3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(11):1932-1942. doi:10.7150/jca.23284 This issue Cite
Research Paper
MLH1 Promoter Methylation and Prediction/Prognosis of Gastric Cancer: A Systematic Review and Meta and Bioinformatic Analysis
Tumor Etiology and Screening Department of Cancer Institute and General Surgery, the First Hospital of China Medical University, and Key Laboratory of Cancer Etiology and Prevention (China Medical University), Liaoning Provincial Education Department, Shenyang 110001, China
Received 2017-10-11; Accepted 2018-2-25; Published 2018-4-30
Abstract
Background: The promoter methylation of MLH1 gene and gastric cancer (GC)has been investigated previously. To get a more credible conclusion, we performed a systematic review and meta and bioinformatic analysis to clarify the role of MLH1 methylation in the prediction and prognosis of GC.
Methods: Eligible studies were targeted after searching the PubMed, Web of Science, Embase, BIOSIS, CNKI and Wanfang Data to collect the information of MLH1 methylation and GC. The link strength between the two was estimated by odds ratio with its 95% confidence interval. The Newcastle-Ottawa scale was used for quantity assessment. Subgroup and sensitivity analysis were conducted to explore sources of heterogeneity. The Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) were employed for bioinformatics analysis on the correlation between MLH1 methylation and GC risk, clinicopathological behavior as well as prognosis.
Results: 2365 GC and 1563 controls were included in the meta-analysis. The pooled OR of MLH1 methylation in GC was 4.895 (95% CI: 3.149-7.611, P<0.001), which considerably associated with increased GC risk. No significant difference was found in relation to Lauren classification, tumor invasion, lymph node/distant metastasis and tumor stage in GC. Analysis based on GEO and TCGA showed that high MLH1 methylation enhanced GC risk but might not related with GC clinicopathological features and prognosis.
Conclusion: MLH1 methylation is an alive biomarker for the prediction of GC and it might not affect GC behavior. Further study could be conducted to verify the impact of MLH1 methylation on GC prognosis.
Keywords: MLH1, methylation, gastric cancer, risk, prognosis
Introduction
DNA methylation is a major epigenetic alteration that plays a key role in the occurrence of cancer [1]. It is a genetically modified style with reversibility and heredity and has important biological significance, manifested in the control of tissue-specific gene expression, maintenance of chromosomal integrity [2]. The methylation of tumor-associated genes has been shown to be one of the important mechanisms involved in the process of gene transcriptional silencing and regulating gene expression then results in tumor suppressor gene inactivation, oncogene activation, eventually leading to tumorigenesis [3, 4]. The feasible technology to detect methylated DNA allows us to use DNA methylation as a molecular biomarker for cancer prediction and prognosis [5-7].
DNA mismatch repair (MMR) system is one of key link in suppressing tumor formation, which can repair mismatched DNA in DNA replication to maintain genome stability. The MMR system contains a few key genes like MLH1, MSH2, MSH6 and PMS2 etc., from which encoding protein can form heterodimer to identified mismatched bases, together with other repair proteins, to complete DNA repair [8]. It is well accepted that the inactivation of MMR function is derived from germline mutation, somatic mutations or epigenetic silencing. The abnormalities of MMR can lead to microsatellite instability (MSI), which is short (1-6 base pairs) tandem repeats, spreading throughout the genome, being the identified heteromorphosis related to the occurrence and development of cancer. And colorectal cancers with MSI-H have an improved prognosis [9].MMR preferentially protects genes from mutation and has important consequences for understanding the evolution of genomes during both natural selection and human tumor growth. MMR deficiency disproportionately increases the numbers of single-nucleotide variants in genes [10].
MLH1 gene is localized at chromosome 3p22.2. It is responsible for the replacement of the mispaired nucleotides in the genome during the replication [11]. As a key member of MMR system, MLH1 is epigenetically inactivated via methylation of the gene promoter that lead to the deficiency of MMR. For example, in colorectal cancer (CRC) MSI resulted from methylation of MLH1 gene promoter, can cause its transcriptional silencing and affect other growth regulation and apoptosis-related genes, leading to the carcinogenesis of CRC. The majority of sporadic MSI tumors are caused by an epigenetic inactivation of MLH1 or MSH2. MMR deficient tumors have 10-100 times more somatic mutations than MMR proficient (pMMR) tumors leading to increased neoantigen burden and immunogenicity [12]. Similarly, MLH1 hypermethylation is also preceded by malignant proliferation of other cancers such as endometrial cancer, lung cancer, breast cancer, esophageal cancer and gastric cancer [13-20]. So the detection of MLH1 methylation can be used for prediction of tumorigenesis.
Gastric cancer (GC) is the third major cause of cancer-related deaths in the world [21]. Environmental, genetic, diet and other predisposing factors contribute to the development of gastric cancer. In recent years, more and more evidence shows that methylation of tumor suppressor gene is not be ignored risk factor in gastric carcinogenesis. In 2014, Cancer Genome Atlas Research classified GC into four pathological subtypes, in which MSI type including MMR methylation was proposed for the first time [22]. Along with the popularization of the classification, MMR methylation in gastric cancer had been widely studied. But the relevance between MLH1 methylation and GC, especially the role of MLH1 methylation on the risk prediction and prognosis of GC, remains controversial.
Here, we conducted a systematic review and meta and bioinformatic analysis to evaluate the correlation between MLH1 promoter methylation and GC through comparing cancer with healthy controls. Moreover, we also assessed the correlation between MLH1 promoter methylation and biological behavior as well as prognosis of GC by comparing cancer with different clinical pathological parameters and survival status. This study expects to get more credible information to assess the role of MLH1 methylation in gastric cancer prediction and prognosis.
Methods
Search strategy
Electronic databases, including PubMed, Web of Science, Embase, BIOSIS, Chinese National Knowledge Infrastructure (CNKI), Wanfang Data were used to systematically look for related studies published in English and Chinese until May 1, 2017. The following terms were searched: methylation or DNA methylation or hypermethylation, gastric cancer or gastric carcinoma, and MLH1 or hMLH1. Furthermore, references that were cited in each included study were also searched manually to identify potential relevant studies.
