] | |
| VP: 32 C, 16 HC | ||||||
| 100 C, 50 HC | I-II | miR-21 | Upregulated | miR-16 | [65] | |
| 67 C, 50 HC | NA | miR-144 | Upregulated | NA | [67] | |
| 5 C, 5 HC | NA | miR-196a | Upregulated | miR-16 | [68] | |
| Pancreatic | DP: 8 C, 8 HC VP: 40 C, 20 BT, 40 HC | I-III | miR-3679-5p, miR-940 | Downregulated (miR-3679-5p) Upregulated (miR-940) | U6-snRNA | [28] |
| 30 C, 32 HC | NA | miR-17, miR-21, miR-181b, miR-196a | Upregulated | miR-16 | [69] | |
| 7 C, 4 P, 2 IPMN, 4 HC | NA | miR-21, miR-23a, miR-23b, miR-29c, miR-216 | Upregulated | RQ manager 1.2.1 and Data Assist v3.0 (Applied Biosystems) | [40] | |
| 12 C, 13 HC | II-IV | miR-1246, miR-4644 | Upregulated | U6-snRNA | [70] | |
| Prostatic | NA (Supernatant and pellet) | NA | miR-21, miR-141 | Upregulated | Exogenous miR-21 and miR-141 | [72] |
| Colorectal | 34 CRC, 34 HC | II-IV | miR-21 | Upregulated | U6-snRNA | [73] |
Abbreviations: DP, discovery phase; VP, validation phase; C, cancer; P, pancreatitis; BT, benign tumour; HC, healthy controls; IPMN, intraductal papillary mucinous neoplasia; OSCC, oral squamous cell carcinoma; OSCC-R, OSCC in remission; OPMDs, oral potentially malignant disorders; RAS, recurrent aphthosus stomatitis; OLP, oral lichen planus; OVL, oral verrucous leukoplakia; HNSCC, head and neck squamous cell carcinoma; PGNs, parotid glands neoplasms; TSCC, tongue squamous cell carcinoma; CRC, colorectal carcinoma; NA, not available.
Salivary miRNAs in cancer diagnosis.
Head and neck squamous cell carcinoma
According to National Cancer Institute, head and neck cancers are a group of cancers that start in the tissues and organs of the head and neck, including the oral cavity, pharynx, larynx, paranasal sinuses, nasal cavity and salivary glands. Salazar et al. [46] used a novel saliva-based miRNA biomarker panel to detect head and neck cancers located in the oral cavity, pharynx, oropharynx and larynx. In a discovery phase they selected five miRNAs (miR-9, miR-127, miR-134, miR-191 and miR-222) by microarray analysis. Differential expression of these miRNAs was assessed in confirmation and validation phases in independent cohorts. miR-9, miR-191, miR-127 and miR-222 were over-expressed in the cancer group relative to the control group. An independent validation based on the Cancer Genome Atlas (TCGA) miRNA-seq data from 334 tumours and 39 normal tissues showed that miR-9, miR-191 and miR-222 were significantly over-expressed in tumour samples. The receiver-operating characteristic (ROC) curve analysis indicated high discriminatory power for miR-9, miR-191 and miR-222 with AUC values of 0.85, 0.74 and 0.98, respectively. A salivary panel formed by the combination of these three miRNAs had an AUC of 0.74. The authors also evaluated the power of miRNA expression profiles to discriminate between head and neck squamous cell carcinoma (HNSCC) patients with tumours in different anatomical locations (oral cavity, pharynx, oropharynx and larynx). miR-222, miR-191 and miR-127 differentiated between patients with cancer in the oral cavity and pharynx, miR-9 (AUC = 0.77) differentiated between oropharynx and larynx cancer patients, miR-191 differentiated between oropharynx and pharynx patients (AUC = 0.94), and miR-127 differentiated between pharynx and larynx cancer patients (AUC = 1.00). Although this study encompassed several types of head and neck cancers, oral squamous cell cancer (OSCC) and parotid gland tumours are the head and neck cancers most commonly investigated using saliva.
Oral Squamous Cell Cancer
The first study to identify miRNAs in saliva was performed in OSCC [41]. Park et al. [41] analyzed saliva supernatant from OSCC patients and observed group differences in the level of expression of miR-200a and miR-125a. Salivary miRNAs were detected in both whole and saliva supernatant, although a more diverse miRNA population was observed in total saliva, probably because whole saliva contains miRNAs from desquamated oral epithelial cells [41]. In addition, a variety of miRNAs have been reported in metastatic (miR-181 and miR-296) and non-metastatic (miR-31 and miR-130b) OSCC samples [47]. Liu et al. [48] reported that salivary levels of miR-31 were significantly upregulated in pre-surgery OSCC patients compared with the levels in healthy individuals. In this study, not significant differences were observed between oral verrucous leukoplakia patients and control group, but a significant lower value was observed respect to OSCC patients [48]. Expression of miR-31 was reduced after tumour resection and, indeed, a high correlation was observed between salivary and plasma levels of miR-31. Interestingly, levels of miR-31 were more abundant in saliva than in plasma, indicating that salivary miR-31 has potential as a tool for diagnosis of pre-cancerous lesions. The reduction in miR-31 salivary levels after surgical resection suggested a relationship between the tumoural tissue and the salivary molecular profile [48].
