3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(14):2549-2558. doi:10.7150/jca.24560 This issue Cite
Review
The emergence of long non-coding RNAs in hepatocellular carcinoma: an update
1. Key Laboratory of Carcinogenesis of the Chinese Ministry of Health and the Key Laboratory of Carcinogenesis and Cancer Invasion of Chinese Ministry of Education, Xiangya Hospital, Central South University, Changsha 410078, P.R. China; Cancer Research Institute, Central South University, Changsha 410078, P.R. China.
2. Guangdong Province Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Research Center of Medicine, Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University, Guangzhou 510120, P.R. China.
3. Guangdong Provincial Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University, Guangzhou 510120, China.
4. Breast Tumor Center, Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University, Guangzhou, Guangdong 510120, China.
5. Department of Oncology, the third Xiangya Hospital, Central South University, Changsha 410013, P.R. China.
* Li Peng and Xiao-Qing Yuan are equally contributed to this work.
Received 2017-12-25; Accepted 2018-3-31; Published 2018-6-22
Abstract
Hepatocellular carcinoma (HCC) accounting for roughly 90% of all primary liver neoplasms is the sixth most frequent neoplasm and the second prominent reason of tumor fatality worldwide. As regulators of diverse biological processes, long non-coding RNAs (lncRNAs) are involved in onset and development of neoplasms. With the continuous booming of well-featured lncRNAs in HCC from 2016 to now, we reviewed the newly-presented comprehension about the relationship between lncRNAs and HCC in this study. To be specific, we summarized the overview function and study tools of lncRNAs, elaborated the roles of lncRNAs in HCC, and sketched the molecule mechanisms of lncRNAs in HCC. In addition, the application of lncRNAs serving as biomarkers in early diagnosis and outcome prediction of HCC patients was highlighted.
Keywords: long non-coding RNAs, hepatocellular carcinoma, function, mechanism, diagnosis, prognostic biomarker
Introduction
Hepatocellular carcinoma (HCC) accounts for roughly 90% of all primary liver neoplasms, and is the sixth most frequent neoplasm and the second prominent reason of tumor fatality worldwide [1]. The highest morbidity of HCC has been found in China and the Asia-Pacific area, which accounts for >50% of HCC cases in the world [2]. Meanwhile, liver cancer is fatal in both developed and developing countries, with the 5-year overall survival rate generally lower than 20% [3]. HCC is the sole solid neoplasm that could be cured by liver transplant which can simultaneously heal the neoplasm and latent cirrhosis, and which is not influenced by the extent of liver function impairment [4]. However, the extreme shortage of liver donors results in procrastination before transplantation, and increasing the risk of tumor progression, and transplantation failure during this period [5]. These suggested that HCC is a devastating disease with disappointing outcomes and limited therapeutic options. Similar to other cancers, HCC is characterized by the gradual accumulation of genetic and epigenetic changes [6-8]. Among these alterations, long non-coding RNAs (lncRNAs) play crucial roles in the initiation and progression of HCC.
LncRNAs are transcripts with more than 200 bp in length, and have incrementally been recognized as the emerging star in neoplasm study as their essential function in tumor biology [9-11]. These transcripts are lack of the protein-coding potential in general and are particularly ubiquitous in almost whole livings [9]. LncRNAs have emerged as regulators in diverse biological processes [12-14] containing cellular proliferation, differentiation, motility, invasiveness, survival and so on [15-17]. Mounting evidences have demonstrated that lncRNAs are involved in tumor onset and development [18, 19], and their expression is frequently deregulated in cancers [20-27]. LncRNAs are strikingly cell type-specificity and cancer type-specificity in expression which are relatively stable [18, 28]. Therefore, lncRNAs stand a chance to serve as a kind of desired indicators with underlying utilizations in neoplasm divination, early-discovery, classification and treatment.
With the continuous booming of well-featured lncRNAs in HCC from 2016 to now, a great quantity of newly-presented researches is coming into being on the relationship between lncRNAs and HCC. Here, we reviewed the present comprehension of lncRNAs in HCC. To be specific, we summarized the overview function and study tools of lncRNAs, elaborated the roles of lncRNAs in HCC, and sketched the molecule mechanisms of lncRNAs in HCC. Meanwhile, the utilization of lncRNAs as markers in HCC early-diagnosis and outcome prediction was also highlighted.
Tools for the research of lncRNAs
A mounting number of databases about lncRNAs are developed to assist in the research of lncRNAs that include the functions of lncRNAs under the physiological state and pathological conditions. Over twenty databases touching upon the biological properties of lncRNAs are outlined in Table 1. These databases will dramatically facilitate a better understanding of lncRNAs which are essential members of epigenetic regulation, and their interaction with other RNAs.
Dysregulation and roles of lncRNAs in HCC
Tumor progression and recurrence
HCC is a complicated disease referred to multiple factors. A growing body of evidence suggested that lncRNAs involved in the occurrence and development of diverse neoplasm containing HCC. Importantly, some could affect the features of neoplasm, including proliferation, apoptosis, motility, invasiveness and angiogenesis. As an example, HOTAIR expression levels increased in both HCC tumor tissues and HepG2 cells, which promoted HCC progression [29] and were bound up with earlier relapse [30, 31]. Those showed the importance of HOTAIR in the evolvement and the relapse of HCC. Furthermore, lncRNAs could be involved in tumor progression and had potential to serve as an attractive target for precision therapy in HCC.
Invasiveness and metastasis
It is well known that invasiveness is an origin in worse disease prognosis and higher recurrence of neoplasm patients [32], which was partly correlated with certain behaviors of cancerous cells. Studies have showed that lncRNAs were related to vital growth-boosting properties and their deregulation dedicated to the survival of cancerous cells. LINC00052 [33], ZEB1-AS1 [34] and LINC01225 [35] was showed to accelerate cellular motility and invasiveness. In short, these lncRNAs could function as oncogenes via facilitating invasion and metastasis of HCC cells. Inversely, it was indicated that CPS1-IT1 [36], lncRNA XIST [37] and lncRNA FTX [38] were down-regulated in HCC samples and inhibited the relapse and metastasis of HCC cells. These lncRNAs act as tumor suppressor genes via restraining invasion and metastasis in HCC.
Proliferation and apoptosis
A flood of literatures have mirrored that lncRNAs participated in the development of HCC via modulating cell proliferation and apoptosis. On the one aspect, some lncRNAs could accelerate cell proliferation while inhibit cell apoptosis in HCC. It was demonstrated that the expression of XIST [39], lncRNA HOST2 [40], HOXA-AS2 [41], CCHE1 [42] and AFAP1-AS1 [43] was significantly elevated in HCC tumor tissues and/or cell lines, which promoted cell proliferation while protected cells from apoptosis in HCC. Conversely, lncRNA AK058003 [44], lincRNA-p21 [45] and lncRNA XIST [37] were revealed to decrease in HCC tumors, and act as a tumor suppressor, suppressing HCC cellular multiplication and clonality while accelerating cellular apoptosis. These results suggested that lncRNAs played important roles in proliferation and apoptosis of HCC and may serve as latent therapeutic targets of HCC.
Chemo-sensitivity and radio-resistance
LncRNAs have also been reported to function in chemo-sensitivity or radio-resistance through arrest of cell cycle, suppression of apoptosis as well as strengthening of DNA injury repair [46, 47]. For instance, lncRNA TUC338 [48] and MALAT1 [49] was involved in HCC evolution and drug-resistance (such as sorafenib). These suggested that lncRNAs have the potential to serve as new targets for exploiting novel strategies of chemotherapy and radiotherapy in HCC patients.
Angiogenesis
Coupling with the increase of neoplasm sizes, angiogenesis is required. In order to supply nutrient substances and O2, angiogenesis permits neoplasm to handle its metabolism waste with following access to blood metastasis process. Frequently, neoplasm cell leads to induction of pro-angiogenic signs or blockage of anti-angiogenic markers, which could open an “on-off” of angiogenesis. LncRNAs are mirrored to serve as a key player in the regulation of angiogenesis. LncRNAs contributed to abnormal hypervascularity of HCC and afforded a novel landscape into the mechanism of neoplasm angiogenesis.