Inclusion and exclusion criteria
Eligible studies had to meet the following inclusion criteria: 1) Research topic focused on the MLH1 methylation and gastric cancer; 2) Case-control or cohort studies; 3) The studies with sufficient data for calculating odds ratios (ORs) and 95% confidence intervals (CIs); 4) Subjects investigated had a defined diagnosis by pathology. 1) Researches not related to methylation; 2) Researches not related to MLH1 methylation or methylation sites were not in the promoter region; 3) Researches not focus on GC, such as gastric ulcer and gastric functional dyspepsia and precancerous lesions; 4) Researches that selected subgroups (such as selected based on age, sex, and tumor stage); 5) Case reports and reviews; 6) Animal and cell studies. 7) Paper with insufficient or duplicated data. For duplicated data, only the most comprehensive studies were included.
Data extraction
Data from the included studies were extracted independently by two authors, Shixuan Shen and Xiaohui Chen. The information was collected from extracted data including: the first author's name, publication year, country where study conducted, detection method, sample type, the frequency of MLH1 methylation in case and control groups, clinicopathological parameters (i.e., Lauren classification, tumor invasion, lymph node status, distant metastasis and tumor stage) and survival status. The two authors reached a consensus on each item. If the data could not be obtained from the original studies, we would contact the corresponding author on reasonable request. If the authors are not convenient or willing to cooperate, we would exclude this study.
Quantity assessment
The Newcastle-Ottawa scale (NOS) with eight items was used to evaluate the quality of the included studies using three parameters: selection (four items, each awarded one star), comparability (one item, which can be awarded up to two stars) and exposure/outcome (three items, each awarded one star) [23]. NOS scores of 1-3, 4-6 and 7-9 were considered low, medium and high quality, respectively. Only studies with scores ≥ 7 were included in the analysis.
Bioinformatical analysis
We screened the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) which is a public repository that archives and freely distributes microarray [24] and use GEO2R (NCBI) to compare the methylation level in GC and normal tissues and then analyzed the association between MLH1 promoter methylation and the GC risk.
The information of 338 GC patients was downloaded from the Cancer Genome Atlas (TCGA) database by TCGA-assembler in R software. We put the raw data into analyzing the role of MLH1 promoter methylation in the GC risk prediction, behavior determination and prognosis evaluation.
Statistical analyses
Stata 11.0 (Stata Corporation, TX, USA) were used in this meta-analysis. The link strength between MLH1 methylation and GC risk or clinicopathologic features was estimated by odds ratio (OR) with its 95% CI. The heterogeneity among the studies was assessed by Q-test and further quantified by the I2 metric [25]. If there was substantial heterogeneity (P<0.05 or I2 > 50%), a random effect model was used to pool the ORs; otherwise, a fixed effect model was employed [26]. P value < 0.05 was considered statistically significant. Egger's linear regression test were applied to examine whether the results existed publication bias [27]. All tests were two-sided, and P<0.05 indicated statistical significance. When heterogeneity was shown, sensitivity analysis was performed to identify heterogeneity sources. Subgroup analysis was carried out to explore the effect of country, ethnicity, and methylation testing methods.
SPSS 22.0 software (SPSS, Chicago, IBM, USA) were used in the current bioinformatics analysis. Person χ2 test was applied to evaluate the association of MLH1 methylation with clinicopathologic features. Kaplan-Meier curves were drew to outline the survival status and the differences between the groups were analyzed using the log-rank test. P values<0.05 were considered statistically significant.
Results
Meta-analysis
Study characteristics
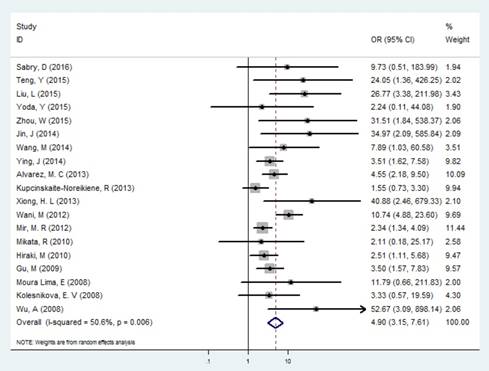
According to the literature selection criteria and search strategy, 26 studies [11, 17, 18, 28-50] were included in the present meta-analysis, including 2365 gastric cancer cases and 1563 nonmalignant controls. The study screening process is shown in Figure 1. Among these studies, 16 studies reporting 1794 cases and 1563 nonmalignant controls were selected to evaluate the relevance between MLH1 methylation and GC risk. Furthermore, 10 studies, including 476 intestinal GC and 290 diffuse GC, estimated the Lauren classification-based association; 9 studies assessed the tumor invasion-based association; 16 studies explored the lymph node metastasis-based association; 8 studies appraised the distant metastasis-based association and 9 studies including 219 stage Ⅰ-Ⅱ patients and 422 stage Ⅲ-Ⅳ patients evaluated the tumor stage-based association. These 26 studies were published between 2008 and 2016. All of them were written in English or Chinese. Of the studies, 10 came from China, 4 came from Brazil ,3 came from Japan, 2 came from Korea, 2 came from India, 1 came from Egypt, 1 came from Iran, 1 came from Lithuania, 1 came from Russia and 1 came from Spain, severally. The basic characteristics of all the included studies were summarized in Table 1. The NOS results showed that all the involved studies were at a higher quality level with scores ≥ 7. Full results of NOS quality assessment were summarized (Table S1).
MLH1 promoter methylation and GC risk
In the identification of GC and controls, slight heterogeneity was existed (I2 = 36.46% and P =0.006), therefore a random effect model was performed. Our results exhibited that the frequency of MLH1 promoter methylation was enhanced in patients with GC compared with control groups (OR= 4.895, 95% CI: 3.149-7.611, P<0.001, Figure 2, Table 2), showing that the MLH1 methylation status was significantly associated with the GC risk. We furthermore performed subgroup analyses stratified by country, ethnicity, testing methods, and materials respectively. Country-specific OR showed an increased risk for individuals with the MLH1 methylation compared with those without MLH1 methylation in China (OR=15.222, 95% CI: 5.395-42.952, P<0.001) and Japan (OR=2.452, 95% CI: 1.158-5.193, P<0.001). Then we calculated the pool OR for MLH1 promoter in the Asian subgroup, that was 5.949 (95% CI: 3.393-10.431, P<0.001) within a random effect model, and that for the Negroid subgroup was 5.017 (95% CI: 2.510-10.027, P<0.001) under a random effect model. But for the Caucasian subgroup, the pool OR was 1.744 (95% CI: 0.871-3.491, P= 0.116), showing no significance with MLH1 promoter methylation. Subgroup analysis based on the testing methods indicated that considerably increased risks were found in both MSP (OR=5.426, 95% CI: 3.215-9.156) and Methylight (OR=3.168, 95% CI:1.521-6.599) groups (Table 2). Testing materials analysis revealed that the pool OR was 4.472 (95% CI: 2.874-6.959, P<0.001) for the tissue and 12.538 (95% CI:1.861-84.463, P=0.009) for the blood. That is, the GC risk was significantly raised in both tissue and blood subgroup.