Deregulation of miRNA expression is a complex process that contributes to the development of human cancer and is associated with the progressive accumulation of multiple genetic abnormalities. In oral carcinogenesis, epigenetic mechanisms such as altered DNA methylation patterns play an important role in silencing tumour-suppressive miRNAs [49]. Wilkund et al. [50] analyzed miRNA expression and changes in DNA methylation in OSCC patients rinsed with a PBS solution (oral rinse) and saliva. Levels of miR-375 were lower in the cancer group than the control group in both types of sample. In oral rinse, patients' miR-200a levels were lower than those of the control group, and miR-200c-141 CpG methylation was significantly higher. The authors suggested that oral rinse could be a more sensitive source of OSCC-specific miRNA and DNA methylation, due to the exposure of the buccal epithelia to environmental factors [50]. Recently (2014), Momen-Heravi et al. [51] reported that 13 miRNAs were deregulated in salivary cancer samples compared to controls; of these 13, only miRNA-24 and miRNA-27b were significantly over-expressed. miR-27b had the best sensitivity and specificity when it came for discriminating OSCC patients from the other groups: OSCC vs. healthy individuals: sensitivity = 85.71%, specificity = 100%; OSCC vs. OSCC in remission (OSCC-R): sensitivity = 85.71%, specificity = 83.33%; OSCC vs. oral lichen planus (OLP): sensitivity = 85.71%, specificity = 100%. Moreover, miR-136 was under-expressed in saliva of OSCC patients relative to healthy controls, and discriminated between them with 88.89% sensitivity and 100% specificity. However, it was also found that plasma and tissue expression of miR-27b was significantly reduced in OSCC patients [52]. The secretion of miRNAs from tumour cells into saliva could lead to a difference in the expression profile for body fluids and cancer tissue. In 2015, Zahran et al. [53] reported that salivary levels of miR-21 and miR-184 levels were significantly higher in OSCC patients and oral potentially malignant disorders (OPMDs) patients than in healthy and disease controls, while salivary miR-145 levels were significantly lower in OSCC and OPMDs. ROC analysis indicated the discriminatory power of miR-21, miR-145 and miR-184 with sensitivity values of 65%, 60% and 80% respectively, and specificity values of 65%, 70% and 75% respectively. A microarray study of tongue squamous cell carcinoma revealed that 183 miRNAs were significantly upregulated in the salivary samples of the cancer group and 236 miRNAs were significantly downregulated. The upregulation of two miRNAs (miR-33a-3p and miR-198) and the downregulation of one miRNA (miR-139-5p) was validated in 50 saliva samples. Salivary expression of miR-139-5p was significantly lower in preoperative patients than controls, but levels were similar in preoperative and postoperative patients. ROC curves showed that miR-139-5p differentiated between preoperative patients and both healthy individuals and postoperative patients, with AUCs of 0.805 and 0.713 respectively [54]. Importantly, miR-139-5p has an anticancer function, playing an important role in cell proliferation, apoptosis [55], lymph node involvement [56], and metastasis [57].
Parotid Gland Tumour
Matse et al. [58] investigated miRNA profiles in whole saliva from patients with malignant or benign parotid gland tumours. They found 57 miRNAs differentially expressed in the benign and malignant tumour groups. Of the 18 verified miRNAs, differential expression of nine (namely miR-140-5p, miR-374, miR-222, miR-15b, let-7g, miR-132, miR-519b-3p, miR-223, and miR-30a-3p) were validated in an independent set of samples that showed increased salivary levels for those miRNAs in patients with a malignant parotid gland tumour. Statistically significant levels were observed for miR-374, miR-222, miR-15b, let-7g, miR-132, and miR-140-5p between benign and malignant parotid gland tumour groups. A logistic model consisting of four miRNAs (miR-132, miR-15b, miR-140, and miR-223) showed 69% sensitivity and 95% specificity with an AUC of 0.90 to identify parotid gland tumours. Recently, the same research group [59] reported new data on miRNA expression profiles for whole saliva from patients with parotid gland neoplasms. Of the 742 miRNAs analyzed in an exploratory phase, eight miRNAs were differently expressed in tumour patients and healthy controls. In the validation phase, seven miRNAs (miR-296-5p, miR-1233, miR-1267, miR-1825, miR-103a-3p, miR-211 and miR-425-5p) were found to be significantly over-expressed relative to controls in whole saliva from patients. A logistic regression model containing a combination of these seven miRNAs yielded an AUC of 0.95, with 93% sensitivity and 86% specificity for predicting the presence of a parotid gland neoplasm. In comparison, a model based on just miR-211 and miR-1233 had sensitivity of 91% and specificity of 86%. The authors also analyzed the expression of the validated miRNAs in a set of 12 parotid saliva samples (affected gland and healthy gland) and in 14 healthy controls (both healthy glands). Only miR-425-5p and miR-1825 were expressed in parotid saliva at similar levels to those in whole saliva. The absence of miR-103a-3p, miR-211, miR-296-5p, miR-1233 and miR-1267 from parotid saliva indicates that stimulated and unstimulated saliva have different miRNA expression profiles.
Oesophageal cancer
Several authors have analyzed the role of miR-21 and miR-144 in the pathogenesis of oesophageal cancer [60-63]. In a two-phase study, Xie et al. [27] analyzed miRNA expression in the saliva of patients with oesophageal cancer. In a discovery phase, ten saliva samples were subjected to microarray analysis and five miRNAs (miR-144, miR-10b, miR-451, miR-486-5p, and miR-634) were selected for the validation phase. miR-21 was also included in the validation phase as several authors had shown that its expression was aberrant in the plasma and tissue of oesophageal cancer patients. Curiously, levels of miR-144, miR-10b and miR-451 in whole saliva were significantly correlated with levels in saliva supernatant. In whole saliva, three miRNAs discriminated between oesophageal cancer patients and controls: miR-10b, miR-144 and miR-451 had sensitivities of 89.7%, 92.3% and 84.6%, and specificities of 57.9%, 47.4% and 57.9%, respectively. In saliva supernatant, four miRNAs discriminated between oesophageal cancer patients and controls: miR-10b, miR-144, miR-21 and miR-451 had sensitivities of 79.5%, 43.6%, 89.7% and 51.3%, and specificities of 57.9%, 89.5%, 47.4% and 84.2%, respectively. Cancer stage, pathology type and presence of nodal metastases were independent factors contributing to variance in salivary levels of these miRNAs.
In a preliminary study, Xie et al. [64] showed significant upregulation of miR-21 in saliva supernatant from oesophagal cancer patients (sensitivity = 87.5%, specificity = 62.5%). Salivary and plasma levels of miR-21 have also been evaluated [65]. miR-21 plasma and salivary levels were significantly higher in cancer samples and there was a positive correlation between levels in these two body fluids. Interestingly, high plasma levels of miR-21 have been associated with poor survival [66]. In another study, Wu et al. [67] evaluated the salivary levels of miR-144 in whole saliva and saliva supernatant from 67 patients and 50 healthy controls. Levels of miR-144 were significantly higher in salivary cancer samples than control samples, indicating moderate diagnostic power with respect to oesophageal cancer, with a sensitivity of 74.6% and specificity of 92% for whole saliva and a sensitivity of 53.7% and specificity of 94% for saliva supernatant.
Recently, Fendereski et al. [68] observed a significant upregulation of miR-196a in tissue tumour samples as compared with the normal matching tissue from the same patients. Besides, a significantly increased expression of miR-196a was found in saliva, showing the potential of this miRNA as a diagnostic biomarker of oesophageal cancer.