Main databases of long noncoding RNAs
| NO. | Database name | Availability | Characteristics # | References |
|---|---|---|---|---|
| 1 | Arraystar | http://www.arraystar.com/ | analysis for expression profiling and for the regulation of RNAs, especially the regulatory ncRNAs | ― |
| 2 | C-It-Loci | http://c-it-loci.uni-frankfurt.de/ | conserved loci and silico screening of tissue-enriched lncRNAs | [83] |
| 3 | ceRDB | http://www.oncomir.umn.edu/cefinder/ | miRNA binding sites for a given mRNA target | [84] |
| 4 | CHIPBase | http://rna.sysu.edu.cn/chipbase/ | the TF binding sites and motifs, co-expression patterns, ChIP-function and genome browser | [85, 86] |
| 5 | Co-LncRNA | http://www.bio-bigdata.com/Co-LncRNA/ | GO annotations and KEGG pathways | [87] |
| 6 | DIANA-LncBase v.2 | http://www.microrna.gr/LncBase | expression regulation and the annotation of MREs on the basis of "ceRNA hypothesis" | [88] |
| 7 | GEPIA | http://gepia.cancer-pku.cn/index.html | customizable functions analysis, such as expression, survival and correlation analysis | [84] |
| 8 | Linc2GO | http://www.bioinfo.tsinghua.edu.cn/~liuke/Linc2GO/index.html | comprehensive functional annotations on the basis of "ceRNA hypothesis" | [90] |
| 9 | Lnc2Cancer | http://www.bio-bigdata.net/lnc2cancer | relationships between lncRNA and human tumors | [91] |
| 10 | LncACTdb | http://www.bio-bigdata.net/LncACTdb/ | interactions and annotations of lncRNA-miRNA-mRNA | [92] |
| 11 | lncATLAS | http://lncatlas.crg.eu/ | subcellular localization of lncRNAs | [93] |
| 12 | lnCeDB | http://gyanxet-beta.com/lncedb/ | lncRNAs acting potentially as ceRNAs | [94] |
| 13 | LNCipedia | http://www.lncipedia.org | basic transcript information and structure; protein coding potential and miRNA binding sites; downloads of lncRNA sequences and structures | [95, 96] |
| 14 | LncRBase | http://bicresources.jcbose.ac.in/zhumur/lncrbase/ | transcript features, microarray probes and lncRNA expression | [97] |
| 15 | LncRNA2Function | http://mlg.hit.edu.cn/lncrna2function/ | functions, annotations, lncRNA expression values, associations between lncRNAs and functional terms, as well as known functions of human lncRNAs | [98] |
| 16 | lncRNAdb | http://www.lncrnadb.org/ | annotations of eukaryotic lncRNAs and references information about these RNAs | [99, 100] |
| 17 | LncRNADisease | http://cmbi.bjmu.edu.cn/lncrnadisease | lncRNA-disease association data, novel lncRNA-disease associations and curated lncRNA interactions | [101] |
| 18 | lncRNASNP | http://bioinfo.life.hust.edu.cn/lncRNASNP/ | resources of SNPs in human/mouse lncRNAs and functional SNP selection | [102, 103] |
| 19 | LncRNAWiki | http://lncrna.big.ac.cn/index.php | as a component of ScienceWikis, it affords community-curated resource of lncRNA knowledge | [104] |
| 20 | LncRNome | http://genome.igib.res.in/lncRNome/ | types, chromosomal locations, descriptions on the biological functions and disease associations, protein-lncRNA interactions, and genomic variations | [105] |
| 21 | miRcode | http://www.mircode.org | Affords "whole transcriptome" human miRNA target predictions based on the GENCODE gene annotation | [106] |
| 22 | NONCODE | http://www.noncode.org, or http://www.bioinfo.org/noncode/) | conservation annotations and lncRNAs-diseases relationships | [107-112] |
| 23 | ncRNA Expression Database (NRED) | http://www.nred.org/, or http://jsm-research.imb.uq.edu.au/NRED | gene expression information and ancillary data for featured ncRNAs | [113] |
| 24 | RegRNA2.0 | http://regrna2.mbc.nctu.edu.tw/ | functional RNA motifs and sites | [114,115] |
| 25 | StarBase v2.0 | http://starbase.sysu.edu.cn/ | miR-function and ceRNA-function web tools | [116, 117] |
| 26 | TANRIC | http://ibl.mdanderson.org/tanric/_design/basic/index.html | the expression profiles of lncRNAs in large patient cohorts of 20 cancer types including TCGA, CCLE and other independent datasets | ― |
| 27 | TPGLDA | https://github.com/USTC-HIlab/TPGLDA | prediction of relationships of lncRNAs and diseases | [118] |
Molecular mechanisms of lncRNAs in HCC
Epigenetic regulatory lncRNAs
LncRNAs have been shown to excert regulatory roles at nearly all stages of gene expression [50], from targeting epigenetic modification, transcriptional regulation, interactions of lncRNAs-proteins, lncRNAs-miRNAs-mRNAs, lncRNA-lncRNA, autophagy as well as signaling pathway to modulation of mRNA stabilization. LncRNA GIHCG promoted HCC progression through epigenetically upregulating H3K27me3 levels and DNA methylation contents [51]. LncRNA ZNFX1-AS1 suppressed HCC cell growth through modulating miR-9 methylation [52]. Linc00441 promoted HCC tumorigenesis in a H3K27 modification-dependent manner [53].
Transcriptional regulatory lncRNAs
Several lncRNAs could serve as transcriptional regulators. It was reported that CCAT2 [54] and HOTAIR [55] accelerated EMT process mediated by transcription factor (TF) Slug and Snail in HCC, respectively. LncRNA ZEB1-AS1 promoted neoplasm invasiveness and indicated an adverse clinical outcome via positively regulating the ZEB1 expression in HCC [34]. Meanwhile, lncRNA CPS1-IT1 suppressed HCC aggressivity via controlling HIF-1α activity [36]. LncRNA uc.338 was found to promote cellular growth via modulating of CDKN1A transcription in HCC [56].
LncRNAs-proteins interactions
LncRNA HNF1A-AS1 repressed NKD1 and p21 expression via interacting with EZH2, and then promoted HCC cell proliferation [57]. LncRNA NEAT1 modulated hnRNP-A2 level to facilitate cellular multiplication and invasiveness in HCC cell lines [58]. LncRNA TUC338 targeted RASAL1 to be involved in sorafenib-sensitized HCC cells [48]. LncRNA BANCR elevated the protein level of VIM while lowered the protein content of E-cad to cripple cellular malignant degree [59]. The lncRNA EGFR-AS1, a target of GHR, up-regulated the expression of EGFR in HCC [60]. To summarize, lncRNAs-proteins interactions functioned as a vital mechanism of lncRNAs in HCC.
LncRNAs-miRNAs-mRNAs interactions
It has been demonstrated that microRNAs (miRNAs) functioned crucially in many caners, including HCC [61-63]. LncRNAs could share miRNA recognition elements (MREs) with mRNAs and modulate the function of mRNAs with miRNAs-mediated mechanisms, where lncRNAs were called competing endogenous RNAs (ceRNAs) [64-66]. For instance, lncRNA SNHG12 - miR-199a/b-5p - MLK3 - NF-κB signaling [67] accelerated tumorigenesis and metastasis in HCC. LncRNA UCA1 - miR-203 - Slug axis [68] and lncRNA Ftx - miR-545 - RIG-I network [69] were involvement in HCC progression. The HIF-2α - MALAT1 - miR-216b axis regulated multi-drug resistance of HCC cells [49]. Overall, lncRNAs could serve as “miRNA sponges” to share the same MREs with mRNAs.
Others
LncRNAs - lncRNAs interactions
LncRNAs functioned vitally in a variety of biology processes. Frequently, the occurrence and development of neoplasm frequently was attributed to the interaction effects of some lncRNAs. LncRNA-lncRNA synergistic networks were beneficial to explore clinically related lncRNAs in neoplasm [70]. Reports suggested that LncRNA HULC cooperated with MALAT1 to promote CSCs proliferation in HCC [71]. And the overexpression of HULC along with MALAT1 increased the binding of RNA pol II, P300, CREPT to TRF2, which triggered the upregulation, phosphorylation and SUMOylation of TRF2 [71]. Indeed, lncRNAs could interact with other lncRNAs.