Forest plot of the correlation between MLH1 methylation and GC.
MLH1 promoter methylation and GC clinicopathologic features
Fixed-effects model was applied for Lauren classification, tumor invasion, distant metastasis status and tumor stage (all Ph > 0.1) and random-effects model was used for lymph node status (Ph < 0.1). There was no significant difference in MLH1 methylation detected in Lauren classification (OR=0.878, 95% CI: 0.619-1.244, P=0.463, Figure 3a), tumor invasion (OR=0.844, 95% CI: 0.568-1.253, P=0.400, Figure 3b), lymph node status (OR=0.929, 95% CI: 0.620-1.390, P=0.720, Figure 3c), distant metastasis status (OR=0.819, 95% CI: 0.481-1.396, P=0.464, Figure 3d) and tumor stage (OR=0.687, 95% CI: 0.455-1.039, P=0.075, Figure 3e) in gastric cancer(Table 3).
Flow chart of literature search and study selection.
Sensitivity analysis
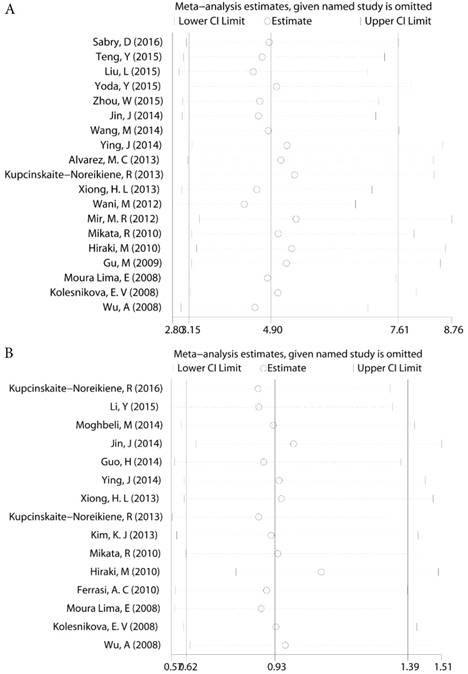
For those groups which existed slight heterogeneity (GC risk I2=36.46%, Ph=0.006, Lymph node status I2=37.30%, Ph=0.072, Figure 4), Sensitivity analysis was subsequently performed to detect the influence of individual study on the pooled estimate by omitting one study from the pooled analysis each time. The exclusion of each single study did not significantly change the pooled OR, suggesting that the results of the meta-analysis were robust.
Publication bias
As indicated in Table 3, slight publication bias was perceived by Egger's test and Begg's test in the contrast of cancer and control groups, and also in distant metastasis as well as tumor stage subgroups. There was no obvious publication bias stated in other analytic subgroups. (all P > 0.05).
Characteristics of the studies included in the meta-analysis
| Author | Year | Country | Ethnicity | Method | Sample type | Case(M/U) | Control(M/U) | Lauren classification (M/U) | Tumor invastion (M/U) | Lymph node status(M/U) | Distant metastasis(M/U) | TNM stage (M/U) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Intestinal | Diffuse | T1-T2 | T3-T4 | Negative | Positive | Negative | Positive | Ⅰ-Ⅱ | Ⅲ-Ⅳ | ||||||||
| Sabry, D | 2016 | Egypt | Negroids | MethyLight | Tissue | 10/0 | 20/9 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Kupcinskaite, R | 2016 | Spain | Caucasians | MSP | Tissue | 25/56 | NA | 13/29 | 12/27 | 7/14 | 18/42 | 11/17 | 14/39 | NA | NA | NA | NA |
| Yoda, Y | 2015 | Japan | Asians | Bead array | Tissue | 7/43 | 0/6 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Teng, Y | 2015 | China | Asians | MSP | Tissue | 13/27 | 0/24 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Liu, L | 2015 | China | Asians | MSP | Tissue | 24/26 | 1/29 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Li, Y | 2015 | China | Asians | MSP | Tissue | 22/80 | NA | 13/51 | 9/24 | 3/10 | 19/70 | 10/26 | 12/54 | 21/73 | 1/7 | 6/13 | 16/67 |
| Zhou, W | 2015 | China | Asians | MSP | Blood | 36/47 | 0/20 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Wang, M | 2014 | China | Asians | MethyLight | Tissue | 20/114 | 1/45 | 5/47 | 12/55 | NA | NA | NA | NA | 18/94 | 2/20 | NA | NA |
| Jin, J | 2014 | China | Asians | MSP | Tissue | 16/267 | 0/283 | NA | NA | 6/116 | 10/151 | 6/152 | 10/115 | 13/245 | 3/22 | NA | NA |
| Moghbeli, M | 2014 | Iran | Asians | MSP | Tissue | 13/38 | NA | 7/29 | 5/8 | 4/15 | 9/23 | 2/6 | 11/32 | NA | NA | 3/17 | 10/21 |
| Guo, H | 2014 | China | Asians | MSP | Tissue | 16/54 | NA | NA | NA | 6/20 | 10/34 | 6/15 | 10/39 | 15/51 | 1/3 | 8/25 | 8/29 |
| Ying, J | 2014 | China | Asians | MSP | Tissue | 29/91 | 10/110 | NA | NA | NA | NA | 3/13 | 26/78 | 22/75 | 7/16 | 3/15 | 26/76 |
| Xiong, H. L | 2013 | China | Asians | MSP | Blood | 19/394 | 0/413 | NA | NA | 8/170 | 11/224 | 9/222 | 10/172 | 15/362 | 4/32 | NA | NA |
| Kupcinskaite, R | 2013 | Lithuania | Caucasians | MSP | Tissue | 22/47 | 16/53 | 10/25 | 11/21 | NA | NA | 10/15 | 11/31 | NA | NA | 4/9 | 17/36 |