Pancreatic cancer
In pancreatic cancer, Gao et al. [69] found miRNAs specifically expressed in three different groups of ZHENG disease conditions (Shi-Re, Pi-Xu and Xue-Yu), which is the cornerstone in traditional Chinese medicine. They found significantly higher levels of miR-17, miR-21 and miR-181b in patients with Shi-Re ZHENG and higher levels of miR-196a in patients with Pi-Xu ZHENG. Interestingly, these findings indicate that miRNAs may have a role as specific biomarkers for different syndromes of traditional Chinese medicine. In 2015, Xie et al. [28] reported two salivary miRNAs (miR-3679-5p and miR-940) as potential non-invasive biomarkers of resectable pancreatic cancer. miR-3679-5p was significantly found downregulated in pancreatic cancer whereas miR-940 was significantly found upregulated. A logistic regression model was used to evaluate the power of miR-3679 and miR-940 to discriminate between various cancer and non-cancer groups. The model had 72.5% sensitivity and 70% specificity when comparing a cancer group and a healthy control group; 62.5% sensitivity and 80% specificity when comparing the cancer group and the benign pancreatic tumour group; and 70% sensitivity and 70% specificity when comparing the cancer group with a combined non-cancer group.
In a recent pilot study, Humeau et al. [40] found that relative to controls, miR-21, miR-23a, miR-23b and miR-29c were significantly upregulated in saliva of pancreatic cancer patients group, with sensitivity values of 71.4 %, 85.7%, 85.7% and 57%, respectively, and specificity of 100% in all cases. In addition, miR-210 and let-7c were upregulated in pancreatitis patients compared with a healthy group, and miR-216 was upregulated in cancer patients relative to the pancreatitis group. In an experimental murine model, differential expression of miR-21 was validated using xerografts of pancreatic cancer cells. The study revealed over-expression of salivary miR-21 in tumour-bearing mice relative to non-cancer mice, indicating that elevation of salivary miR-21 precedes systemic cancer markers.
Recently, Machida et al. [70] analyzed miRNAs in salivary exosomes from patients with pancreatobiliary tract cancer. Levels of miR-1246 and miR-4644 were significantly higher in the cancer group than in the control group. ROC curves were used to evaluate the discriminatory power of miR‑1246 and miR‑464 and showed AUCs of 0.814 and 0.763, respectively. Although further investigation is necessary, salivary exosomal miRNAs has been described as useful biomarkers of pancreatic cancer. In addition, Alemar et al. [71] evaluated the serum expression profile of five oncogenic miRNAs (miR-21, miR-155, miR-196a, miR-200b, and miR-376a) and one tumour suppressive miRNA (miR-34a). miR-21 and miR-34a were over-expressed in cancer samples relative to control samples, with AUCs of 0.889 and 0.865, respectively. Levels of miR-21 and miR-34a were significantly correlated. Serum expression of miR-200a was significantly associated with histological grade and tumour location. However, although these miRNAs were detected in salivary samples, levels were similar in cancer patients and controls.
Prostate cancer
Hizir M.S. and colleagues [72] evaluated the expression of miR-21 and miR-141, two miRNAs over-expressed in early and advanced prostate cancer patients, respectively. They have demonstrated simultaneous detection of endogenous and exogenous miR-21 and miR-141 from human body fluids including blood, urine, and saliva, by using a specially designed biosensing nanographene oxide system, which uses two different wavelengths to detect miR-21 and miR-141 simultaneously (520 and 670 nm, respectively). Their approach is faster and easier than multiple and simultaneous qPCR detection, and can be performed with a portable spectrofluorometer; however, improved sensitivity is still needed for further clinical application.
Colorectal cancer
miR-21 was the first miRNA analyzed in salivary samples from colorectal cancer (CRC) patients [73]. Several cancer-related genes such as genes for phosphatase and tensin homologue (PTEN), tropomyosin 1 (TPM1) and programmed cell death-4 (PDCD4) are regulated by miR-21, which is an oncogenic miRNA [74]. Indeed, numerous studies have reported increased expression of miR-21 in plasma and serum from CRC patients [75-78]. In serum, high levels of miR-21 were associated with large tumour size, distant metastasis and advanced TNM stage [79]. Recently, Sazanov et al. [73] evaluated expression levels of miR-21 in plasma and saliva samples from CRC patients. In both body fluids, median levels of miR-21 were significantly higher in the cancer group than in the control group, but there were no differences between patients at stages II, III and IV. In addition, levels of miR-21 in the cancer and control groups were independent of gender and age. However, no significant individual correlation and linear regression of miR-21 expression levels in the plasma and saliva were detected. Importantly, miR-21 levels in plasma had a sensitivity of 65% and a specificity of 85% for discriminating between CRC patients and controls, whereas in saliva miR-21 had a sensitivity of 97% and specificity of 91%. The high sensitivity and specificity values for salivary miR-21 reflected the potential use of this saliva-based test as a method of CRC diagnosis.
MiRNAs normalization strategies in saliva
Normalization is important for accurate quantification of RNA levels using reverse transcription-PCR (qRT-PCR). The most commonly used normalization strategies for expression of circulating miRNAs are Global Mean Expression [80], exogenous (spiked-in) miRNAs and endogenous controls [81]. The most widely used method is normalization based on endogenous controls such as miRNA, small nuclear RNA (snRNA) and small nucleolar RNA (snoRNA) [82]. However, suitable reference genes should be selected in order to reduce the technical variation and avoid the misinter-pretation of the data [80, 82, 83], but there is no consensus on which miRNA is best for normalization. Researchers select reference genes based on other studies [27, 28, 48, 59, 73], or searching stable miRNAs in their own samples [51], following the strategy described by Vandesompele et al. [84], where stable genes across the samples (among other factors) make it suitable for their usage as a normalization standards.
The reference genes most commonly used for normalization of miRNA expression in saliva are miR-16 and miR-191 [27, 48, 50, 51, 64, 69], but also two snoRNAs (SNORD68 and SNORD96A) [46, 53] and one snRNA (U6) [28, 41, 54, 58, 59, 70, 73]. miR-191 has been used as a reference gene for the miRNA normalization in oral cancer patients [50, 51]. In this sense, Momen-Heravi et al. [51] evaluated three miRNAs (miR-16, miR-191 and miR-484) generally considered to be stable using the NormFinder algorithm. Although miR-16 and miR-484 had high stability values, miR-191 had the lowest intragroup and intergroup variability according to NormFinder, so miR-191 was used as reference internal control in their research.
miR-16 has been selected for miRNAs normalization in serum [85] and plasma [76, 86] due to its high stability and abundance [33, 76, 86] and is the internal control most commonly used for analyses of saliva [27, 48, 64, 65, 69]. However, its suitability as a reference gene for normalization in saliva has never been systematically evaluated. In addition, Chen et al. [36] observed that miR-16 and other normalizers such as 5S rRNA and U6 were degraded in serum samples. In their study levels of the analyzed miRNAs were directly normalized to the total RNA amount. Other authors have also reported that the levels of U6 are unstable, decreasing over freeze-thaw cycles relative to miR-16 and miR-24 [87].