Autophagy
Autophagy is a cellular degradation pathway that is essential to maintain cell physiology progress, and its disruption leads to multiple diseases in humans [72]. Recently, macro-autophagy/autophagy has emerged as a promising therapeutic target in various types of solid tumor treatment [73]. LncRNAs exerted crucial roles in the regulation of autophagy lately, which made them act as potential biomarkers of disease phenotypes [74]. LncRNA HULC could induce a protective autophagy [75]. LncRNA HNF1A-AS1 served as a autophagic accelerator in HCC [76]. And the lncRNA HOTAIR activated autophagy [30]. These studies indicated that the lncRNAs-autophagy networks are likely to supply additional insights into treatment interventions and markers evaluation in human diseases, especially in cancers.
Signaling pathway
As we all know, signaling pathways functioned considerably in the development of HCC and presented some therapeutic strategies based on in vivo and in vitro findings. LncRNA T-UCR severed as a potential growth driver gene modulated by the Wnt/β-catenin signaling in hepatobiliary carcinogenesis [77, 164]. LncRNA MEG3 could activate ER stress and p53 signaling related to NF-κB pathway, which followed by suppressing cell multiplication and promoting apoptosis [78]. Lnc-DILC was downregulated in liver cancer stem cells and mediated intrahepatic inflammation via governing the cross-linking of TNF-α - NF-κB pathway with IL-6 - STAT3 pathway [79].
mRNA stabilization
Transcription activity and post-transcription mechanisms involved in lncRNAs could modulate ribonucleotide reductase followed by altering the stability of message RNAs [80]. In turn, mRNA steady state levels could directly influence their expression [80]. The lncRNA MALAT1 accelerated arsenite-induced glycolysis, which was mediated via HIF-1α mRNA stabilization in human liver L-02 cell lines [81]. LncRNA AK058003 could reduce the expression of mRNA stabilizing protein HuR and act as a precursor of miR-15a to suppress γ-synuclein-mediated cell proliferation and the metastasis of HCC [44]. In addition, lncRNA HULC triggered autophagy via stabilizing Sirt1 and attenuated the chemosensitivity of HCC cells [75]. These studies suggested that some lncRNAs participating biological processes may be mediated by selective stabilization of mRNAs.
Potential clinical application of lncRNAs in HCC
LncRNAs for diagnosis of HCC
A growing amount of evidence supports that benefiting from their attractive characteristics, lncRNAs are potential indicators for human diseases including HCC. The majority of lncRNAs displayed the trait of strict tissue-specific and neoplasm-specific expression. Frequently, distinct upregulation or downregulation of lncRNA was found in neoplasm tissues in comparison with normal samples. Meanwhile, a few lncRNAs were declared to be existed in urine, blood and other body fluids, which could be easily acquired through the non-invasive way as much as possible. What's more, utilizing lncRNAs as indicators of neoplasm states was superior to protein-coding RNAs, due to the more representative expression of lncRNAs [82]. Compared with current varying greatly biomarkers in protein coding genes, lncRNAs are likely to be advantageous indicators on the strength of their specificity and easy acquirement.
The pooled sensitivity and specificity of HCC patients predicted by lncRNAs were showed as Table 2. These lncRNAs are likely to act as valuable indicators for neoplasm diagnosis and have potential application value for clinic diagnosis of HCC patients.
LncRNAs as prognostic biomarkers for HCC
Accumulating reports declared that lncRNAs have potential to serve as prognostic predictors in neoplasms containing HCC patients. A variety of lncRNAs were demonstrated to exhibit abnormal expression in HCC, which were significantly correlated with the survival time and were independent outcome predictors in HCC patients (Table 3). Therefore, lncRNAs might be acted as potential and useful prognostic indicators in HCC.
Conclusion and future prospective
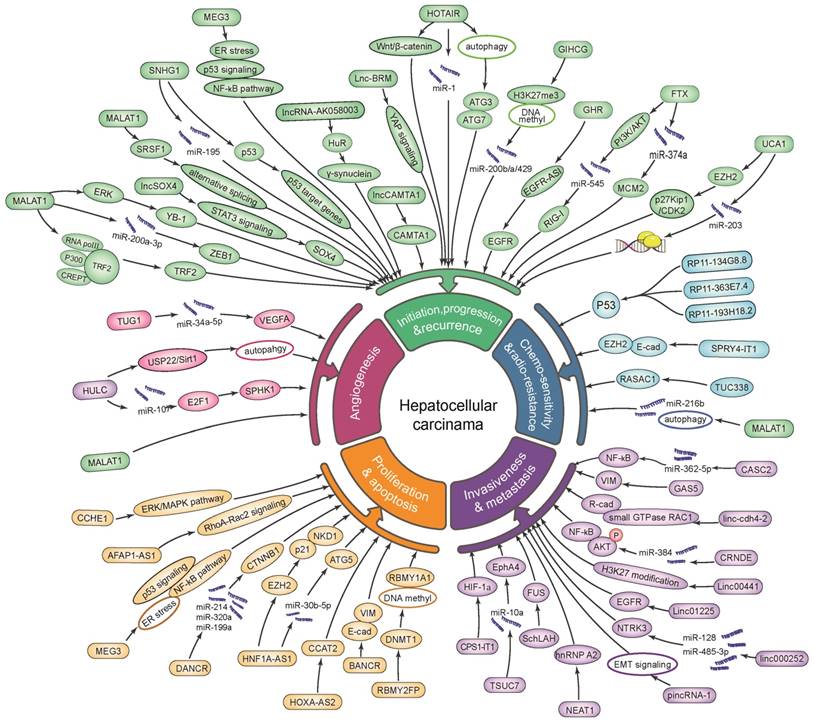
In a nutshell, lncRNAs have the potential to serve as promising biomarkers for tumor progression and recurrence (lncRNA HOTAIR, SNHG1, HULC, MALAT1, CRNDE, GIHCG, UCA1, lncSox4 and lncBRM), as well as for invasion and metastasis (SNHG20, CCAT2, HOST2, Linc-cdh4-2, LINC00052, AFAP1-AS1, ZEB1-AS1, ZEB2-AS1, LINC01225, UC001kfo, SPRY4-IT1, Unigene56159, HULC, plncRNA-1, SchLAH, lncRNA-AK058003, CPS1-IT1, XIST, FTX, TUSC7 and GAS5). Besides, lncRNAs are involved in HCC through affecting on cell proliferation and apoptosis (UC001kfo, SPRY4-IT1, ZEB2-AS1, RBMY2FP, uc.338, UCA1, XIST, HOST2, HOXA-AS2, CCHE1, CCAT2, SNHG1, HNF1A-AS1, PCAT-1, AFAP1-AS1, HULC, MALAT1, lncCAMTA1, lnc-DILC, lncRNA-AK058003, ZNFX1-AS1, lincRNA-p21, XIST, FTX and GAS5), as well as angiogenesis (lncRNA TUG1 and HULC). Additionally, lncRNAs serve as a conceivable indicator for chemo-sensitivity and radio-resistance (HULC, RP11-134G8.8, RP11-363E7.4, RP1-193H18.2, TUC338 and MALAT1; Figure 1, Table S1). Meanwhile, lncRNAs exerted their efficient and effective actions in mechanisms of interaction with proteins/ miRNAs/ mRNAs/ lncRNAs, in epigenetic regulation/transcriptional regulation, as well as in regulation of autophagy, signaling pathway and mRNA stabilization in HCC on the horizon of the current studies (Figure 1, Table S1). Furthermore, these lncRNAs might be promising indicators for disease diagnosis (Table 2), prognosis and recurrence prediction (Table 3), and even be new up-and-coming targets for therapeutic intervention of HCC.
Yet, the precise biological function and molecule mechanisms of lncRNAs in HCC remained uncharacterized. Thus, further exploration and validation researches are required for illuminating the intricate mechanisms (especially in epigenetic and transcriptional modulation of lncRNAs) as well as the clinical utilizations of lncRNAs in HCC.
Dysregulation and functional roles of lncRNAs in hepatocellular carcinoma.