| Alvarez, M. C | 2013 | Brazil | Negroids | MSP | Tissue | 36/56 | 12/85 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Kim, K. J | 2013 | South Korea | Asians | MethyLight | Tissue | 80/22 | NA | 72/17 | 1/3 | 19/9 | 61/13 | 33/9 | 47/13 | NA | NA | 47/15 | 33/7 |
| Wani, M | 2012 | India | Asians | MSP | Tissue | 51/19 | 14/56 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Mir, M. R | 2012 | India | Asians | MSP | Tissue | 104/26 | 82/48 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Alves, M. K | 2011 | Brazil | Negroids | MSP | Tissue | 25/51 | NA | 15/33 | 10/18 | NA | NA | NA | NA | NA | NA | NA | NA |
| Mikata, R | 2010 | Japan | Asians | MSP | Tissue | 2/19 | 1/20 | NA | NA | NA | NA | 0/6 | 2/13 | NA | NA | 0/8 | 2/11 |
| Hiraki, M | 2010 | Japan | Asians | MethyLight | Tissue | 32/17 | 21/28 | 17/10 | 7/15 | NA | NA | 7/13 | 25/4 | NA | NA | 12/12 | 20/5 |
| Ferrasi, A. C | 2010 | Brazil | Negroids | MSP | Tissue | 27/62 | NA | 19/38 | 8/23 | NA | NA | 6/11 | 21/50 | NA | NA | NA | NA |
| Gu, M | 2009 | Korea | Asians | MSP | Tissue | 39/15 | 23/31 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Moura Lima, E | 2008 | Brazil | Negroids | MSP | Tissue | 10/36 | 0/20 | 6/20 | 4/16 | 3/9 | 7/27 | 5/11 | 5/25 | 10/31 | 0/5 | NA | NA |
| Kolesnikova, E. V | 2008 | Russia | Caucasians | MSP | Blood | 5/15 | 2/20 | NA | NA | 2/8 | 3/7 | 2/7 | 3/8 | 4/12 | 1/3 | NA | NA |
| Wu, A | 2008 | China | Asians | MSP | Tissue | 18/42 | 0/60 | NA | NA | NA | NA | 14/37 | 4/5 | NA | NA | 5/17 | 13/25 |
Abbreviations: M, methylations; U, unmethylations; MSP, methylation-specifc PCR; NA, not available
Bioinformatical analysis
MLH1 promoter methylation and GC risk
We extracted 2 GEO series (GSEs) within 180 GEO series both related to gastric cancer and methylation. All two GSEs were solely derived from human tissues and used methylation probe to detect the methylation rate. We screened two methylation location, cg18320188 and cg02279071 on the CpG island of MLH1 DNA sense strand. They located on chromosome 3 (37008972 - 37010459). Their sequences were showed in Table S2. After analyze the data using GEO2R, we found that MLH1 promoter methylation showed a high level in GC compared to normal tissues (P=0.0149 from GSE30601 probe cg18320188, P=0.0442 from GSE25869 probe cg02279071).
Then we search data covering DNA methylation and gene expression on the website of MethHC (A database of DNA Methylation and gene expression in Human Cancer http://methhc.mbc.nctu.edu.tw/php/index.php) and the TCGA (The Cancer Genome Atlas) [51]. The data showed that there were significant differences in methylation levels between cancer and normal tissues (P<0.005). All the results expounded that MLH1 methylation status was considerably related with the GC risk.
Subgroup analysis of MLH1 promoter methylation in gastric cancers compared with controls.
| Studies | Heterogeneity test | Test for overall effect | ||||
|---|---|---|---|---|---|---|
| I2 (%) | Ph | OR (95% CI) | P-value | |||
| Gastric cancer risk | 19 | 36.46% | 0.006 | 4.895 (3.149-7.611) | <0.001 | |
| Subgroup | ||||||
| Country | ||||||
| China | 8 | 46.20% | 0.072 | 15.222 (5.395-42.952) | <0.001 | |
| Japan | 3 | 0.00% | 0.989 | 2.452(1.158-5.193 ) | <0.001 | |
| Ethnicity | ||||||
| Asians | 14 | 55.70% | 0.006 | 5.949 (3.393-10.431) | <0.001 | |
| Caucasians | 2 | 0.00% | 0.436 | 1.744 (0.871-3.491) | 0.116 | |
| Negroids | 3 | 0.00% | 0.729 | 5.017 (2.510-10.027) | <0.001 | |
| Methods | ||||||
| MSP | 15 | 59.20% | 0.002 | 5.426 (3.215-9.156) | <0.001 | |
| Methylight | 3 | 0.00% | 0.408 | 3.168 (1.521-6.599) | 0.002 | |
| Bead array | 1 | NA | NA | 2.241 (0.114-44.084) | 0.595 | |
| Materials | ||||||
| Tissue | 16 | 50.30% | 0.011 | 4.472 (2.874-6.959) | <0.001 | |
| Blood | 3 | 46.00% | 0.157 | 12.538 (1.861-84.463) | 0.009 | |
Note: Values in bold indicate statistical significance.
Abbreviations: CI, confidence interval; Ph, P-value of Q test for heterogeneity among studies; OR, odds ratio; NA, not available
Association of MLH1 promoter methylation with clinicopathologic features in gastric cancer.