Due to technical variations in sample processing and extracellular RNA (exRNA) extraction and analysis, proper normalization is critical for consistent detection of true biological differences between samples, an issue that was also discussed by the NIH exRNA consortium [88]. They present a preliminary study suggesting that it may be possible, at least in plasma, to identify a set of exRNAs with relatively stable abundances, which may be used as internal reference standards for exRNA quantification. Overall, there is no consensus on validated internal controls for accurate quantification of the RNA levels. This represents a serious problem as many researchers use reference genes that have not been validated beforehand.
Salivary miRNAs: onset hypothesis
Many diagnostic miRNAs have been discovered through deep sequencing technologies, and although further research is needed to elucidate the complexity of the heterogeneous origin of miRNAs in body fluids, they provide precise information that could lead to early detection and recognition of cancer [89]. miRNAs are small RNA molecules that are stable in circulation and importantly are protected from RNases degradation [90]. Several mechanisms have been proposed for circulatory miRNA protection against degradation, such as by forming ribonucleoproteins complexes (i.e. with AGO2 or HDL proteins) or by being incorporated into extracellular vesicles (apoptotic bodies, shedding vesicles and exosomes) [89].
Saliva is a proximal body fluid in the oral cavity and therefore is intuitively sound for detection of oral diseases. Currently, there are several theories and studies (including pre-clinical models) supporting why saliva is termed as “liquid biopsy”, useful for detection of other systemic diseases as well. Some studies hypothesize that molecules from the primary tumour can be carried in the bloodstream and appear altered in saliva from cancer patients compared to healthy patients (or patients with benign diseases). Gao et al. [91] used induced tumour-bearing mice models of melanoma and lung cancer, and defined a salivary transcriptome profile associated to each tumour-type. Lau et al. [92], by using a pancreatic tumour-bearing mice model, revealed the basic mechanisms underlying the rationale of salivary biomarkers through the hypothesis that exosome-like vesicles carry, drive, and deliver tumour markers into the saliva. Yet, the lack of certainty and detailed mechanistic insights showing how salivary biomarkers can reflect disease states elsewhere in the body has compromised the scientific acceptance of this emerging field in the clinics. The clinical and scientific references to saliva for systemic disease detection will represent an important step forward that will transform molecular diagnostics globally.
Salivary miRNAs as point-of-care systems
Despite we are far from fully understanding the fundamental biology of salivary biomarkers, some achievements have been made aiming at developing a device for specific salivary biomarkers' detection. In particular, the “Oral Fluid NanoSensor Test (OFNASET)” is a prototype nanotechnology point-of-care sensor that has the ability to detect multiplexed analytes in saliva for oral cancer detection, through the combination of mRNA and protein electrochemical detection [93]. In addition, a device able to detect EGFR mutation at DNA level by using patients' saliva for lung cancer diagnosis, in order to reduce the current invasive procedures and reduce unnecessary biopsies was developed [94]. More recently, another device for oral cancer detection was published based on a novel optical microfluidic biosensor with highly sensitive organic photodetectors for absorbance-based detection of salivary protein biomarkers [95]. The results of measuring IL-8 and IL-1E protein levels were in agreement with those provided by two commercial assays of ELISA thereby offering an attractive and cost-effective tool for diagnostic or screening purposes at the point of care. Several biosensors allowed the detection of multiple analytes including DNA, RNA and proteins for both oral and systemic diseases [96], including the unique study that developed a biosensor system to detect salivary miRNAs for prostate cancer detection [72]. Continuous research on the field is needed to translate salivary diagnostics into a routinely clinical practice to improve cancer diagnosis and reduce mortality. In turn, the generation of point-of-care systems will benefit health system and quality life of patients.
Other sources of extracellular miRNAs
In addition to saliva, circulating cell-free miRNAs previously identified in cells and tissues have been also identified in urine, plasma, serum and stool as potential diagnostic and prognostic cancer biomarkers. In prostate and bladder cancer, numerous studies have reported the utility of urine-circulating miRNAs as potential biomarkers [97, 98]. In addition, urinary miRNAs has been purposed as biomarkers for breast, ovarian, hepatocellular and pancreatic cancer [99-102]. In plasma and serum, numerous miRNAs have been found significantly differentially expressed in patients compared with healthy controls in several cancers, such as oral [103, 104], breast [105, 106], prostate [107, 108] and gastric [109, 110] cancers, as well as in oesophageal carcinoma [111, 112], colorectal [78, 113], pancreatic [114, 115] and lung [116, 117] cancers and hepatocellular carcinoma [118, 119]. Nowadays, only few cancer studies have evaluated the expression profiles of circulating miRNAs in saliva compared to serum or plasma [48, 71, 73]. In serum and saliva, Alemar et al. [71] have evaluated the expression levels of six pancreatic ductal adenocarcinoma-associated miRNAs (miR-21, miR-34a, miR-155, miR-196a, miR-200b, and miR-376a). Only significant differences were observed for serum miR-21 and miR-34a. Conversely, in saliva only miR-196a was significantly higher in the control group, showing a heterogeneous and almost undetectable expression [71]. In OSCC, miR-31 salivary levels revealed a strong correlation with miR-31 serum levels. Besides, saliva presented greater amount of miR-31 than plasma, indicating the enormous value of saliva as diagnostic biofluid [48]. Recently, expression levels of miR-21 were analyzed in plasma and saliva of CRC patients. Although, no significant individual correlation and linear regression of miR-21 expression levels in the plasma and saliva were observed, saliva showed a higher sensitivity and specificity than plasma for CRC. Thus, the diagnostic sensitivity of miR-21 expression in the plasma was 65% and specificity was 85% whereas in the saliva was 97% and 91%, respectively [73]. Overall, miRNAs have demonstrated to be potential biomarkers in cancer diagnosis and prognosis, although research efforts should be made to improve our knowledge about miRNAs biology among different body fluids.