Application as diagnostic index of lncRNA in hepatocellular carcinoma patients
| LncRNAs | Functions | AUC | Sensitivity | Specificity | References |
|---|---|---|---|---|---|
| 2-lncRNA signature (PVT1 and uc002mbe.2) | distinguishes HCC patients from the healthy population | 0.764 (0.684-0.833) | 60.56 % | 90.62 % | [119] |
| CCHE1 | discriminates tumor tissues from normal tissues | 0.9262 | ― | ― | [42] |
| CRNDE | distinguishes tumor tissues from adjacent normal tissues in HCC | 0.699 | ― | ― | [120] |
| DANCR | differentiates patients with HCC from HVs and patients with CHB and cirrhosis | 0.868 | 83.8 % | 72.7 % | [121] |
| differentiates patients with HCC from CHB and cirrhosis | 0.864 | 80.8 % | 84.3 % | [121] | |
| DGCR5 | discriminates tumor tissues from normal tissues | 0.782 | 63.3% | 83.3% | [122] |
| LINC RP1130-1 | distinguishes HCC from adjacent normal tissues | 0.74 | ― | ― | [123] |
| MALAT1 (plasma) | discriminates HCC patients and hepatic disease patients | 0.66 | 51.1 % | 89.3 % | [124] |
| JPX | discriminates between HCC patients and controls | 0.814 | 100.0 % | 52.4 % | [125] |
| JPX and AFP | discriminates between HCC patients and controls | 0.905 | 97.1 % | 72.2 % | [125] |
| SPRY4-IT1 | differentiates HCC patients from controls | 0.702 (0.609-0.796) | 87.3 % | 50.0 % | [126] |
| SPRY4-IT1 and AFP | differentiates HCC patients from controls | 0.800 (0.706-0.874) | 87.3 % | 65.0 % | [126] |
| UCA1 | discriminates HCC patients from healthy controls | 0.91 | 91.4 % | 88.6 % | [127] |
Prognostic abilities of lncRNAs for hepatocellular carcinoma patients in Cox proportional hazards model
| LncRNAs | Univariate analysis | Multivariate analysis | References | ||
|---|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | ||
| OS | |||||
| JPX (low vs. high) | 2.283 (1.211-4.304) | 0.010 | 2.294 (1.178-4.468) | 0.015 | [125] |
| XIST (low vs. high) | 2.155 (1.136-4.088) | 0.003 | 2.207 (1.152-4.225) | 0.017 | [125] |
| CCHE1 expression (Low vs. High) | 2.246 (0.605-5.172) | 0.027 | 0.977 (0.312-3.434) | 0.041 | [42] |
| lncRNA SNHG15 (High vs. Low) | 3.017 (1.448-6.221) | 0.018 | 2.247 (1.331-6.255) | 0.001 | [128] |
| SNHG20 | 4.440 (2.254-8.743) | 0.000 | 3.985 (1.981-8.017) | 0.000 | [129] |
| lncRNA GAS5 (Low vs. High) | ― | <0.0001 | 3.257 (2.418 - 8.839) | <0.001 | [130] |
| CARLo-5 (Low vs. High) | 3.267 (1.620-6.271) | 0.014 | 2.981 (1.316-3.952) | 0.011 | [131] |
| SNHG3 | 4.442 (2.368-8.332) | 0.000 | 3.464 (1.820-6.594) | 0.000 | [132] |
| CCAT2 expression (high vs low) | 2.118 (1.245-3.603) | 0.006 | 1.849 (1.064-3.213) | 0.029 | [54] |
| UC001kfo (High vs. Low) | 1.876 (1.098-3.207) | 0.021 | 1.751 (1.017-3.015) | 0.043 | [133] |
| lnc-DILC (low vs. high) | 2.618 (1.136-6.036) | 0.024 | 2.465 (1.03-5.9) | 0.043 | [79] |
| RFS | |||||
| CARLo-5 (Low vs. High) | 2.873 (1.669-5.852) | 0.005 | 1.810 (1.562-5.177) | 0.010 | [131] |
| lnc-DILC (low vs. high) | 2.142 (1.025-4.477) | 0.043 | 1.499 (0.651-3.451) | 0.342 | [79] |
| PFS | |||||
| UC001kfo (High vs. Low) | 2.147 (1.260-3.658) | 0.005 | 1.975 (1.149-3.394) | 0.014 | [133] |
Supplementary Material
Supplementary table.
Acknowledgements
This work was supported by the Open-End Fund for the Valuable and Precision Instruments of Central South University (CSUZC201746 for Li Peng), Hunan Provincial Innovation Foundation for Postgraduates (CX2016B056 for Li Peng), Fundamental Research Funds for the Central Universities of Central South University (2015zzts096 for Li Peng), and the Science and Technology Department Research Foundation of Hunan province (12JJ2052 for Guan-Cheng Li).
Authors' contributions
LP collected the references and drafted the manuscript. GCL and XQY participated in the design of the review and helped to draft the manuscript. CYZ, JYP, YQZ and XP revised critically the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Granito A, Bolondi L. Non-transplant therapies for patients with hepatocellular carcinoma and Child-Pugh-Turcotte class B cirrhosis. Lancet Oncol. 2017;18:e101-e12
2. Yuen MF, Hou JL, Chutaputti A. Hepatocellular carcinoma in the Asia pacific region. J Gastroenterol Hepatol. 2009;24:346-53
3. Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang XS. et al. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet. 2015;385:977-1010
4. Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245-55
5. Freeman RB, Edwards EB, Harper AM. Waiting list removal rates among patients with chronic and malignant liver diseases. Am J Transplant. 2006;6:1416-21
6. Lin DC, Mayakonda A, Dinh HQ, Huang P, Lin L, Liu X. et al. Genomic and Epigenomic Heterogeneity of Hepatocellular Carcinoma. Cancer Res. 2017;77:2255-65
7. Hou Y, Guo H, Cao C, Li X, Hu B, Zhu P. et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016;26:304-19
8. Kanda M, Sugimoto H, Kodera Y. Genetic and epigenetic aspects of initiation and progression of hepatocellular carcinoma. World J Gastroenterol. 2015;21:10584-97
9. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391-407
10. Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M. et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20:908-13
11. Li J, Han L, Roebuck P, Diao L, Liu L, Yuan Y. et al. TANRIC: An Interactive Open Platform to Explore the Function of lncRNAs in Cancer. Cancer Res. 2015;75:3728-37
12. Castellanos-Rubio A, Fernandez-Jimenez N, Kratchmarov R, Luo X, Bhagat G, Green PH. et al. A long noncoding RNA associated with susceptibility to celiac disease. Science. 2016;352:91-5
13. Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S. et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310-3
14. Lee S, Kopp F, Chang TC, Sataluri A, Chen B, Sivakumar S. et al. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell. 2016;164:69-80
15. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155-9
16. Di Gesualdo F, Capaccioli S, Lulli M. A pathophysiological view of the long non-coding RNA world. Oncotarget. 2014;5:10976-96
17. Sun L, Luo H, Liao Q, Bu D, Zhao G, Liu C. et al. Systematic study of human long intergenic non-coding RNAs and their impact on cancer. Sci China Life Sci. 2013;56:324-34
18. Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD. et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell. 2015;28:529-40
19. Wong CM, Tsang FH, Ng IO. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol. 2018 doi: 10.1038/nrgastro.2017.169. [Epub ahead of print]
20. Yang CM, Wang TH, Chen HC, Li SC, Lee MC, Liou HH. et al. Aberrant DNA hypermethylation-silenced SOX21-AS1 gene expression and its clinical importance in oral cancer. Clin Epigenetics. 2016;8:129
21. Martinez-Fernandez M, Feber A, Duenas M, Segovia C, Rubio C, Fernandez M. et al. Analysis of the Polycomb-related lncRNAs HOTAIR and ANRIL in bladder cancer. Clin Epigenetics. 2015;7:109