| Clinicopathological features | Studies | Heterogeneity test | Statistical model | Test for overall effect | Begg's test | Egger's test | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| I2 (%) | Ph | OR (95% CI) | P-value | z-value | P-value | t-value | P-value | |||
| Gastric cancer risk | 19 | 36.46% | 0.006 | R | 4.895 (3.149-7.611) | <0.001 | 0.190 | 0.234 | 3.110 | 0.006 |
| Lauren classification | 10 | 1.30% | 0.426 | F | 0.878 (0.619-1.244) | 0.463 | 0.000 | 1.000 | 1.470 | 0.180 |
| Tumor invasion | 9 | 0.00% | 0.949 | F | 0.844 (0.568-1.253) | 0.400 | 0.310 | 0.754 | 0.250 | 0.806 |
| Lymph node status | 15 | 37.30% | 0.072 | R | 0.929 (0.620-1.390) | 0.720 | 0.990 | 0.322 | -1.050 | 0.314 |
| Distant metastasis | 8 | 0.00% | 0.479 | F | 0.819 (0.481-1.396) | 0.464 | 1.360 | 0.174 | 2.050 | 0.087 |
| Tumor stage | 9 | 0.70% | 0.428 | F | 0.687 (0.455-1.039) | 0.075 | 0.620 | 0.536 | -0.160 | 0.877 |
Abbreviations: R, random effect model; F, fixed effect model
Association of MLH1 promoter methylation with clinicopathologic features in gastric cancer based on bioinfromatic analysis
| Clinical features | Methylation status | ||||||
|---|---|---|---|---|---|---|---|
| cg18320188 | cg02279071 | ||||||
| M | U | P value | M | U | P value | ||
| Lauren classification | Intestinal | 85 | 68 | 0.57 | 82 | 71 | 0.27 |
| Diffuse | 36 | 34 | 32 | 38 | |||
| Tumor invastion | T1-T2 | 42 | 43 | 0.90 | 50 | 35 | 0.05 |
| T3-T4 | 127 | 126 | 118 | 135 | |||
| Lymph node status | Negative | 55 | 50 | 0.56 | 58 | 47 | 0.19 |
| Positive | 111 | 116 | 108 | 119 | |||
| Distant metastasis | Negative | 152 | 149 | 0.79 | 155 | 146 | 0.43 |
| Positive | 9 | 10 | 8 | 11 | |||
| TNM stage | stage Ⅰ-Ⅱ | 79 | 75 | 0.90 | 83 | 71 | 0.18 |
| stage Ⅲ-Ⅳ | 88 | 86 | 81 | 93 | |||
MLH1 promoter methylation and GC clinicopathologic features
We analyzed the association between MLH1 methylation and clinicopathologic features such as Lauren classification, tumor invasion, distant metastasis status and tumor stage using the same methylation probe as GSE30601 cg18320188 and GSE25869 cg02279071. No correlation was found between the two. (Table 4).
MLH1 promoter methylation and GC prognosis
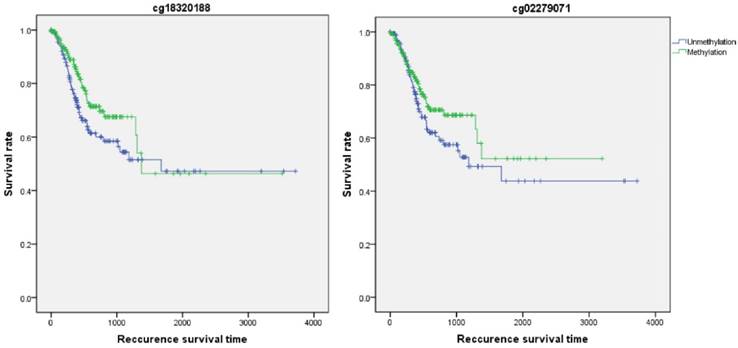
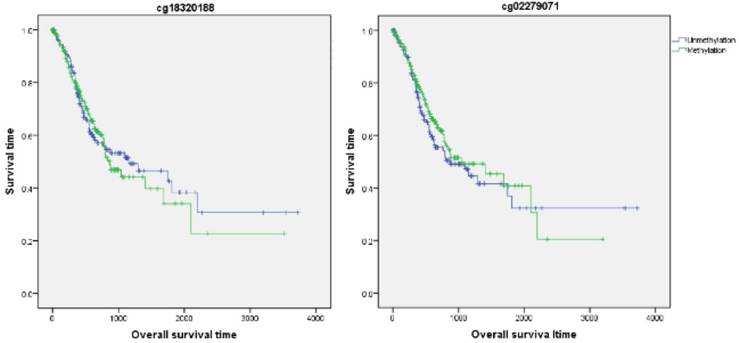
Firstly, the influence of MLH1 methylation on recurrence free survival (RFS) time was assessed. A total of 338 patients with recurrence free survival time related data were enrolled in this section. Analytic results of Kaplan-Meier curve and Log-Rank test suggested that MLH1 methylation was not significantly associated with RFS (Table 5, Figure 5). And then, the association between MLH1 methylation and overall survival time (OS) was also evaluated. Similar to RFS, the results did not show any correlation between MLH1 methylation and OS of GC (Table 5, Figure 5).
Forest plot of the correlation between MLH1 methylation and GC clinicopathologic features. a. Forest plot of the correlation between MLH1 methylation and Lauren classification. b. Forest plot of the correlation between MLH1 methylation and tumor invasion. c. Forest plot of the correlation between MLH1 methylation and lymph node status. d. Forest plot of the correlation between MLH1 methylation and distant metastasis status. e. Forest plot of the correlation between MLH1 methylation and tumor stage.

Association of MLH1 promoter methylation with prognosis in gastric cancer based on bioinfromatic analysis.
| Methylation probe | RFS | OS | |||||
|---|---|---|---|---|---|---|---|
| Median survival time | X2 value | P value | Median survival time | X2 value | P value | ||
| cg18320188 | Unmethylation | 1676 | 3.09 | 0.08 | 1153 | 0.06 | 0.81 |
| Methylation | 1376 | 869 | |||||
| cg02279071 | Unmethylation | 1184 | 2.21 | 0.14 | 869 | 0.63 | 0.43 |
| Methylation | NA | 1043 | |||||
Discussion
The methylation frequency of the MLH1 promoter was inconsistent in GC with a range from 4% to 100% [37]. Thus, the association between MLH1 promoter methylation and GC exists controversy. To get a more credible conclusion, we performed a systematic review and meta and bioinformatic analysis using the previously published studies and database to assess the relevance between MLH1 methylation and GC. Our study showed a strong correlation between MLH1 methylation and GC risk, indicating that MLH1 methylation could predict the occurrence of gastric cancer as a convincing biomarker. No significant correlation was found between MLH1 methylation and GC clinicopathological behavior as well as prognosis.
Sensitivity analysis a. The sensitivity analysis of MLH1 methylation and GC b. The sensitivity analysis of MLH1 methylation and lymph node status.