Analysis of microRNAs in body fluids: advantages and disadvantages
MiRNAs in body fluids can provide great insights in the advance towards personalized medicine. Although there is a long way to go in the investigation of extracellular miRNAs till they could be used in clinical practice, it seems that nowadays the advantages are superior to the disadvantages. One of the advantages is that miRNAs present a robust stability and resistance to degradation in body fluids, showing its potential as tumour biomarkers [120]. The non-invasiveness collection and easy reproducibility are key factors [121], reinforcing the high value of circulating miRNAs as biomarkers for the cancer patients' management. Thus, in human cancer, aberrant circulating miRNA profiles allow to know the clinicopathological features of the tumour non-invasively [122]. In this way, a molecular classification based on miRNAs expression profiles could be of a great interest for the pathologic classification of each tumour [123]. Furthermore, miRNA expression has been evaluated in cancer with the goal to provide promising biomarkers for the early diagnosis [124], prognosis [125], therapy and outcome prediction [126, 127]. Nevertheless, one of the main handicaps of using circulating miRNA as a liquid biopsy is their unknown origin since they can come from other sources than tumour cells. Besides, non-cancer conditions (diet, exercise, infection and hypoxia) can affected to the expression levels of circulating miRNAs [114-116]. Fundamental efforts should be addressed to develop clinically relevant experimental research on miRNA and clinical trials with reliability and reproducibility results, taking into account the lack of concordance between the labs and inability to validate studies reported by other labs [128]. To achieve this purpose, standard methods of sample collection, storage, RNA isolation, sequencing and data evaluation are necessary to translate laboratory findings into tangible clinical applications [129]. In this sense, in spite of constant revolutionary advances in miRNA research, new horizons on the field of cancer research must be pursued to establish circulating miRNAs as cornerstone of precision medicine.
Future Perspectives
In recent years, the promise of saliva as a diagnostic tool has aroused great interest in oncological research. The salivary secretome represents an attractive source of potential novel tumour markers for diagnosis and monitoring of cancer. MiRNAs together with the proteome, transcriptome, metabolome and microbiome represent the five diagnostic alphabets of saliva. Salivary miRNAs have emerged as specific group of biomolecules with high value as diagnostic salivary biomarkers of cancer. Noticeably, further research on salivary miRNAs is necessary to establish an effective saliva-based diagnostic test. Future medical research should address the following objectives: 1) identify miRNAs with high sensitivity and specificity; 2) analyse miRNAs in well-designed studies (discovery, verification and validation phases) with well characterised and well-matched clinical groups; 3) establish a gold standard normalization method; 4) evaluate the expression profiles of salivary miRNAs in other body fluids (e.g. serum, plasma and urine); and 5) evaluate potential uses of salivary miRNAs in establishing prognosis and monitoring disease as well as in diagnosis. In conclusion, future research should be directed towards finding specific salivary miRNAs that can be used to individualise the management of cancer patients.
Acknowledgements
The authors would like to thank the Eugenio Rodríguez Pascual Foundation and the Fundación Dexeus Salud de la Mujer, for supporting the authors' studies in the field of salivary biomarkers.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Falco M, Palma G, Rea D. et al. Tumour biomarkers: homeostasis as a novel prognostic indicator. Open Biol. 2016;6(12pii):160254
2. Kishore L, Kaur N, Singh R. Distinct biomarkers for early diagnosis of diabetic nephropathy. Curr Diabetes Rev. 2017;13(6):598-605 [Epub ahead of print]
3. Bidard FC, Proudhon C, Pierga JY. Circulating tumor cells in breast cancer. Mol Oncol. 2016;10(3):418-30
4. Song Y, Zhang W, Zhang L. et al. Cerebrospinal fluid IL-10 and IL-10/IL-6 as accurate diagnostic biomarkers for primary central nervous system large B-cell lymphoma. Sci Rep. 2016;6:38671
5. Lee YH, Wong DT. Saliva: an emerging biofluid for early detection of diseases. Am J Dent. 2009;22(4):241-8
6. Bonne NJ, Wong DT. Salivary biomarker development using genomic, proteomic and metabolomic approaches. Genome Med. 2012;4(10):82
7. Malathi N, Mythili S, Vasanthi HR. Salivary diagnostics: a brief review. ISRN Dent. 2014;2014:158786
8. Zhang L, Huang J, Yang N. et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci. 2006;103(24):9136-41
9. Lopez-Serra P, Esteller M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene. 2011;31(13):1609-22
10. Lujambio A, Calin G a, Villanueva A. et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105(36):13556-61
11. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509-24
12. Guo X, Liao Q, Chen P. et al. The microRNA-processing enzymes: Drosha and Dicer can predict prognosis of nasopharyngeal carcinoma. J Cancer Res Clin Oncol. 2012;138(1):49-56
13. O'Donnell KA, Wentzel EA, Zeller KI. et al. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839-43
14. He L, He X, Lim LP. et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130-4
15. Di Leva G, Garofalo M, Croce CM. microRNAs in cancer. Annu Rev Pathol Mech Dis. 2104; 9: 287-314.
16. Shah MY, Ferrajoli A, Sood AK. et al. microRNA therapeutics in cancer - an emerging concept. EBioMedicine. 2016;12:34-42
17. Calin GA, Croce CM. Chronic lymphocytic leukemia: Interplay between noncoding RNAs and protein-coding genes. Blood. 2009;114(23):4761-70
18. Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9(10):775-89
19. Krützfeldt J, Rajewsky N, Braich R. et al. Silencing of microRNAs in vivo with “antagomirs.” Nature. 2005; 438(7068): 685-9.