22. Tao H, Yang JJ, Zhou X, Deng ZY, Shi KH, Li J. Emerging role of long noncoding RNAs in lung cancer: Current status and future prospects. Respir Med. 2016;110:12-9
23. Garzon R, Volinia S, Papaioannou D, Nicolet D, Kohlschmidt J, Yan PS. et al. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc Natl Acad Sci U S A. 2014;111:18679-84
24. Reon BJ, Anaya J, Zhang Y, Mandell J, Purow B, Abounader R. et al. Expression of lncRNAs in Low-Grade Gliomas and Glioblastoma Multiforme: An In Silico Analysis. PLoS Med. 2016;13:e1002192
25. Sand M, Bechara FG, Sand D, Gambichler T, Hahn SA, Bromba M. et al. Expression profiles of long noncoding RNAs in cutaneous squamous cell carcinoma. Epigenomics. 2016;8:501-18
26. Ni S, Zhao X, Ouyang L. Long non-coding RNA expression profile in vulvar squamous cell carcinoma and its clinical significance. Oncol Rep. 2016;36:2571-8
27. Li JP, Liu LH, Li J, Chen Y, Jiang XW, Ouyang YR. et al. Microarray expression profile of long noncoding RNAs in human osteosarcoma. Biochem Biophys Res Commun. 2013;433:200-6
28. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915-27
29. Su DN, Wu SP, Chen HT, He JH. HOTAIR, a long non-coding RNA driver of malignancy whose expression is activated by FOXC1, negatively regulates miRNA-1 in hepatocellular carcinoma. Oncol Lett. 2016;12:4061-7
30. Yang L, Zhang X, Li H, Liu J. The long noncoding RNA HOTAIR activates autophagy by upregulating ATG3 and ATG7 in hepatocellular carcinoma. Mol Biosyst. 2016;12:2605-12
31. Gao JZ, Li J, Du JL, Li XL. Long non-coding RNA HOTAIR is a marker for hepatocellular carcinoma progression and tumor recurrence. Oncol Lett. 2016;11:1791-8
32. Malek E, Jagannathan S, Driscoll JJ. Correlation of long non-coding RNA expression with metastasis, drug resistance and clinical outcome in cancer. Oncotarget. 2014;5:8027-38
33. Xiong D, Sheng Y, Ding S, Chen J, Tan X, Zeng T. et al. LINC00052 regulates the expression of NTRK3 by miR-128 and miR-485-3p to strengthen HCC cells invasion and migration. Oncotarget. 2016;7:47593-608
34. Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z. et al. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2016;35:1575-84
35. Wang X, Zhang W, Tang J, Huang R, Li J, Xu D. et al. LINC01225 promotes occurrence and metastasis of hepatocellular carcinoma in an epidermal growth factor receptor-dependent pathway. Cell Death Dis. 2016;7:e2130
36. Wang TH, Yu CC, Lin YS, Chen TC, Yeh CT, Liang KH. et al. Long noncoding RNA CPS1-IT1 suppresses the metastasis of hepatocellular carcinoma by regulating HIF-1alpha activity and inhibiting epithelial-mesenchymal transition. Oncotarget. 2016;7:43588-603
37. Zhuang LK, Yang YT, Ma X, Han B, Wang ZS, Zhao QY. et al. MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death Dis. 2016;7:e2203
38. Liu F, Yuan JH, Huang JF, Yang F, Wang TT, Ma JZ. et al. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene. 2016;35:5422-34
39. Mo Y, Lu Y, Wang P, Huang S, He L, Li D. et al. Long non-coding RNA XIST promotes cell growth by regulating miR-139-5p/PDK1/AKT axis in hepatocellular carcinoma. Tumour Biol. 2017;39:1010428317690999
40. Liu RT, Cao JL, Yan CQ, Wang Y, An CJ, Lv HT. Effects of LncRNA-HOST2 on cell proliferation, migration, invasion and apoptosis of human hepatocellular carcinoma cell line SMMC-7721. Biosci Rep. 2017;37:BSR20160532
41. Wang F, Yang H, Deng Z, Su Y, Fang Q, Yin Z. HOX Antisense lincRNA HOXA-AS2 Promotes Tumorigenesis of Hepatocellular Carcinoma. Cell Physiol Biochem. 2016;40:287-96
42. Peng W, Fan H. Long noncoding RNA CCHE1 indicates a poor prognosis of hepatocellular carcinoma and promotes carcinogenesis via activation of the ERK/MAPK pathway. Biomed Pharmacother. 2016;83:450-5
43. Zhang JY, Weng MZ, Song FB, Xu YG, Liu Q, Wu JY. et al. Long noncoding RNA AFAP1-AS1 indicates a poor prognosis of hepatocellular carcinoma and promotes cell proliferation and invasion via upregulation of the RhoA/Rac2 signaling. Int J Oncol. 2016;48:1590-8
44. He X, Zheng Y, Zhang Y, Gan Y, Zhou Y, Liang H. et al. Long non-coding RNA AK058003, as a precursor of miR-15a, interacts with HuR to inhibit the expression of gamma-synuclein in hepatocellular carcinoma cells. Oncotarget. 2017;8:9451-65
45. Jia M, Jiang L, Wang YD, Huang JZ, Yu M, Xue HZ. lincRNA-p21 inhibits invasion and metastasis of hepatocellular carcinoma through Notch signaling-induced epithelial-mesenchymal transition. Hepatol Res. 2016;46:1137-44
46. Harries LW. Long non-coding RNAs and human disease. Biochem Soc Trans. 2012;40:902-6
47. Lipovich L, Johnson R, Lin CY. MacroRNA underdogs in a microRNA world: evolutionary, regulatory, and biomedical significance of mammalian long non-protein-coding RNA. Biochim Biophys Acta. 2010;1799:597-615
48. Jin W, Chen L, Cai X, Zhang Y, Zhang J, Ma D. et al. Long non-coding RNA TUC338 is functionally involved in sorafenib-sensitized hepatocarcinoma cells by targeting RASAL1. Oncol Rep. 2017;37:273-80
49. Yuan P, Cao W, Zang Q, Li G, Guo X, Fan J. The HIF-2alpha-MALAT1-miR-216b axis regulates multi-drug resistance of hepatocellular carcinoma cells via modulating autophagy. Biochem Biophys Res Commun. 2016;478:1067-73
50. Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300-7
51. Sui CJ, Zhou YM, Shen WF, Dai BH, Lu JJ, Zhang MF. et al. Long noncoding RNA GIHCG promotes hepatocellular carcinoma progression through epigenetically regulating miR-200b/a/429. J Mol Med (Berl). 2016;94:1281-96
52. Wang T, Ma S, Qi X, Tang X, Cui D, Wang Z. et al. Long noncoding RNA ZNFX1-AS1 suppresses growth of hepatocellular carcinoma cells by regulating the methylation of miR-9. Onco Targets Ther. 2016;9:5005-14
53. Tang J, Xie Y, Xu X, Yin Y, Jiang R, Deng L. et al. Bidirectional transcription of Linc00441 and RB1 via H3K27 modification-dependent way promotes hepatocellular carcinoma. Cell Death Dis. 2017;8:e2675
54. Xu Y, Wang B, Zhang F, Wang A, Du X, Hu P. et al. Long non-coding RNA CCAT2 is associated with poor prognosis in hepatocellular carcinoma and promotes tumor metastasis by regulating Snail2-mediated epithelial-mesenchymal transition. Onco Targets Ther. 2017;10:1191-8
55. Battistelli C, Cicchini C, Santangelo L, Tramontano A, Grassi L, Gonzalez FJ. et al. The Snail repressor recruits EZH2 to specific genomic sites through the enrollment of the lncRNA HOTAIR in epithelial-to-mesenchymal transition. Oncogene. 2017;36:942-55
56. Bo C, Li N, Li X, Liang X, An Y. Long noncoding RNA uc.338 promotes cell proliferation through association with BMI1 in hepatocellular carcinoma. Hum Cell. 2016;29:141-7
57. Wang C, Mou L, Chai HX, Wang F, Yin YZ, Zhang XY. Long non-coding RNA HNF1A-AS1 promotes hepatocellular carcinoma cell proliferation by repressing NKD1 and P21 expression. Biomed Pharmacother. 2017;89:926-32
58. Mang Y, Li L, Ran J, Zhang S, Liu J, Chen Y. et al. Long noncoding RNA NEAT1 promotes cell proliferation and invasion by regulating hnRNP A2 expression in hepatocellular carcinoma cells. Onco Targets Ther. 2017;10:1003-16