In the present meta-analysis, 2365 GC cases and 1563 control samples, from 26 studies were selected totally. Compared with the controls, the accumulated OR of MLH1 methylation in GC patients was 4.895 (95% CI: 3.149-7.611, P<0.001). It was in accordance with earlier studies in which the frequency of MLH1 promoter methylation in GC was enhanced compared with control groups [30, 34, 37]. Our bioinformatics analysis based on GEO and TCGA also showed that MLH1 promoter methylation sustained a high level in GC compared to normal tissues (P=0.0149, P=0.0442 respectively). The results drew from both meta and bioinformatics analysis suggested that the methylation of MLH1 was significantly associated with increased GC risk. The consequence may be caused by two main reasons. Firstly, Mismatch repair (MMR) deficiency leads to a tumour phenotype known as microsatellite instability (MSI), in which cells accumulate genetic errors [19]. MLH1 is a functional member of DNA MMR system, which is responsible for the replacement of the mispaired nucleotides in the genome during the DNA replication [11]. When performing mismatch repair function, the heteroduplex composed of MLH1 and PMS2 could combine with DNA fragment thereby trigger the repair process. In addition to that it reactivates cell cycle arrest and caspase-mediated apoptosis in response to DNA damage, promotes cell mobility and interacts with other significant cell signaling proteins [52-55]. The aberration of the MLH1 function could lead to the dysfunction of DNA MMR system therefore result in the GC carcinogenesis [56]. Secondly, MLH1 is also a tumor suppressor gene, which expression is repressed by promoter methylation. And that's exactly one of the key features of cancer [57]. As a tumor suppressor gene MLH1 silencing mediated by aberrant promoter DNA hypermethylation could lead to the tumor information [58]. Based on the present results and analysis, we could conclude that MLH1 methylation significantly elevated the risk of GC and might be a probable biomarker for the prediction of GC.
The current meta and bioinformatic analysis revealed that no significant difference of MLH1 methylation in relation to clinicopathological features, such as Lauren classification, tumor invasion, lymph node status, distant metastasis and tumor stage in GC (all P > 0.1), suggesting that the methylation status of MLH1 promoter may not affect the biological behavior of GC. The phenomenon that MLH1 methylation increased the risk of GC but not related with clinicopathological features hinted that DNA methylation occurs early in the multistep process of gastric carcinogenesis. Bischoff et al. reported that MLH1 methylation with a consequent protein decrease occurred early during endometrial carcinogenesis [13]. And the coherent conclusions were also elucidated in lung cancer and breast cancer. [14-16]. Thus, we can infer that MLH1 methylation may contribute to initial carcinogenesis but not progression of GC.
It has been reported that MLH1 methylation are associated with poor prognosis in cancers, such as in non-small cell lung cancer and ovarian cancer after chemotherapy [59, 60]. But the current bioinformatics analysis revealed that no relationship between MLH1 methylation with the prognosis of GC including RFS and OS based on the data from TCGA database. Only one study in the meta-analysis revealed that among oxaliplatin-treated patients, OS was longer in the MLH1 unmethylated group than in the MLH1 methylated group [32]. The phenomenon suggested that MLH1 methylation may not affect the prognosis of GC. The different effects of MLH1 methylation on prognosis in different tumors may be due to the organ specificity. There may exist some gastric specific indicators commonly affected the consequence of MLH1 methylation and prognosis [61]. Further expanding of the sample size could be conduct to verify the impact of MLH1 methylation on GC prognosis.
Recurrence survival time analysis of GC correlated with MLH1 methylation.
Overall survival time analysis of GC correlated with MLH1 methylation.
Our study had some limitations. First, our meta-analysis could not adjust for confounding factors such as age, sex, smoking behavior, or H.pylori infection due to some relevant data could not be extracted. Second, the studies included was only searched by English and Chinese, the other language of studies were not included therefore some important researches may be omitted. Third, up to now, few studies reported the association of MLH1 methylation with prognosis of GC. On this point, we did only the bioinformatics analysis and failed to meta-analysis. There is a need to strengthen the prognosis-based association study in the future. Fourth, Heterogeneity existed in our meta-analysis. Although we try to eliminate the heterogeneity by subgroup analysis according to the potential heterogeneous factors, such as geographic region, ethnicity, testing methods and materials, there is still some heterogeneity in this meta-analysis because some original studies did not provide the necessary information.
In summary, this systematic review and meta and bioinformatic analysis showed a strong correlation between MLH1 methylation and GC risk and no significant correlation was found between MLH1 methylation and GC clinicopathological behavior as well as prognosis. The present results suggest that MLH1 methylation can be used as a favorable molecular marker for the prediction of GC and it might not affect GC behavior. Further study could be conducted to verify the impact of MLH1 methylation on GC prognosis.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work is supported by grants from the 13th five years for the National Key Research and Development Program & Key Special Project (2016YFC1303202).
Author Contributions
Conceived and designed the experiments: Yuan Yuan and Liping Sun. Analyzed the data: Shixuan Shen, Xiaohui Chen and Hao Li. Wrote the paper: Shixuan Shen. Revised the manuscript: Yuan Yuan.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683-92
2. Qureshi I, Mehler M. Emerging role of epigenetics in stroke: part 1: DNA methylation and chromatin modifications. Arch Neurol. 2010;67:1316-22
3. Issa JP. Opinion - CpG island methylator phenotype in cancer. Nature Reviews Cancer. 2004;4:988-93
