3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(16):2963-2972. doi:10.7150/jca.25638 This issue Cite
Review
ESR1 PvuII (rs2234693 T>C) polymorphism and cancer susceptibility: Evidence from 80 studies
1. Department of Pharmacy, Guangdong General Hospital, Guangdong Academy of Medical Sciences, Guangzhou 510080, Guangdong, China
2. Department of Pharmacy, The First Affiliated Hospital, Jinan University, Guangzhou 510630, Guangdong, China
3. Laboratory Animal Management Office, Public Service Platform for Science and Technology, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen 518055, Guangdong, China
*These authors contributed equally to this work.
Received 2018-2-21; Accepted 2018-6-9; Published 2018-7-30
Abstract
Emerging epidemiological researches have been performed to assess the association of ESR1 PvuII (rs2234693 T>C) polymorphism with the risk of cancer, yet with conflicting conclusions. Therefore, this updated meta-analysis was performed to make a more accurate evaluation of such relationship. We adopted EMBASE, PubMed, CNKI, and WANFANG database to search relevant literature before January 2018. Odds ratios (ORs) and 95% confidence intervals (CIs) were employed to estimate the relationship strengths. In final, 80 studies (69 publications) involving 26428 cases and 43381 controls were enrolled. Our results failed to provide significant association between overall cancer risk and PvuII polymorphism under homozygous (TT vs. CC) and heterozygous (TT vs. CT) models. Statistically significant relationship was only observed for PvuII polymorphism in allele model T vs. C (OR=0.95, 95% CI=0.91-0.99). Stratification analysis by cancer type suggested that T genotype significantly decreased prostate cancer risk (TT vs. CC: OR=0.79, 95% CI=0.66-0.94; T vs. C: OR=0.89, 95% CI=0.82-0.98), Leiomyoma risk (T vs. C: OR=0.82, 95% CI=0.68-0.98), and HCC risk (TT vs. CC: OR=0.45, 95% CI=0.28-0.71; T vs. C: OR=0.67, 95% CI=0.47-0.95). Furthermore, significantly decreased risk was also found for Africans, population-based and hospital-based studies in the stratified analyses. These results suggest that ESR1 PvuII (rs2234693 T>C) polymorphism may only have little impact on cancer susceptibility. In the future, large-scale epidemical studies are warranted to verify these results.
Keywords: meta-analysis, ESR1, PvuII, polymorphism, cancer risk
Introduction
Worldwide, cancer still ranks the number one killer that threatens people's life. Approximately 14.1 million new cancer cases and 8.2 million cancer-caused deaths occurred globally in 2013 [1]. In 2018, 1,735,350 new cancer cases and 609,640 cancer deaths are projected to occur in the United States [2]. By now, the definitive etiology of cancer remains unknown. However, a myriad of evidence has suggested that cancer is a complex disease caused by both genetic and environmental factors [3, 4]. Numerous functional polymorphisms have been found to be implicated in the development of cancers [5-7].
Previous researches have reported that hormonal factors play crucial roles in the development of some cancers. Common genetic variants in hormonal-related genes were associated with cancer susceptibility [8]. Among them, estrogen receptor (ER) was the most related-hormone in cancer risk. Estrogen receptor (ER) has two forms, which is alpha and beta [9]. Estrogen receptor-α plays a critical role in mediating hormonal response in estrogen-sensitive tissues. It consists of several domains important for hormone regulation, activation of transcription and DNA binding. Evidence points to estrogen receptor-α as the main receptor correlated to initiation of cancer [10]. Estrogen receptor-α, a transcription factor, is encoded by the ESR1 gene.
The ESR1 gene, comprises of 8 exons and 7 introns, is located on chromosome 6q25.1. Several SNPs of ESR1 gene have been identified to influence the risk of cancer, but the most popular studied SNP is ESR1 PvuII (rs2234693 T>C) polymorphism [11]. Although increasing studies have been performed, the conclusions of the roles of ESR1 PvuII (T>C) polymorphism in cancer risk are conflicting. The inconsistent conclusions between ESR1 PvuII (rs2234693 T>C) polymorphism and cancer risk may be due to the limitations in the sample size of the corresponding studies or the inadequate statistical power in genetic studies with complex characteristics. Several meta-analyses regarding this issue have been performed to resolve the conflicting situation but somehow failed. With the aim to solve such embarrassment, we conducted this comprehensive meta-analysis by adopting all published articles.