59. Zhou T, Gao Y. Increased expression of LncRNA BANCR and its prognostic significance in human hepatocellular carcinoma. World J Surg Oncol. 2016;14:8
60. Qi HL, Li CS, Qian CW, Xiao YS, Yuan YF, Liu QY. et al. The long noncoding RNA, EGFR-AS1, a target of GHR, increases the expression of EGFR in hepatocellular carcinoma. Tumour Biol. 2016;37:1079-89
61. Ren FH, Yang H, He RQ, Lu JN, Lin XG, Liang HW. et al. Analysis of microarrays of miR-34a and its identification of prospective target gene signature in hepatocellular carcinoma. BMC Cancer. 2018;18:12
62. Pinto Y, Buchumenski I, Levanon EY, Eisenberg E. Human cancer tissues exhibit reduced A-to-I editing of miRNAs coupled with elevated editing of their targets. Nucleic Acids Res. 2018;46:71-82
63. Kim SS, Cho HJ, Nam JS, Kim HJ, Kang DR, Won JH. et al. Plasma MicroRNA-21, 26a, and 29a-3p as Predictive Markers for Treatment Response Following Transarterial Chemoembolization in Patients with Hepatocellular Carcinoma. J Korean Med Sci. 2018;33:e6
64. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-8
65. Peng L, Yuan XQ, Li GC. The emerging landscape of circular RNA ciRS-7 in cancer. Oncol Rep. 2015;33:2669-74
66. Peng L, Yuan X, Jiang B, Tang Z, Li GC. LncRNAs: key players and novel insights into cervical cancer. Tumour Biol. 2016;37:2779-88
67. Lan T, Ma W, Hong Z, Wu L, Chen X, Yuan Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes tumorigenesis and metastasis by targeting miR-199a/b-5p in hepatocellular carcinoma. J Exp Clin Cancer Res. 2017;36:11
68. Xiao JN, Yan TH, Yu RM, Gao Y, Zeng WL, Lu SW. et al. Long non-coding RNA UCA1 regulates the expression of Snail2 by miR-203 to promote hepatocellular carcinoma progression. J Cancer Res Clin Oncol. 2017;143:981-90
69. Liu Z, Dou C, Yao B, Xu M, Ding L, Wang Y. et al. Ftx non coding RNA-derived miR-545 promotes cell proliferation by targeting RIG-I in hepatocellular carcinoma. Oncotarget. 2016;7:25350-65
70. Li Y, Chen J, Zhang J, Wang Z, Shao T, Jiang C. et al. Construction and analysis of lncRNA-lncRNA synergistic networks to reveal clinically relevant lncRNAs in cancer. Oncotarget. 2015;6:25003-16
71. Wu M, Lin Z, Li X, Xin X, An J, Zheng Q. et al. HULC cooperates with MALAT1 to aggravate liver cancer stem cells growth through telomere repeat-binding factor 2. Sci Rep. 2016;6:36045
72. Gallolu Kankanamalage S, Lee AY, Wichaidit C, Lorente-Rodriguez A, Shah AM, Stippec S. et al. WNK1 is an unexpected autophagy inhibitor. Autophagy. 2017:1-2
73. Chen X, Clark J, Wunderlich M, Fan C, Davis A, Chen S. et al. Autophagy is dispensable for Kmt2a/Mll-Mllt3/Af9 AML maintenance and anti-leukemic effect of chloroquine. Autophagy. 2017:1-12
74. Xu Z, Yan Y, Qian L, Gong Z. Long non-coding RNAs act as regulators of cell autophagy in diseases (Review). Oncol Rep. 2017;37:1359-66
75. Xiong H, Ni Z, He J, Jiang S, Li X, Gong W. et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36:3528-40
76. Liu Z, Wei X, Zhang A, Li C, Bai J, Dong J. Long non-coding RNA HNF1A-AS1 functioned as an oncogene and autophagy promoter in hepatocellular carcinoma through sponging hsa-miR-30b-5p. Biochem Biophys Res Commun. 2016;473:1268-75
77. Fernandez-Barrena MG, Perugorria MJ, Banales JM. Novel lncRNA T-UCR as a potential downstream driver of the Wnt/beta-catenin pathway in hepatobiliary carcinogenesis. Gut. 2017;66:1177-8
78. Chen RP, Huang ZL, Liu LX, Xiang MQ, Li GP, Feng JL. et al. Involvement of endoplasmic reticulum stress and p53 in lncRNA MEG3-induced human hepatoma HepG2 cell apoptosis. Oncol Rep. 2016;36:1649-57
79. Wang X, Sun W, Shen W, Xia M, Chen C, Xiang D. et al. Long non-coding RNA DILC regulates liver cancer stem cells via IL-6/STAT3 axis. J Hepatol. 2016;64:1283-94
80. Burton TR, Kashour T, Wright JA, Amara FM. Cellular signaling pathways affect the function of ribonucleotide reductase mRNA binding proteins: mRNA stabilization, drug resistance, and malignancy (Review). Int J Oncol. 2003;22:21-31
81. Luo F, Liu X, Ling M, Lu L, Shi L, Lu X. et al. The lncRNA MALAT1, acting through HIF-1alpha stabilization, enhances arsenite-induced glycolysis in human hepatic L-02 cells. Biochim Biophys Acta. 2016;1862:1685-95
82. Hauptman N, Glavac D. Long non-coding RNA in cancer. Int J Mol Sci. 2013;14:4655-69
83. Weirick T, John D, Dimmeler S, Uchida S. C-It-Loci: a knowledge database for tissue-enriched loci. Bioinformatics. 2015;31:3537-43
84. Sarver AL, Subramanian S. Competing endogenous RNA database. Bioinformation. 2012;8:731-3
85. Zhou KR, Liu S, Sun WJ, Zheng LL, Zhou H, Yang JH. et al. ChIPBase v2.0: decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data. Nucleic Acids Res. 2017;45:D43-D50
86. Yang JH, Li JH, Jiang S, Zhou H, Qu LH. ChIPBase: a database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic Acids Res. 2013;41:D177-87
87. Zhao Z, Bai J, Wu A, Wang Y, Zhang J, Wang Z. et al. Co-LncRNA: investigating the lncRNA combinatorial effects in GO annotations and KEGG pathways based on human RNA-Seq data. Database (Oxford). 2015. 2015
88. Paraskevopoulou MD, Georgakilas G, Kostoulas N, Reczko M, Maragkakis M, Dalamagas TM. et al. DIANA-LncBase: experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res. 2013;41:D239-45
89. Tang Z, Li C, Kang B, Gao G, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102
90. Liu K, Yan Z, Li Y, Sun Z. Linc2GO: a human LincRNA function annotation resource based on ceRNA hypothesis. Bioinformatics. 2013;29:2221-2
91. Ning S, Zhang J, Wang P, Zhi H, Wang J, Liu Y. et al. Lnc2Cancer: a manually curated database of experimentally supported lncRNAs associated with various human cancers. Nucleic Acids Res. 2016;44:D980-5
92. Wang P, Ning S, Zhang Y, Li R, Ye J, Zhao Z. et al. Identification of lncRNA-associated competing triplets reveals global patterns and prognostic markers for cancer. Nucleic Acids Res. 2015;43:3478-89
93. Mas-Ponte D, Carlevaro-Fita J, Palumbo E, Hermoso Pulido T, Guigo R, Johnson R. LncATLAS database for subcellular localization of long noncoding RNAs. RNA. 2017;23:1080-7
94. Chakraborty S, Deb A, Maji RK, Saha S, Ghosh Z. LncRBase: an enriched resource for lncRNA information. PLoS One. 2014;9:e108010
95. Das S, Ghosal S, Sen R, Chakrabarti J. lnCeDB: database of human long noncoding RNA acting as competing endogenous RNA. PLoS One. 2014;9:e98965
96. Volders PJ, Helsens K, Wang X, Menten B, Martens L, Gevaert K. et al. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2013;41:D246-51
97. Volders PJ, Verheggen K, Menschaert G, Vandepoele K, Martens L, Vandesompele J. et al. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015;43:D174-80
98. Jiang Q, Ma R, Wang J, Wu X, Jin S, Peng J. et al. LncRNA2Function: a comprehensive resource for functional investigation of human lncRNAs based on RNA-seq data. BMC Genomics. 2015;16(Suppl 3):S2
99. Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39:D146-51
100. Quek XC, Thomson DW, Maag JL, Bartonicek N, Signal B, Clark MB. et al. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015;43:D168-73
101. Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X. et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983-6
102. Gong J, Liu W, Zhang J, Miao X, Guo AY. lncRNASNP: a database of SNPs in lncRNAs and their potential functions in human and mouse. Nucleic Acids Res. 2015;43:D181-6
103. Miao YR, Liu W, Zhang Q, Guo AY. lncRNASNP2: an updated database of functional SNPs and mutations in human and mouse lncRNAs. Nucleic Acids Res. 2018;46:D276-D80
104. Ma L, Li A, Zou D, Xu X, Xia L, Yu J. et al. LncRNAWiki: harnessing community knowledge in collaborative curation of human long non-coding RNAs. Nucleic Acids Res. 2015;43:D187-92
105. Bhartiya D, Pal K, Ghosh S, Kapoor S, Jalali S, Panwar B. et al. lncRNome: a comprehensive knowledgebase of human long noncoding RNAs. Database (Oxford). 2013;2013:bat034
106. Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28:2062-3
107. Zhao Y, Yuan J, Chen R. NONCODEv4: Annotation of Noncoding RNAs with Emphasis on Long Noncoding RNAs. Methods Mol Biol. 2016;1402:243-54
108. Zhao Y, Li H, Fang S, Kang Y, Wu W, Hao Y. et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016;44:D203-8
109. Xie C, Yuan J, Li H, Li M, Zhao G, Bu D. et al. NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2014;42:D98-103
110. Bu D, Yu K, Sun S, Xie C, Skogerbo G, Miao R. et al. NONCODE v3.0: integrative annotation of long noncoding RNAs. Nucleic Acids Res. 2012;40:D210-5
111. He S, Liu C, Skogerbo G, Zhao H, Wang J, Liu T. et al. NONCODE v2.0: decoding the non-coding. Nucleic Acids Res. 2008;36:D170-2
112. Liu C, Bai B, Skogerbo G, Cai L, Deng W, Zhang Y. et al. NONCODE: an integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 2005;33:D112-5
113. Dinger ME, Pang KC, Mercer TR, Crowe ML, Grimmond SM, Mattick JS. NRED: a database of long noncoding RNA expression. Nucleic Acids Res. 2009;37:D122-6
114. Chang TH, Huang HY, Hsu JB, Weng SL, Horng JT, Huang HD. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics. 2013;14(Suppl 2):S4
115. Huang HY, Chien CH, Jen KH, Huang HD. RegRNA: an integrated web server for identifying regulatory RNA motifs and elements. Nucleic Acids Res. 2006;34:W429-34
116. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92-7
117. Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202-9
118. Ding L, Wang M, Sun D, Li A. TPGLDA: Novel prediction of associations between lncRNAs and diseases via lncRNA-disease-gene tripartite graph. Sci Rep. 2018;8:1065
119. Yu J, Han J, Zhang J, Li G, Liu H, Cui X. et al. The long noncoding RNAs PVT1 and uc002mbe.2 in sera provide a new supplementary method for hepatocellular carcinoma diagnosis. Medicine (Baltimore). 2016;95:e4436
120. Chen Z, Yu C, Zhan L, Pan Y, Chen L, Sun C. LncRNA CRNDE promotes hepatic carcinoma cell proliferation, migration and invasion by suppressing miR-384. Am J Cancer Res. 2016;6:2299-309
121. Ma X, Wang X, Yang C, Wang Z, Han B, Wu L. et al. DANCR Acts as a Diagnostic Biomarker and Promotes Tumor Growth and Metastasis in Hepatocellular Carcinoma. Anticancer Res. 2016;36:6389-98
122. Huang R, Wang X, Zhang W, Zhangyuan G, Jin K, Yu W. et al. Down-Regulation of LncRNA DGCR5 Correlates with Poor Prognosis in Hepatocellular Carcinoma. Cell Physiol Biochem. 2016;40:707-15
123. Xiao C, Wang C, Cheng S, Lai C, Zhang P, Wang Z. et al. The significance of low levels of LINC RP1130-1 expression in human hepatocellular carcinoma. Biosci Trends. 2016;10:378-85
124. Konishi H, Ichikawa D, Yamamoto Y, Arita T, Shoda K, Hiramoto H. et al. Plasma level of metastasis-associated lung adenocarcinoma transcript 1 is associated with liver damage and predicts development of hepatocellular carcinoma. Cancer Sci. 2016;107:149-54
125. Ma W, Wang H, Jing W, Zhou F, Chang L, Hong Z. et al. Downregulation of long non-coding RNAs JPX and XIST is associated with the prognosis of hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2017;41:163-70
126. Jing W, Gao S, Zhu M, Luo P, Jing X, Chai H. et al. Potential diagnostic value of lncRNA SPRY4-IT1 in hepatocellular carcinoma. Oncol Rep. 2016;36:1085-92
127. El-Tawdi AH, Matboli M, El-Nakeep S, Azazy AE, Abdel-Rahman O. Association of long noncoding RNA and c-JUN expression in hepatocellular carcinoma. Expert Rev Gastroenterol Hepatol. 2016;10:869-77
128. Zhang JH, Wei HW, Yang HG. Long noncoding RNA SNHG15, a potential prognostic biomarker for hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2016;20:1720-4
129. Zhang D, Cao C, Liu L, Wu D. Up-regulation of LncRNA SNHG20 Predicts Poor Prognosis in Hepatocellular Carcinoma. J Cancer. 2016;7:608-17
130. Chang L, Li C, Lan T, Wu L, Yuan Y, Liu Q. et al. Decreased expression of long non-coding RNA GAS5 indicates a poor prognosis and promotes cell proliferation and invasion in hepatocellular carcinoma by regulating vimentin. Mol Med Rep. 2016;13:1541-50
131. Wang F, Xie C, Zhao W, Deng Z, Yang H, Fang Q. Long non-coding RNA CARLo-5 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Clin Exp Med. 2017;17:33-43
132. Zhang T, Cao C, Wu D, Liu L. SNHG3 correlates with malignant status and poor prognosis in hepatocellular carcinoma. Tumour Biol. 2016;37:2379-85
133. Pan Y, Qin T, Yin S, Zhang X, Gao X, Mu L. Long non-coding RNA UC001kfo promotes hepatocellular carcinoma proliferation and metastasis by targeting alpha-SMA. Biomed Pharmacother. 2017;87:669-77
134. Zeng T, Wang D, Chen J, Chen K, Yu G, Chen Q. et al. AF119895 regulates NXF3 expression to promote migration and invasion of hepatocellular carcinoma through an interaction with miR-6508-3p. Exp Cell Res. 2018;363:129-39
135. Lu X, Zhou C, Li R, Liang Z, Zhai W, Zhao L. et al. Critical role for the long non-coding RNA AFAP1-AS1 in the proliferation and metastasis of hepatocellular carcinoma. Tumour Biol. 2016;37:9699-707
136. Wen J, Xu J, Sun Q, Xing C, Yin W. Upregulation of long non coding RNA PCAT-1 contributes to cell proliferation, migration and apoptosis in hepatocellular carcinoma. Mol Med Rep. 2016;13:4481-6
137. Ma J, Li T, Han X, Yuan H. Knockdown of LncRNA ANRIL suppresses cell proliferation, metastasis, and invasion via regulating miR-122-5p expression in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2018;144:205-14
138. Zhao L, Zhang Y. Long noncoding RNA CASC2 regulates hepatocellular carcinoma cell oncogenesis through miR-362-5p/Nf-kappaB axis. J Cell Physiol. 2018
139. Zhou N, Si Z, Li T, Chen G, Zhang Z, Qi H. Long non-coding RNA CCAT2 functions as an oncogene in hepatocellular carcinoma, regulating cellular proliferation, migration and apoptosis. Oncol Lett. 2016;12:132-8
140. Ye Y, Xu Y, Lai Y, He W, Li Y, Wang R. et al. Long non-coding RNA cox-2 prevents immune evasion and metastasis of hepatocellular carcinoma by altering M1/M2 macrophage polarization. J Cell Biochem. 2018;119:2951-63