20. Vester B, Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43(42):13233-41
21. Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4(9):721-6
22. Gumireddy K, Young DD, Xiong X. et al. Small-molecule inhibitors of microRNA miR-21 function. Angew Chemie Int Ed Engl. 2008;47(39):7482-4
23. Esposito CL, Cerchia L, Catuogno S. et al. Multifunctional aptamer-miRNA conjugates for targeted cancer therapy. Mol Ther. 2014;22(6):1151-63
24. Spielmann N, Wong DT. Saliva: diagnostics and therapeutic perspectives. Oral Dis. 2011;17(4):345-54
25. Yoshizawa JM, Schafer CA, Schafer JJ. et al. Salivary biomarkers: toward future clinical and diagnostic utilities. Clin Microbiol Rev. 2013;26(4):781-91
26. Rapado-Gonzalez O, Majem B, Muinelo-Romay L. et al. Cancer salivary biomarkers for tumours distant to the oral cavity. Int J Mol Sci. 2016:17 (9pii)(E): 1531
27. Xie Z, Chen G, Zhang X. et al. Salivary microRNAs as promising biomarkers for detection of esophageal cancer. PLoS One. 2013;8(4e):57502
28. Xie Z, Yin X, Gong B. et al. Salivary microRNAs show potential as a noninvasive biomarker for detecting resectable pancreatic cancer. Cancer Prev Res (Phila). 2015;8(2):165-73
29. Michael A, Bajracharya SD, Yuen PS. et al. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16(1):34-8
30. Montani F, Bianchi F. Circulating cancer biomarkers: the macro-revolution of the micro-RNA. EBioMedicine. 2016;5:4-6
31. Cekaite L, Eide PW, Lind GE. et al. MicroRNAs as growth regulators, their function and biomarker status in colorectal cancer. Oncotarget. 2016;7(6):6476-505
32. He M, Zhou W, Li C. et al. MicroRNAs, DNA damage response, and cancer treatment. Int J Mol Sci. 2016;17(12pii):E2087
33. Giráldez MD, Lozano JJ, Ramírez G. et al. Circulating microRNAs as biomarkers of colorectal cancer: Results from a genome-wide profiling and validation study. Clin Gastroenterol Hepatol. 2013;11(6):681-8
34. Lan H, Lu H, Wang X. et al. MicroRNAs as potential biomarkers in cancer: opportunities and challenges. Biomed Res Int. 2015;2015:125094
35. Weber JA, Baxter DH, Zhang S. et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56(11):1733-41
36. Chen X, Ba Y, Ma L. et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997-1006
37. Kishikawa T, Otsuka M, Ohno M. et al. Circulating RNAs as new biomarkers for detecting pancreatic cancer. World J Gastroenterol. 2015;21(28):8527-40
38. Yang Q, Diamond MP, Al-Hendy A. The emerging role of extracellular vesicle-derived miRNAs: implication in cancer progression and stem cell related diseases. J Clin Epigenet. 2016;2(1pii):13
39. D'Angelo B, Benedetti E, Cimini A. et al. MicroRNAs: A Puzzling tool in cancer diagnostics and therapy. Anticancer Res. 2016;36(11):5571-6
40. Humeau M, Vignolle-Vidoni A, Sicard F. et al. Salivary microRNA in pancreatic cancer patients. PLoS One. 2015;10(6e):0130996
41. Park NJ, Zhou H, Elashoff D. et al. Salivary microRNA: Discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res. 2009;15(17):5473-7
42. Vlassov AV, Magdaleno S, Setterquist R. et al. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820(7):940-8
43. Lässer C, Alikhani VS, Ekström K. et al. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J Transl Med. 2011;9(1):9
44. Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C. et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282
45. Larrea E, Sole C, Manterola L. et al. New concepts in cancer biomarkers: Circulating miRNAs in liquid biopsies. Int J Mol Sci. 2016;17(5pii):E627
46. Salazar C, Nagadia R, Pandit P. et al. A novel saliva-based microRNA biomarker panel to detect head and neck cancers. Cell Oncol. 2014;37(5):331-8
47. Severino P, Oliveira LS, Andreghetto FM. et al. Small RNAs in metastatic and non-metastatic oral squamous cell carcinoma. BMC Med Genomics. 2015;8:31
48. Liu CJ, Lin SC, Yang CC. et al. Exploiting salivary miR-31 as a clinical biomarker of oral squamous cell carcinoma. Head Neck. 2012;34(2):219-24
49. Kozaki KI, Imoto I, Mogi S. et al. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68(7):2094-105
50. Wiklund ED, Gao S, Hulf T. et al. MicroRNA alterations and associated aberrant DNA methylation patterns across multiple sample types in oral squamous cell carcinoma. PLoS One. 2011;6(11e):27840
51. Momen-Heravi F, Trachtenberg AJ, Kuo WP. et al. Genomewide study of salivary microRNAs for detection of oral cancer. J Dent Res. 2014;93(Suppl 7):S86-S93
52. Lo WY, Wang HJ, Chiu CW. et al. MiR-27b-regulated TCTP as a novel plasma biomarker for oral cancer: From quantitative proteomics to post-transcriptional study. J Proteomics. 2012;77:154-66
53. Zahran F, Ghalwash D, Shaker O. et al. Salivary microRNAs in oral cancer. Oral Dis. 2015;21(6):739-47
54. Duz MB, Karatas OF, Guzel E. et al. Identification of miR-139-5p as a saliva biomarker for tongue squamous cell carcinoma: a pilot study. Cell Oncol (Dordr). 2016;39(2):187-93
55. Sun C, Sang M, Li S. et al. Hsa-miR-139-5p inhibits proliferation and causes apoptosis associated with down-regulation of c-Met. Oncotarget. 2015;6(37):39756-92
56. Liu R, Yang M, Meng Y. et al. Tumor-suppressive function of miR-139-5p in esophageal squamous cell carcinoma. PLoS One. 2013;8(10e):77068
57. Krishnan K, Steptoe AL, Martin HC. et al. miR-139-5p is a regulator of metastatic pathways in breast cancer. RNA. 2013;19(12):1767-80
58. Matse JH, Yoshizawa J, Wang X. et al. Discovery and prevalidation of salivary extracellular microRNA biomarkers panel for the noninvasive detection of benign and malignant parotid gland tumors. Clin Cancer Res. 2013;19(11):3032-8
59. Matse JH, Yoshizawa J, Wang X. et al. Human salivary micro-RNA in patients with parotid salivary gland neoplasms. PLoS One. 2015;10(11):e0142264
60. Kurashige J, Kamohara H, Watanabe M. et al. Serum microRNA-21 is a novel biomarker in patients with esophageal squamous cell carcinoma. J Surg Oncol. 2012;106(2):188-92