Materials and methods
Publication search
We first inputted the following key words: “single nucleotide polymorphism or polymorphism or variant or SNP” and “ESR1 or ESRα or Estrogen Receptor α or Estrogen Receptor 1”, and “cancer or tumor or neoplasm or carcinoma)” in database of PubMed and EMBASE. In addition, we also searched the Chinese database CNKI and WANFANG to include more eligible studies. Further, additional studies were also manually extracted from the references of the above obtained publications. The date of the final literature search was set on January 2018. We did not set any language publication restrictions here. The article will be considered as different studies if it contains more than two ethnicities. If the searched articles have overlapping data, the largest one will be selected.
Eligibility criteria
The evaluating publications in this meta-analysis should fulfill all the following requirements: 1) unrelated case-control studies; 2) original epidemiological studies; 3) analyzing the relationship between ESR1 PvuII (rs2234693 T>C) polymorphism and cancer risk; 4) enough data to obtain odds ratios (ORs) and 95% confidence intervals (CIs); 5) articles written in English or in Chinese.
Data extraction
Two authors separately extracted data by screening all eligible studies. They collected the information regarding first author's surname, country, publication year, ethnicity, genotyping methods, the source of controls, and numbers of cases and controls with CC, CT and TT genotypes. All the disagreed information was settle down after fully discussed by the two authors.
Statistical methods
Hardy-Weinberg equilibrium (HWE) in the controls was determined using goodness-of-fit χ2 test. P<0.05 was considered as departure from HWE. Three genetic models, homozygous model (TT vs. CC), heterozygous model (TT vs. CT), and allele comparison (T vs. C), were applied to assess the association between ESR1 PvuII (rs2234693 T>C) polymorphism and cancer risk. The strength of such association was assessed by calculating ORs with the corresponding 95% CIs. Stratification analyses were also conducted by ethnicity, cancer type, source of control, and HWE in controls, in all studies. Chi square-based Q-test was adopted to monitor between-study heterogeneity. The fixed-effects model (the Mantel-Haenszel method) was chosen to estimate the pooled OR, if the studies were homogeneous (P>0.10 for the Q test). Otherwise, the random-effects model (the DerSimonian and Laird method) was used. Sensitivity analysis was conducted by excluding each study individually and re-calculating the ORs and 95% CIs. Begg's funnel plot and Egger's linear regression were used to evaluate whether there exists publication bias [12, 13]. The asymmetric plot and P value less than 0.5 was considered as the existence of publication bias. We also conducted quality assessment to detect the quality of each study using the quality assessment criteria [14]. The version 11.0 STATA software was adopted to perform all statistical analysis (Stata Corporation, College Station, TX). All the statistics were two-sided with P value of <0.05 as significant findings.
Results
Study characteristics
Our first research in databases identified 185 candidate publications. After screening the title and abstract, we kept 64 publication s in the analysis [15-78]. Moreover, we further extracted 5 articles from the references of the retrieval articles [79-83]. The flow chart of screening process was graphically shown in Figure 1. In final, 80 studies (69 publications) with 26428 cases and 43381 controls were included in the pooled analysis (Table 1). Among them, 38 studies focused on Asians, 36 on Caucasians, 3 on Africans, 1 on Hispanics and 1 on non-Hispanic Caucasians, 1 on Hispanic Caucasians. 44 studies were hospital-based design, 36 were population-based design. The controls' genotype frequencies were in agreement with HWE (P>0.05) in 74 studies, except for 6 studies.
Flowchart of study selection process.