4. Calcagno DQ, Gigek CO, Chen ES, Burbano RR, Smith Mde A. DNA and histone methylation in gastric carcinogenesis. World journal of gastroenterology. 2013;19:1182-92
5. Tahara T, Arisawa T. DNA methylation as a molecular biomarker in gastric cancer. Epigenomics. 2015;7:475-86
6. Yuan W, Chen J, Shu Y, Liu S, Wu L, Ji J. et al. Correlation of DAPK1 methylation and the risk of gastrointestinal cancer: A systematic review and meta-analysis. PloS one. 2017;12:e0184959
7. Zhang Z, Xin S, Gao M, Cai Y. Promoter hypermethylation of MGMT gene may contribute to the pathogenesis of gastric cancer: A PRISMA-compliant meta-analysis. Medicine. 2017;96:e6708
8. Ryan E, Sheahan K, Creavin B, Mohan HM, Winter DC. The current value of determining the mismatch repair status of colorectal cancer: A rationale for routine testing. Critical reviews in oncology/hematology. 2017;116:38-57
9. Zhu L, Li Z, Wang Y, Zhang C, Liu Y, Qu X. Microsatellite instability and survival in gastric cancer: A systematic review and meta-analysis. Molecular and clinical oncology. 2015;3:699-705
10. Belfield E, Ding Z, Jamieson F, Visscher A, Zheng S, Mithani A. et al. DNA mismatch repair preferentially protects genes from mutation. Genome Res. 2017
11. Moghbeli M, Moaven O, Memar B, Raziei HR, Aarabi A, Dadkhah E. et al. Role of hMLH1 and E-cadherin promoter methylation in gastric cancer progression. Journal of gastrointestinal cancer. 2014;45:40-7
12. Kim S, Klempner S, Park S, Park J, Park Y, Lim H. et al. Correlating programmed death ligand 1 (PD-L1) expression, mismatch repair deficiency, and outcomes across tumor types: implications for immunotherapy. Oncotarget. 2017;8:77415-23
13. Bischoff J, Ignatov A, Semczuk A, Schwarzenau C, Ignatov T, Krebs T. et al. hMLH1 promoter hypermethylation and MSI status in human endometrial carcinomas with and without metastases. Clinical & experimental metastasis. 2012;29:889-900
14. Pastuszak-Lewandoska D, Kordiak J, Antczak A, Migdalska-Sek M, Czarnecka KH, Gorski P. et al. Expression level and methylation status of three tumor suppressor genes, DLEC1, ITGA9 and MLH1, in non-small cell lung cancer. Medical oncology (Northwood, London, England). 2016;33:75
15. Geng X, Wang F, Zhang L, Zhang WM. Loss of heterozygosity combined with promoter hypermethylation, the main mechanism of human MutL Homolog (hMLH1) gene inactivation in non-small cell lung cancer in a Chinese population. Tumori. 2009;95:488-94
16. Alkam Y, Mitomi H, Nakai K, Himuro T, Saito T, Takahashi M. et al. Protein expression and methylation of DNA repair genes hMLH1, hMSH2, MGMT and BRCA1 and their correlation with clinicopathological parameters and prognosis in basal-like breast cancer. Histopathology. 2013;63:713-25
17. Kupcinskaite-Noreikiene R, Ugenskiene R, Noreika A, Rudzianskas V, Gedminaite J, Skieceviciene J. et al. Gene methylation profile of gastric cancerous tissue according to tumor site in the stomach. BMC cancer. 2016;16:40
18. Sabry D, Ahmed R, Abdalla S, Fathy W, Eldemery A, Elamir A. Braf, Kras and Helicobacter pylori epigenetic changes-associated chronic gastritis in Egyptian patients with and without gastric cancer. World journal of microbiology & biotechnology. 2016;32:92
19. Baniak N, Senger JL, Ahmed S, Kanthan SC, Kanthan R. Gastric biomarkers: a global review. World journal of surgical oncology. 2016;14:212
20. Nakamura J, Tanaka T, Kitajima Y, Noshiro H, Miyazaki K. Methylation-mediated gene silencing as biomarkers of gastric cancer: a review. World J Gastroenterol. 2014;20:11991-2006
21. Karimi P, Islami F, Anandasabapathy S, Freedman N, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23:700-13
22. Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-9
23. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. European journal of epidemiology. 2010;25:603-5
24. Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M. et al. NCBI GEO: archive for functional genomics data sets-update. Nucleic acids research. 2013;41:D991-5
25. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Bmj. 2003;327:557-60
26. Zintzaras E, Ioannidis JP. Heterogeneity testing in meta-analysis of genome searches. Genetic epidemiology. 2005;28:123-37
27. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Bmj. 1997;315:629-34
28. Yoda Y, Takeshima H, Niwa T, Kim JG, Ando T, Kushima R. et al. Integrated analysis of cancer-related pathways affected by genetic and epigenetic alterations in gastric cancer. Gastric cancer: official journal of the International Gastric Cancer Association and the Japanese Gastric Cancer Association. 2015;18:65-76
29. Teng Y, Dai D, Shen W, Liu H. Effect of methylation of hMLH1 gene promotor on stage tumorigenesis and progression of human gastric cancer. Zhonghua wei chang wai ke za zhi = Chinese journal of gastrointestinal surgery. 2015;18:166-70
30. Liu L, Yang X. Implication of Reprimo and hMLH1 gene methylation in early diagnosis of gastric carcinoma. International journal of clinical and experimental pathology. 2015;8:14977-82
31. Zhou W, Lu S, Wei HY, Miao ZG. Value of P16、E-cad and hMLH1 Gene Promoter Methylation in Early Detection of Gastric Carcinoma. The Practical Journal of Cancer. 2015;30:1758-60
32. Li Y, Yang Y, Lu Y, Herman JG, Brock MV, Zhao P. et al. Predictive value of CHFR and MLH1 methylation in human gastric cancer. Gastric cancer: official journal of the International Gastric Cancer Association and the Japanese Gastric Cancer Association. 2015;18:280-7
33. Wang M, Li Y, Gao J, Li Y, Zhou J, Gu L. et al. p16 Methylation is associated with chemosensitivity to fluorouracil in patients with advanced gastric cancer. Medical oncology (Northwood, London, England). 2014;31:988
34. Jin J, Xie L, Xie CH, Zhou YF. Aberrant DNA methylation of MGMT and hMLH1 genes in prediction of gastric cancer. Genetics and molecular research: GMR. 2014;13:4140-5
35. Guo H, Yan W, Yang Y, Guo M. Promoter region methylation of DNA damage repair genes in human gastric cancer. Zhonghua yi xue za zhi. 2014;94:2193-6
36. Ying J, Pan HM. Association between promoter methylaiton and protein expression of tumor suppressor genes with clinical characteristics in gastric carcinoma and its effect on the biological function of gastric cancer. ZHejiang University Master's degree paper. 2014