61. Kestens C, Siersema PD, van Baal JW. Current understanding of the functional roles of aberrantly expressed microRNAs in esophageal cancer. World J Gastroenterol. 2016;22(1):1-7
62. Sharma P, Saraya A, Sharma R. Potential diagnostic implications of miR-144 overexpression in human oesophageal cancer. Indian J Med Res. 2016;143(Supplement):S91-S103
63. Shao Y, Li P, Zhu ST. et al. MiR-26a and miR-144 inhibit proliferation and metastasis of esophageal squamous cell cancer by inhibiting cyclooxygenase-2. Oncotarget. 2016;7(12):15173-86
64. Xie ZJ, Chen G, Zhang XC. et al. Saliva supernatant miR-21: a novel potential biomarker for esophageal cancer detection. Asian Pac J Cancer Prev. 2012;13(12):6145-9
65. Ye M, Ye P, Zhang W. et al. [Diagnostic values of salivary versus and plasma microRNA-21 for early esophageal cancer]. Nan Fang Yi Ke Da Xue Xue Bao. 2014;34(6):885-9
66. Li BX, Yu Q, Shi ZL. et al. Circulating microRNAs in esophageal squamous cell carcinoma: Association with locoregional staging and survival. Int J Clin Exp Med. 2015;8(5):7241-50
67. Wu W, Hou W, Wu Z. et al. [miRNA-144 in the saliva is a genetic marker for early diagnosis of esophageal cancer]. Nan Fang Yi Ke Da Xue Xue Bao. 2013;33(12):1783-6
68. Fendereski M, Zia MF, Shafiee M. et al. MicroRNA-196a as a potential diagnostic biomarker for esophageal squamous cell carcinoma. Cancer Invest. 2017;35(2):78-84
69. Gao S, Chen LY, Wang P. et al. MicroRNA expression in salivary supernatant of patients with pancreatic cancer and its relationship with ZHENG. Biomed Res Int. 2014;2014:756347
70. Machida T, Tomofuji T, Maruyama T. et al. miR-1246 and miR-4644 in salivary exosome as potential biomarkers for pancreatobiliary tract cancer. Oncol Rep. 2016;36(4):2375-81
71. Alemar B, Izetti P, Gregório C. et al. miRNA-21 and miRNA-34a are potential minimally invasive biomarkers for the diagnosis of pancreatic ductal adenocarcinoma. Pancreas. 2016;45(1):84-92
72. Hizir MS, Balcioglu M, Rana M. et al. Simultaneous detection of circulating oncomiRs from body fluids for prostate cancer staging using nanographene oxide. ACS Appl Mater Interfaces. 2014;6(17):14772-8
73. Sazanov AA, Kiselyova EV, Zakharenko AA. et al. Plasma and saliva miR-21 expression in colorectal cancer patients. J Appl Genet. 2017;58(2):231-7
74. Yu W, Wang Z, Shen L. et al. Circulating microRNA-21 as a potential diagnostic marker for colorectal cancer: A meta-analysis. Mol Clin Oncol. 2016;4(2):237-44
75. Kanaan Z, Rai SN, Eichenberger MR. et al. Plasma miR-21: a potential diagnostic marker of colorectal cancer. Ann Surg. 2012;256(3):544-51
76. Luo X, Stock C, Burwinkel B. et al. Identification and evaluation of plasma microRNAs for early detection of colorectal cancer. PLoS One. 2013;8(5):e62880
77. Basati G, Emami Razavi A, Abdi S. et al. Elevated level of microRNA-21 in the serum of patients with colorectal cancer. Med Oncol. 2014;31(10):205
78. Yamada A, Horimatsu T, Okugawa Y. et al. Serum miR-21, miR-29a, and miR-125b are promising biomarkers for the early detection of colorectal neoplasia. Clin Cancer Res. 2015;21(18):4234-42
79. Toiyama Y, Takahashi M, Hur K. et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105(12):849-59
80. Mestdagh P, Van Vlierberghe P, De Weer A. et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10(6R):64
81. Yoshizawa JM, Wong DT. Salivary microRNAs and oral cancer detection. Methods Mol Biol. 2013;936:313-24
82. Bignotti E, Calza S, Tassi RA. et al. Identification of stably expressed reference small non-coding RNAs for microRNA quantification in high-grade serous ovarian carcinoma tissues. J Cell Mol Med. 2016;20(12):2341-8
83. Wang L, Liu Y, Du L. et al. Identification and validation of reference genes for the detection of serum microRNAs by reverse transcription-quantitative polymerase chain reaction in patients with bladder cancer. Mol Med Rep. 2015;12(1):615-22
84. Vandesompele J, De Preter K, Pattyn F. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034
85. Liu GH, Zhou ZG, Chen R. et al. Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumor Biol. 2013;34(4):2175-81
86. Huang Z, Huang D, Ni S. et al. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127(1):118-26
87. Xiang M, Zeng Y, Yang R. et al. U6 is not a suitable endogenous control for the quantification of circulating microRNAs. Biochem Biophys Res Commun. 2014;454(1):210-4
88. Laurent LC, Abdel-Mageed AB, Adelson PD. et al. Meeting report: discussions and preliminary findings on extracellular RNA measurement methods from laboratories in the NIH Extracellular RNA Communication Consortium. J Extracell Vesicles. 2015;4:26533
89. Schwarzenbach H, Nishida N, Calin GA. et al. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11(3):145-56
90. Mitchell PS, Parkin RK, Kroh EM. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105(30):10513-8
91. Gao K, Zhou H, Zhang L. et al. Systemic disease-induced salivary biomarker profiles in mouse models of melanoma and non-small cell lung cancer. PLoS One. 2009;4(6e):5875
92. Lau C, Kim Y, Chia D. et al. Role of pancreatic cancer-derived exosomes in salivary biomarker development. J Biol Chem. 2013;288(37):26888-97
93. Gau V, Wong D. Oral Fluid Nanosensor Test (OFNASET) with advanced electrochemical-based molecular analysis platform. Ann N Y Acad Sci. 2007;1098:401-10
94. Wei F, Lin CC, Joon A. et al. Noninvasive saliva-based EGFR gene mutation detection in patients with lung cancer. Am J Respir Crit Care Med. 2014;190(10):1117-26
95. Dong T, Matos Pires NM. Immunodetection of salivary biomarkers by an optical microfluidic biosensor with polyethylenimine-modified polythiophene-C 70 organic photodetectors. Biosens Bioelectron. 2017;94:321-7
96. Mishra S, Saadat D, Kwon O. et al. Recent advances in salivary cancer diagnostics enabled by biosensors and bioelectronics. Biosens Bioelectron. 2016;81:181-97