The baseline characteristics of all qualified studies in this meta-analysis
| Surname | Year | Country | Ethnicity | Cancer type | Control Source | Genotype method | Case | Control | HWE | Score | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TT | CT | CC | All | TT | CT | CC | All | |||||||||
| Modugno | 2001 | USA | Caucasian | Prostate | PB | PCR | 26 | 34 | 21 | 81 | 85 | 109 | 43 | 237 | 0.438 | 8 |
| Massart | 2001 | Italy | Caucasian | Leiomyoma | HB | PCR | 35 | 57 | 27 | 119 | 46 | 77 | 33 | 156 | 0.941 | 5 |
| Suzuki | 2003 | Japan | Asian | Prostate | PB | PCR | 46 | 43 | 12 | 101 | 29 | 59 | 26 | 114 | 0.702 | 9 |
| Massart | 2003 | Italy | Caucasian | Leiomyoma | HB | PCR-RFLP | 54 | 91 | 43 | 188 | 66 | 111 | 48 | 225 | 0.917 | 5 |
| Iwamoto | 2003 | Japan | Asian | Endometrial | HB | PCR-RFLP | 25 | 54 | 13 | 92 | 25 | 28 | 12 | 65 | 0.408 | 4 |
| Shin | 2003 | Korea | Asian | Breast | PB | PCR-RFLP | 75 | 91 | 35 | 201 | 64 | 105 | 26 | 195 | 0.095 | 8 |
| Tanaka | 2003 | Japan | Asian | Prostate | HB | PCR | 23 | 63 | 29 | 115 | 39 | 113 | 48 | 200 | 0.061 | 6 |
| Cai | 2003 | China | Asian | Breast | PB | PCR-RFLP | 415 | 516 | 138 | 1069 | 430 | 546 | 190 | 1166 | 0.452 | 12 |
| Fukatsu | 2004 | Japan | Asian | Prostate | HB | PCR-RFLP | 37 | 57 | 22 | 116 | 81 | 110 | 47 | 238 | 0.384 | 6 |
| wedren | 2004 | Sweden | Caucasian | Breast | PB | PCR-RFLP | 390 | 634 | 268 | 1292 | 384 | 651 | 313 | 1348 | 0.248 | 10 |
| Lu | 2005 | China | Asian | Breast | HB | PCR-RFLP | 54 | 65 | 19 | 138 | 50 | 69 | 21 | 140 | 0.723 | 78 |
| Modugno | 2005 | USA | Caucasian | Breast | PB | PCR-RFLP | 53 | 115 | 80 | 248 | 819 | 1810 | 1272 | 3901 | 0.000 | 6 |
| Onland-Moret | 2005 | Netherlands | Caucasian | Breast | PB | PCR-RFLP | 89 | 150 | 69 | 308 | 88 | 153 | 96 | 337 | 0.093 | 9 |
| Low | 2006 | UK | Caucasian | Prostate | PB | TaqMan | 13 | 41 | 21 | 75 | 49 | 84 | 25 | 158 | 0.266 | 2 |
| Al-Hendy | 2006 | USA | African | Leiomyoma | HB | PCR-RFLP | 22 | 34 | 36 | 92 | 9 | 9 | 3 | 21 | 0.760 | 3 |
| Al-Hendy | 2006 | USA | Caucasian | Leiomyoma | HB | PCR-RFLP | 21 | 23 | 17 | 61 | 57 | 99 | 1 | 157 | 0.000 | 2 |
| Al-Hendy | 2006 | USA | Hispanic | Leiomyoma | HB | PCR-RFLP | 14 | 23 | 8 | 45 | 27 | 18 | 6 | 51 | 0.284 | 11 |
| Zhai | 2006 | China | Asian | HCC | PB | PCR-RFLP | 74 | 117 | 53 | 244 | 91 | 116 | 30 | 237 | 0.457 | 6 |
| Chen | 2006 | China | Asian | Leiomyoma | HB | PCR-RFLP | 35 | 37 | 11 | 83 | 31 | 38 | 9 | 78 | 0.604 | 5 |