37. Xiong HL, Liu XQ, Sun AH, He Y, Li J, Xia Y. Aberrant DNA methylation of P16, MGMT, hMLH1 and hMSH2 genes in combination with the MTHFR C677T genetic polymorphism in gastric cancer. Asian Pacific journal of cancer prevention: APJCP. 2013;14:3139-42
38. Kupcinskaite-Noreikiene R, Skieceviciene J, Jonaitis L, Ugenskiene R, Kupcinskas J, Markelis R. et al. CpG island methylation of the MLH1, MGMT, DAPK, and CASP8 genes in cancerous and adjacent noncancerous stomach tissues. Medicina (Kaunas, Lithuania). 2013;49:361-6
39. Alvarez MC, Santos JC, Maniezzo N, Ladeira MS, da Silva AL, Scaletsky IC. et al. MGMT and MLH1 methylation in Helicobacter pylori-infected children and adults. World journal of gastroenterology. 2013;19:3043-51
40. Kim KJ, Lee TH, Cho NY, Yang HK, Kim WH, Kang GH. Differential clinicopathologic features in microsatellite-unstable gastric cancers with and without MLH1 methylation. Human pathology. 2013;44:1055-64
41. Wani M, Afroze D, Makhdoomi M, Hamid I, Wani B, Bhat G. et al. Promoter methylation status of DNA repair gene (hMLH1) in gastric carcinoma patients of the Kashmir valley. Asian Pacific journal of cancer prevention: APJCP. 2012;13:4177-81
42. Mir MR, Shabir N, Wani KA, Shaff S, Hussain I, Banday MA. et al. Association between p16, hMLH1 and E-cadherin promoter hypermethylation and intake of local hot salted tea and sun-dried foods in Kashmiris with gastric tumors. Asian Pacific journal of cancer prevention: APJCP. 2012;13:181-6
43. Alves MK, Ferrasi AC, Lima VP, Ferreira MV, de Moura Campos Pardini MI, Rabenhorst SH. Inactivation of COX-2, HMLH1 and CDKN2A gene by promoter methylation in gastric cancer: relationship with histological subtype, tumor location and Helicobacter pylori genotype. Pathobiology: journal of immunopathology, molecular and cellular biology. 2011;78:266-76
44. Mikata R, Fukai K, Imazeki F, Arai M, Fujiwara K, Yonemitsu Y. et al. BCL2L10 is frequently silenced by promoter hypermethylation in gastric cancer. Oncology reports. 2010;23:1701-8
45. Hiraki M, Kitajima Y, Sato S, Mitsuno M, Koga Y, Nakamura J. et al. Aberrant gene methylation in the lymph nodes provides a possible marker for diagnosing micrometastasis in gastric cancer. Annals of surgical oncology. 2010;17:1177-86
46. Ferrasi AC, Pinheiro NA, Rabenhorst SH, Caballero OL, Rodrigues MA, de Carvalho F. et al. Helicobacter pylori and EBV in gastric carcinomas: methylation status and microsatellite instability. World journal of gastroenterology. 2010;16:312-9
47. Gu M, Kim D, Bae Y, Choi J, Kim S, Song S. Analysis of microsatellite instability, protein expression and methylation status of hMLH1 and hMSH2 genes in gastric carcinomas. Hepato-gastroenterology. 2009;56:899-904
48. Moura Lima E, Ferreira Leal M, Cardoso Smith Mde A, Rodriguez Burbano R, Pimentel de Assumpcao P, Bello MJ. et al. DNA mismatch repair gene methylation in gastric cancer in individuals from northern Brazil. Biocell: official journal of the Sociedades Latinoamericanas de Microscopia Electronica et al. 2008;32:237-43
49. Kolesnikova EV, Tamkovich SN, Bryzgunova OE, Shelestyuk PI, Permyakova VI, Vlassov VV. et al. Circulating DNA in the blood of gastric cancer patients. Annals of the New York Academy of Sciences. 2008;1137:226-31
50. Wu AC, Liu CS. The association between mismatch repair gene hMLH1 methylation and gastric cancer. Qingdao University Master's degree paper. 2008
51. Huang WY, Hsu SD, Huang HY, Sun YM, Chou CH, Weng SL. et al. MethHC: a database of DNA methylation and gene expression in human cancer. Nucleic acids research. 2015;43:D856-61
52. Hinrichsen I, Ernst BP, Nuber F, Passmann S, Schafer D, Steinke V. et al. Reduced migration of MLH1 deficient colon cancer cells depends on SPTAN1. Molecular cancer. 2014;13:11
53. Siehler SY, Schrauder M, Gerischer U, Cantor S, Marra G, Wiesmuller L. Human MutL-complexes monitor homologous recombination independently of mismatch repair. DNA repair. 2009;8:242-52
54. McDaid JR, Loughery J, Dunne P, Boyer JC, Downes CS, Farber RA. et al. MLH1 mediates PARP-dependent cell death in response to the methylating agent N-methyl-N-nitrosourea. British journal of cancer. 2009;101:441-51
55. Kanao R, Hanaoka F, Masutani C. A novel interaction between human DNA polymerase eta and MutLalpha. Biochemical and biophysical research communications. 2009;389:40-5
56. Torgovnick A, Schumacher B. DNA repair mechanisms in cancer development and therapy. Frontiers in genetics. 2015;6:157
57. Springuel L, Losdyck E, Saussoy P, Turcq B, Mahon FX, Knoops L. et al. Loss of mutL homolog-1 (MLH1) expression promotes acquisition of oncogenic and inhibitor-resistant point mutations in tyrosine kinases. Cellular and molecular life sciences: CMLS. 2016;73:4739-48
58. Guerrero-Preston R, Michailidi C, Marchionni L, Pickering CR, Frederick MJ, Myers JN. et al. Key tumor suppressor genes inactivated by "greater promoter" methylation and somatic mutations in head and neck cancer. Epigenetics. 2014;9:1031-46
59. Seng TJ, Currey N, Cooper WA, Lee CS, Chan C, Horvath L. et al. DLEC1 and MLH1 promoter methylation are associated with poor prognosis in non-small cell lung carcinoma. British journal of cancer. 2008;99:375-82
60. Gifford G, Paul J, Vasey PA, Kaye SB, Brown R. The acquisition of hMLH1 methylation in plasma DNA after chemotherapy predicts poor survival for ovarian cancer patients. Clinical cancer research: an official journal of the American Association for Cancer Research. 2004;10:4420-6
61. Balgkouranidou I, Matthaios D, Karayiannakis A, Bolanaki H, Michailidis P, Xenidis N. et al. Prognostic role of APC and RASSF1A promoter methylation status in cell free circulating DNA of operable gastric cancer patients. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2015;778:46-51
Author contact
![]() Corresponding author: Dr. Yuan Yuan, Tumor Etiology and Screening Department of Cancer Institute and General Surgery, The First Hospital of China Medical University, North Nanjing Street 155#, Heping District, Shenyang110001, China. Telephone: +86-024-83282153; Fax: +86-024-83282383. Email: yuanyuanedu.cn
Corresponding author: Dr. Yuan Yuan, Tumor Etiology and Screening Department of Cancer Institute and General Surgery, The First Hospital of China Medical University, North Nanjing Street 155#, Heping District, Shenyang110001, China. Telephone: +86-024-83282153; Fax: +86-024-83282383. Email: yuanyuanedu.cn

 Global reach, higher impact
Global reach, higher impact