97. Stuopelyte K, Daniunaite K, Bakavicius A. et al. The utility of urine-circulating miRNAs for detection of prostate cancer. Br J Cancer. 2016;115(6):707-15
98. De Long J, Sullivan TB, Humphrey J. et al. A non-invasive miRNA based assay to detect bladder cancer in cell-free urine. Am J Transl Res. 2015;7(11):2500-9
99. Zhou J, Gong G, Tan H. et al. Urinary microRNA-30a-5p is a potential biomarker for ovarian serous adenocarcinoma. Oncol Rep. 2015;33(6):2915-23
100. Erbes T, Hirschfeld M, Rücker G. et al. Feasibility of urinary microRNA detection in breast cancer patients and its potential as an innovative non-invasive biomarker. BMC Cancer. 2015;15:193
101. Gasparri ML, Casorelli A, Bardhi E. et al. Beyond circulating microRNA biomarkers: urinary microRNAs in ovarian and breast cancer. Tumor Biol. 2017;39(5):1010428317695525
102. Abdalla M, Haj-Ahmad Y. Promising candidate urinary microrna biomarkers for the early detection of hepatocellular carcinoma among high-risk hepatitis C virus Egyptian patients. J Cancer. 2012;3:19-31
103. Tachibana H, Sho R, Takeda Y. et al. Circulating miR-223 in oral cancer: its potential as a novel diagnostic biomarker and therapeutic target. PLoS One. 2016;11(7e):0159693
104. Xu H, Yang Y, Zhao H. et al. Serum miR-483-5p: a novel diagnostic and prognostic biomarker for patients with oral squamous cell carcinoma. Tumor Biol. 2016;37(1):447-53
105. Hannafon BN, Trigoso YD, Calloway CL. et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016;18(1):90
106. Shimomura A, Shiino S, Kawauchi J. et al. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Sci. 2016;107(3):326-34
107. Srivastava A, Goldberger H, Dimtchev A. et al. Circulatory miR-628-5p is downregulated in prostate cancer patients. Tumor Biol. 2014;35(5):4867-73
108. Kachakova D, Mitkova A, Popov E. et al. Combinations of serum prostate-specific antigen and plasma expression levels of let-7c, miR-30c, miR-141, and miR-375 as potential better diagnostic biomarkers for prostate cancer. DNA Cell Biol. 2015;34(3):189-200
109. Shin VY, Ng EKO, Chan VW. et al. A three-miRNA signature as promising non-invasive diagnostic marker for gastric cancer. Mol Cancer. 2015;14:202
110. Hou CG, Luo XY, Li G. Diagnostic and prognostic value of serum microRNA-206 in patients with gastric cancer. Cell Physiol Biochem. 2106; 39(4): 1512-20.
111. Warnecke-Eberz U, Chon SH, Hölscher AH. et al. Exosomal onco-miRs from serum of patients with adenocarcinoma of the esophagus: comparison of miRNA profiles of exosomes and matching tumor. Tumor Biol. 2015;36(6):4643-53
112. Li SP, Su HX, Zhao D. et al. Plasma miRNA-506 as a prognostic biomarker for esophageal squamous cell carcinoma. Med Sci Monit. 2016;22:2195-201
113. Chang PY, Chen CC, Chang YS. et al. MicroRNA-223 and microRNA-92a in stool and plasma samples act as complementary biomarkers to increase colorectal cancer detection. Oncotarget. 2016;7(9):10663-75
114. Madhavan B, Yue S, Galli U. et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer. 2015;136(11):2616-27
115. Abue M, Yokoyama M, Shibuya R. et al. Circulating miR-483-3p and miR-21 is highly expressed in plasma of pancreatic cancer. Int J Oncol. 2015;46(2):539-47
116. Fan L, Qi H, Teng J. et al. Identification of serum miRNAs by nano-quantum dots microarray as diagnostic biomarkers for early detection of non-small cell lung cancer. Tumor Biol. 2016;37(6):7777-84
117. Sun L, Chen Y, Su Q. et al. Increased plasma miRNA-30a as a biomarker for non-small cell lung cancer. Med Sci Monit. 2016;22:647-55
118. Sohn W, Kim J, Kang SH. et al. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med. 2015;47(e):184
119. Wen Y, Han J, Chen J. et al. Plasma miRNAs as early biomarkers for detecting hepatocellular carcinoma. Int J Cancer. 2015;137(7):1679-90
120. Liang Y, Ridzon D, Wong L. et al. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166
121. He Y, Lin J, Kong D. et al. Current state of circulating microRNAs as cancer biomarkers. Clin Chem. 2015;61(9):1138-55
122. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857-66
123. Ferracin M, Pedriali M, Veronese A. et al. MicroRNA profiling for the identification of cancers with unknown primary tissue-of-origin. J Pathol. 2011;225(1):43-53
124. Zhu C, Ren C, Han J. et al. A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br J Cancer. 2014;110(9):2291-9
125. Summerer I, Unger K, Braselmann H. et al. Circulating microRNAs as prognostic therapy biomarkers in head and neck cancer patients. Br J Cancer. 2015;113(1):76-82
126. Summerer I, Niyazi M, Unger K. et al. Changes in circulating microRNAs after radiochemotherapy in head and neck cancer patients. Radiat Oncol. 2013;8:296
127. Benson EA, Skaar TC, Liu Y. et al. Carboplatin with decitabine therapy, in recurrent platinum resistant ovarian cancer, alters circulating miRNAs concentrations: A pilot study. PLoS One. 2015;10(10e):0141279
128. Shalaby T, Fiaschetti G, Baumgartner M. et al. MicroRNA signatures as biomarkers and therapeutic target for CNS embryonal tumors: the pros and the cons. Int J Mol Sci. 2014;15(11):21554-86
129. Liu X, Liu X, Wu Y. et al. MicroRNAs in biofluids are novel tools for bladder cancer screening. Oncotarget. 2017;8(19):32370-9
Author contact
![]() Corresponding author: María Mercedes Suárez-Cunqueiro, Associate Professor, Surgery and Medical Surgical Specialties, Medicine and Dentistry School, University of Santiago de Compostela, C/ Entrerrios S/N 15872, Santiago de Compostela, Spain. Tel.: + 34 881812437; e-mail: mariamercedes.suarezes
Corresponding author: María Mercedes Suárez-Cunqueiro, Associate Professor, Surgery and Medical Surgical Specialties, Medicine and Dentistry School, University of Santiago de Compostela, C/ Entrerrios S/N 15872, Santiago de Compostela, Spain. Tel.: + 34 881812437; e-mail: mariamercedes.suarezes

 Global reach, higher impact
Global reach, higher impact