| Denschlag | 2006 | Germany | Caucasian | Leiomyoma | PB | PCR | 33 | 66 | 31 | 130 | 40 | 59 | 40 | 139 | 0.075 | 9 |
| Hernandez | 2006 | USA | Caucasian | Prostate | PB | TaqMan | 47 | 55 | 18 | 120 | 129 | 131 | 43 | 303 | 0.300 | 11 |
| Hernandez | 2006 | USA | Caucasian | Prostate | PB | TaqMan | 115 | 216 | 100 | 431 | 154 | 296 | 132 | 582 | 0.653 | 9 |
| Hernandez | 2006 | USA | African | Prostate | PB | TaqMan | 9 | 22 | 16 | 47 | 50 | 113 | 50 | 213 | 0.373 | 11 |
| Shen | 2006 | China | Asian | Breast | PB | PCR-RFLP | 98 | 120 | 29 | 247 | 107 | 124 | 43 | 274 | 0.480 | 10 |
| Cunningham | 2007 | Minnesota | Caucasian | Prostate | PB | PCR | 257 | 454 | 213 | 924 | 120 | 249 | 120 | 489 | 0.684 | 9 |
| Berndt | 2007 | USA | Caucasian | Prostate | HB | PCR | 121 | 238 | 111 | 470 | 152 | 316 | 135 | 603 | 0.230 | 9 |
| Hsieh | 2007 | China | Asian | Leiomyoma | PB | PCR-RFLP | 25 | 75 | 6 | 106 | 60 | 44 | 6 | 110 | 0.571 | 7 |
| Hu | 2007 | China | Asian | Breast | HB | PCR-RFLP | 39 | 58 | 16 | 113 | 49 | 45 | 19 | 113 | 0.128 | 7 |
| Kadiyska | 2007 | Bulgaria | Caucasian | Colorectal | HB | PCR-RFLP | 34 | 79 | 27 | 140 | 23 | 35 | 19 | 77 | 0.438 | 11 |
| Kjaergaard | 2007 | Danmark | Caucasian | Prostate | PB | TaqMan | 35 | 55 | 26 | 116 | 1203 | 1972 | 830 | 4005 | 0.676 | 11 |
| Kjaergaard | 2007 | Danmark | Caucasian | Breast | PB | TaqMan | 398 | 613 | 245 | 1256 | 727 | 1225 | 537 | 2489 | 0.621 | 7 |
| Wang | 2007 | USA | Caucasian | Breast | PB | PCR | 117 | 188 | 87 | 392 | 214 | 393 | 176 | 783 | 0.862 | 4 |
| Onsory | 2008 | India | Asian | Prostate | HB | PCR-RFLP | 28 | 54 | 18 | 100 | 42 | 48 | 10 | 100 | 0.487 | |
| González-Mancha | 2008 | Spain | Caucasian | Breast | PB | PCR-RFLP | 153 | 209 | 82 | 444 | 193 | 361 | 150 | 704 | 0.435 | 6 |
| Sobti | 2008 | India | Asian | Prostate | HB | PCR | 52 | 77 | 28 | 157 | 64 | 90 | 16 | 170 | 0.050 | 6 |
| Gonzalez-Zuloeta | 2008 | Netherlands | Caucasian | Breast | PB | PCR-RFLP | 72 | 94 | 24 | 190 | 1602 | 1648 | 453 | 3703 | 0.359 | 6 |
| Dunning | 2009 | UK | Caucasian | Breast | PB | TaqMan | 1260 | 2164 | 938 | 4362 | 1318 | 2296 | 934 | 4548 | 0.253 | 8 |
| Ashton | 2009 | Australia | Caucasian | Endometrial | PB | PCR-RLFP | 39 | 95 | 57 | 191 | 96 | 129 | 65 | 290 | 0.088 | 11 |
| Iwasaki | 2009 | Japan | Asian | Breast | HB | TaqMan | 144 | 180 | 64 | 388 | 115 | 196 | 77 | 388 | 0.692 | 10 |