141. Yuan SX, Wang J, Yang F, Tao QF, Zhang J, Wang LL. et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016;63:499-511
142. Jiangzheng Z, Cai X, Hao X, Huang F, He Z, Sun H. et al. LncRNA FUNDC2P4 down-regulation promotes epithelial-mesenchymal transition by reducing E-cadherin expression in residual hepatocellular carcinoma after insufficient radiofrequency ablation. Int J Hyperthermia. 2018:1-17
143. Hu L, Ye H, Huang G, Luo F, Liu Y, Yang X. et al. Long noncoding RNA GAS5 suppresses the migration and invasion of hepatocellular carcinoma cells via miR-21. Tumour Biol. 2016;37:2691-702
144. Zhu XT, Yuan JH, Zhu TT, Li YY, Cheng XY. Long noncoding RNA glypican 3 (GPC3) antisense transcript 1 promotes hepatocellular carcinoma progression via epigenetically activating GPC3. FEBS J. 2016;283:3739-54
145. Quagliata L, Quintavalle C, Lanzafame M, Matter MS, Novello C, di Tommaso L. et al. High expression of HOXA13 correlates with poorly differentiated hepatocellular carcinomas and modulates sorafenib response in in vitro models. Lab Invest. 2018;98:95-105
146. Li D, Liu X, Zhou J, Hu J, Zhang D, Liu J. et al. Long noncoding RNA HULC modulates the phosphorylation of YB-1 through serving as a scaffold of extracellular signal-regulated kinase and YB-1 to enhance hepatocarcinogenesis. Hepatology. 2017;65:1612-27
147. Li SP, Xu HX, Yu Y, He JD, Wang Z, Xu YJ. et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget. 2016;7:42431-46
148. Lu Z, Xiao Z, Liu F, Cui M, Li W, Yang Z. et al. Long non-coding RNA HULC promotes tumor angiogenesis in liver cancer by up-regulating sphingosine kinase 1 (SPHK1). Oncotarget. 2016;7:241-54
149. Gao Y, Wang G, Zhang C, Lin M, Liu X, Zeng Y. et al. Long non-coding RNA linc-cdh4-2 inhibits the migration and invasion of HCC cells by targeting R-cadherin pathway. Biochem Biophys Res Commun. 2016;480:348-54
150. Liang WC, Ren JL, Wong CW, Chan SO, Waye MM, Fu WM. et al. LncRNA-NEF antagonized epithelial to mesenchymal transition and cancer metastasis via cis-regulating FOXA2 and inactivating Wnt/beta-catenin signaling. Oncogene. 2018;37:1445-56
151. Zhu P, Wang Y, Wu J, Huang G, Liu B, Ye B. et al. LncBRM initiates YAP1 signalling activation to drive self-renewal of liver cancer stem cells. Nat Commun. 2016;7:13608
152. Ding LJ, Li Y, Wang SD, Wang XS, Fang F, Wang WY. et al. Long Noncoding RNA lncCAMTA1 Promotes Proliferation and Cancer Stem Cell-Like Properties of Liver Cancer by Inhibiting CAMTA1. Int J Mol Sci. 2016;17:E1617
153. Chen ZZ, Huang L, Wu YH, Zhai WJ, Zhu PP, Gao YF. LncSox4 promotes the self-renewal of liver tumour-initiating cells through Stat3-mediated Sox4 expression. Nat Commun. 2016;7:12598
154. Malakar P, Shilo A, Mogilevsky A, Stein I, Pikarsky E, Nevo Y. et al. Long Noncoding RNA MALAT1 Promotes Hepatocellular Carcinoma Development by SRSF1 Upregulation and mTOR Activation. Cancer Res. 2017;77:1155-67
155. Chen L, Yao H, Wang K, Liu X. Long Non-Coding RNA MALAT1 Regulates ZEB1 Expression by Sponging miR-143-3p and Promotes Hepatocellular Carcinoma Progression. J Cell Biochem. 2017;118:4836-43
156. Dong L, Ni J, Hu W, Yu C, Li H. Upregulation of Long Non-Coding RNA PlncRNA-1 Promotes Metastasis and Induces Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma. Cell Physiol Biochem. 2016;38:836-46
157. Wang P, Cui J, Wen J, Guo Y, Zhang L, Chen X. Cisplatin induces HepG2 cell cycle arrest through targeting specific long noncoding RNAs and the p53 signaling pathway. Oncol Lett. 2016;12:4605-12
158. Ge Z, Cheng Z, Yang X, Huo X, Wang N, Wang H. et al. Long noncoding RNA SchLAH suppresses metastasis of hepatocellular carcinoma through interacting with fused in sarcoma. Cancer Sci. 2017;108:653-62
159. Zhang H, Zhou D, Ying M, Chen M, Chen P, Chen Z. et al. Expression of Long Non-Coding RNA (lncRNA) Small Nucleolar RNA Host Gene 1 (SNHG1) Exacerbates Hepatocellular Carcinoma Through Suppressing miR-195. Med Sci Monit. 2016;22:4820-9
160. Zhang M, Wang W, Li T, Yu X, Zhu Y, Ding F. et al. Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis. Biomed Pharmacother. 2016;80:73-9
161. Liu J, Lu C, Xiao M, Jiang F, Qu L, Ni R. Long non-coding RNA SNHG20 predicts a poor prognosis for HCC and promotes cell invasion by regulating the epithelial-to-mesenchymal transition. Biomed Pharmacother. 2017;89:857-63
162. Cao C, Zhang T, Zhang D, Xie L, Zou X, Lei L. et al. The long non-coding RNA, SNHG6-003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma. Oncogene. 2017;36:1112-22
163. Zhou M, Zhang XY, Yu X. Overexpression of the long non-coding RNA SPRY4-IT1 promotes tumor cell proliferation and invasion by activating EZH2 in hepatocellular carcinoma. Biomed Pharmacother. 2017;85:348-54
164. Carotenuto P, Fassan M, Pandolfo R, Lampis A, Vicentini C, Cascione L. et al. Wnt signalling modulates transcribed-ultraconserved regions in hepatobiliary cancers. Gut. 2017;66:1268-77
165. Dong R, Liu GB, Liu BH, Chen G, Li K, Zheng S. et al. Targeting long non-coding RNA-TUG1 inhibits tumor growth and angiogenesis in hepatoblastoma. Cell Death Dis. 2016;7:e2278
166. Wang Y, Liu Z, Yao B, Dou C, Xu M, Xue Y. et al. Long non-coding RNA TUSC7 acts a molecular sponge for miR-10a and suppresses EMT in hepatocellular carcinoma. Tumour Biol. 2016;37:11429-41
167. Hu JJ, Song W, Zhang SD, Shen XH, Qiu XM, Wu HZ. et al. HBx-upregulated lncRNA UCA1 promotes cell growth and tumorigenesis by recruiting EZH2 and repressing p27Kip1/CDK2 signaling. Sci Rep. 2016;6:23521
168. Lv J, Fan HX, Zhao XP, Lv P, Fan JY, Zhang Y. et al. Long non-coding RNA Unigene56159 promotes epithelial-mesenchymal transition by acting as a ceRNA of miR-140-5p in hepatocellular carcinoma cells. Cancer Lett. 2016;382:166-75
169. Lan T, Chang L, Wu L, Yuan Y. Downregulation of ZEB2-AS1 decreased tumor growth and metastasis in hepatocellular carcinoma. Mol Med Rep. 2016;14:4606-12
Author contact
![]() Corresponding author: Prof. Guan-Cheng Li, Key Laboratory of Carcinogenesis of the Chinese Ministry of Health and the Key Laboratory of Carcinogenesis and Cancer Invasion of Chinese Ministry of Education, Xiangya Hospital, Central South University; Cancer Research Institute, Central South University, 110 Xiangya Road, Changsha, Hunan 410078, P.R. China. Tel: +86 731 84805445; Fax: +86 731 82355042; E-mail: ligc61edu.cn (G.-C. Li)
Corresponding author: Prof. Guan-Cheng Li, Key Laboratory of Carcinogenesis of the Chinese Ministry of Health and the Key Laboratory of Carcinogenesis and Cancer Invasion of Chinese Ministry of Education, Xiangya Hospital, Central South University; Cancer Research Institute, Central South University, 110 Xiangya Road, Changsha, Hunan 410078, P.R. China. Tel: +86 731 84805445; Fax: +86 731 82355042; E-mail: ligc61edu.cn (G.-C. Li)

 Global reach, higher impact
Global reach, higher impact