| Iwasaki | 2009 | Japan | Asian | Breast | HB | TaqMan | 25 | 39 | 15 | 79 | 22 | 43 | 14 | 79 | 0.374 | 9 |
| Iwasaki | 2009 | Japan | Asian | Breast | HB | TaqMan | 107 | 187 | 85 | 379 | 122 | 194 | 63 | 379 | 0.338 | 10 |
| Sonestedt | 2009 | Sweden | Caucasian | Breast | PB | MassARRAY | 158 | 273 | 108 | 539 | 316 | 539 | 218 | 1073 | 0.667 | 10 |
| Beuten | 2009 | USA | non-Hispanic Caucasians | Prostate | PB | PCR | 167 | 304 | 138 | 609 | 222 | 421 | 200 | 843 | 0.988 | 7 |
| Beuten | 2009 | USA | Hispanic Caucasians | Prostate | PB | PCR | 75 | 92 | 28 | 195 | 186 | 246 | 82 | 514 | 0.964 | 7 |
| Beuten | 2009 | USA | African | Prostate | PB | PCR | 18 | 41 | 23 | 82 | 54 | 105 | 50 | 209 | 0.940 | 7 |
| Anghel | 2009 | Romania | Caucasian | Bladder | HB | PCR | 0 | 6 | 9 | 15 | 18 | 48 | 48 | 114 | 0.309 | 5 |
| Anghel | 2009 | Romania | Caucasian | Colorectal | HB | PCR | 2 | 13 | 3 | 18 | 18 | 48 | 48 | 114 | 0.309 | 5 |
| Anghel | 2009 | Romania | Caucasian | AML | HB | PCR | 0 | 5 | 10 | 15 | 18 | 48 | 48 | 114 | 0.309 | 5 |
| Anghel | 2009 | Romania | Caucasian | HCC | HB | PCR | 2 | 6 | 4 | 12 | 18 | 48 | 48 | 114 | 0.309 | 5 |
| Anghel | 2009 | Romania | Caucasian | Breast | HB | PCR | 4 | 65 | 32 | 101 | 15 | 38 | 37 | 90 | 0.333 | 6 |
| Wang JY | 2010 | China | Asian | Leiomyoma | HB | PCR-RFLP | 24 | 46 | 22 | 92 | 51 | 100 | 42 | 193 | 0.592 | 6 |
| Wang XL | 2010 | China | Asian | Leiomyoma | HB | PCR-RFLP | 42 | 48 | 12 | 102 | 35 | 49 | 16 | 100 | 0.867 | 6 |
| Gupta | 2010 | India | Asian | Prostate | HB | PCR-RFLP | 52 | 77 | 28 | 157 | 64 | 90 | 16 | 170 | 0.049 | 6 |
| Park | 2010 | China | Asian | Gallbladder | PB | PCR-RFLP | 41 | 100 | 94 | 235 | 108 | 356 | 314 | 778 | 0.658 | 11 |
| Sonoda | 2010 | Japan | Asian | Prostate | HB | PCR | 60 | 89 | 31 | 180 | 61 | 87 | 29 | 177 | 0.828 | 5 |
| Sakoda | 2011 | China | Asian | Breast | PB | PCR | 229 | 290 | 93 | 612 | 327 | 427 | 120 | 874 | 0.298 | 12 |
| Deng | 2011 | China | Asian | Breast | HB | PCR-RFLP | 42 | 63 | 23 | 128 | 52 | 61 | 17 | 130 | 0.892 | 7 |
| Wang | 2011 | China | Asian | Cervical | HB | PCR-RFLP | 39 | 45 | 18 | 102 | 32 | 52 | 18 | 102 | 0.692 | 6 |
| Sissung | 2011 | USA | Caucasian | Prostate | PB | TaqMan | 25 | 75 | 28 | 128 | 46 | 60 | 20 | 126 | 0.952 | 3 |
| de Giorgi | 2011 | Italy | Caucasian | Melanoma | HB | PCR-RFLP | 32 | 49 | 31 | 112 | 56 | 98 | 41 | 195 | 0.876 | 6 |
| Balistreri | 2011 | Italy | Caucasian | Prostate | HB | PCR-RFLP | 37 | 11 | 2 | 50 | 84 | 7 | 0 | 91 | 0.702 | 4 |
| Han | 2011 | China | Asian | Breast | PB | TaqMan | 353 | 399 | 107 | 859 | 324 | 402 | 151 | 877 | 0.171 | 9 |
| Szendroi | 2011 | Hungary | Caucasian | Prostate | HB | PCR-RFLP | 43 | 122 | 39 | 204 | 31 | 47 | 25 | 103 | 0.392 | 7 |
| Lundie | 2012 | USA | Caucasian | Endometrial | PB | PCR | 116 | 184 | 91 | 391 | 194 | 369 | 146 | 709 | 0.223 | 9 |
| Srivastava | 2012 | India | Asian | Gallbladder | PB | PCR-RFLP | 59 | 218 | 133 | 410 | 19 | 110 | 91 | 220 | 0.075 | 12 |
| Safarinejad | 2012 | Iran | Asian | Prostate | PB | PCR-RFLP | 11 | 94 | 57 | 162 | 65 | 169 | 90 | 324 | 0.373 | 6 |
| Chang | 2012 | China | Asian | Lung | HB | PCR-RFLP | 21 | 60 | 3 | 84 | 62 | 132 | 40 | 234 | 0.034 | 4 |
| Tang | 2013 | China | Asian | Breast | HB | MALDI-TOF | 293 | 374 | 127 | 794 | 334 | 375 | 136 | 845 | 0.076 | 9 |
| Jurecekova | 2013 | Slovak | Caucasian | Prostate | HB | PCR | 78 | 154 | 79 | 311 | 81 | 126 | 49 | 256 | 1 | 5 |
| Pazarbasi | 2013 | Turkey | Caucasian | Prostate | HB | PCR | 14 | 14 | 6 | 34 | 10 | 7 | 10 | 27 | 0.012 | 3 |
| Ramalhinho | 2013 | Portugal | Caucasian | Breast | HB | PCR-RFLP | 28 | 60 | 19 | 107 | 45 | 60 | 16 | 121 | 0.566 | 7 |
| Liu | 2014 | China | Asian | HCC | HB | PCR | 34 | 54 | 19 | 107 | 57 | 38 | 10 | 105 | 0.331 | 6 |
| Chattopadhyay | 2014 | India | Asian | Breast | PB | PCR-RFLP | 157 | 164 | 39 | 360 | 136 | 162 | 62 | 360 | 0.252 | 11 |
| Lu | 2014 | China | Asian | Breast | HB | PCR-RFLP | 227 | 258 | 57 | 542 | 425 | 454 | 137 | 1016 | 0.368 | 5 |
| Madeira | 2014 | Brazil | Asian | Breast | HB | PCR-RFLP | 6 | 49 | 9 | 64 | 25 | 39 | 8 | 72 | 0.211 | 6 |
| Taghizade | 2014 | Iran | Asian | Leiomyoma | HB | PCR-RFLP | 78 | 133 | 65 | 276 | 50 | 74 | 33 | 157 | 0.563 | 7 |
| Cao | 2014 | China | Asian | Breast | HB | PCR-RFLP | 70 | 109 | 42 | 221 | 79 | 124 | 49 | 252 | 0.978 | 7 |
| Lu | 2015 | Japan | Asian | Prostate | HB | TaqMan | 67 | 191 | 94 | 352 | 80 | 175 | 97 | 352 | 0.949 | 7 |
| Nyante | 2015 | USA | Caucasian | Breast | PB | PCR | 518 | 984 | 470 | 1972 | 469 | 908 | 398 | 1775 | 0.297 | 11 |
| Han | 2017 | China | Asian | Prostate | HB | PCR | 94 | 102 | 48 | 244 | 92 | 112 | 28 | 232 | 0.492 | 8 |
Abbreviations: HB, hospital based; PB, population based; PCR, polymerase chain reaction; PCR-RFLP, PCR-restriction fragment length polymorphism; HCC, hepatocarcinoma; AML, acute myeloid leukemia; HWE, Hardy-Weinberg equilibrium.
Meta-analysis results
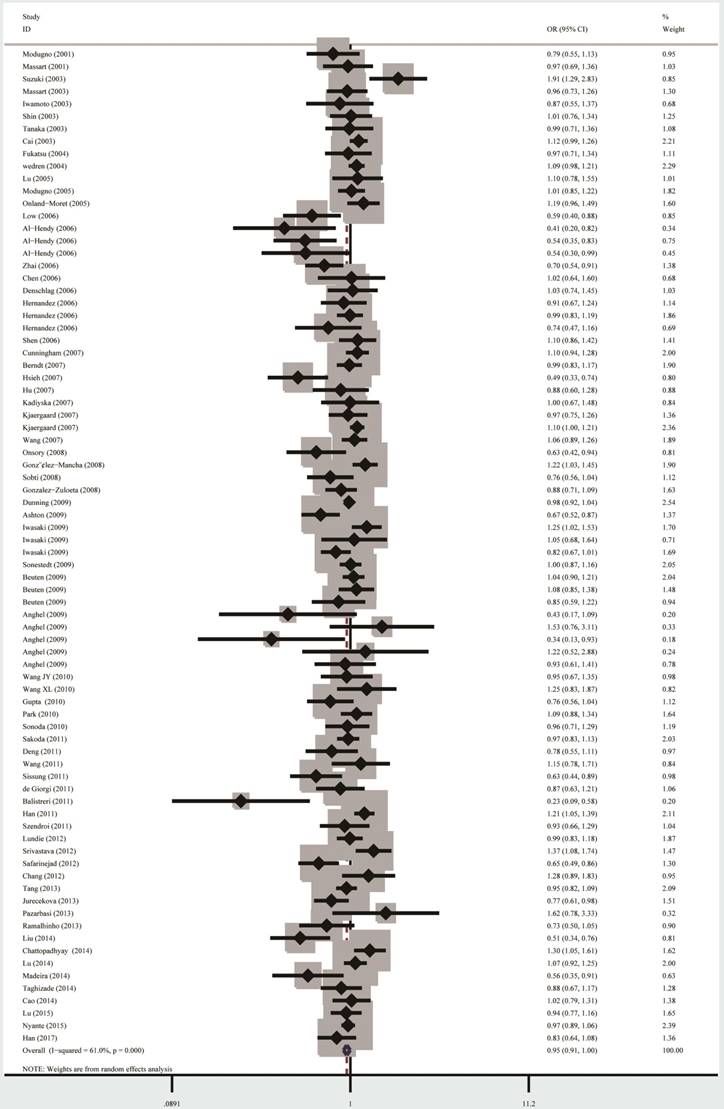
The summary results of meta-analysis were presented in Table 2 and Figure 2. In all, no significant association between the ESR1 PvuII (rs2234693 T>C) polymorphism and cancer risk was observed under homozygous model (TT vs. CC: OR=0.92, 95% CI=0.84-1.01) and heterozygous model (TT vs. CT: OR=0.94, 95% CI=0.88-1.001). Statistically significant relationship was only observed for PvuII in allele model T vs. C (OR=0.95, 95% CI=0.91-0.99).
In subgroup analysis by cancer type, we found that the T genotype significantly decreased prostate cancer risk (TT vs. CC: OR=0.79, 95% CI=0.66-0.94; T vs. C: OR=0.89, 95% CI=0.82-0.98), Leiomyoma risk (T vs. C: OR=0.82, 95% CI=0.68-0.98), and HCC risk (TT vs. CC: OR=0.45, 95% CI=0.28-0.71; T vs. C: OR=0.67, 95% CI=0.47-0.95). However, no relationship between ESR1 PvuII polymorphism and any other types of cancer was observed. Ethnicity subgroup analysis revealed that significant association between ESR1 PvuII genotype and cancer risk was detected among African (TT vs. CC: OR=0.54, 95% CI=0.30-0.98), and Hispanics (TT vs. CT: OR=0.41, 95% CI=0.17-0.99; T vs. C: OR=0.55, 95% CI=0.30-0.99). Such association was not observed for the Asians and Caucasians. In terms of source of controls, we found that the ESR1 PvuII T genotype help to decrease cancer risk in hospital-based group (T vs. C: OR=0.89, 95% CI=0.83-0.96) and in population-based group (TT vs. CC: OR=0.81, 95% CI=0.70-0.94; TT vs. CT: OR=0.86, 95% CI=0.78-0.96). Further subgroup analysis by HWE in controls also failed to detect positive association, except for heterogenous model in HWE>0.05 subgroup (TT vs. CT: OR=0.94, 95% CI=0.88-1.00). Subgroup analysis of quality revealed that ESR1 PvuII T genotype help to decrease cancer risk in group with quality score ≤9.
Heterogeneity and sensitivity analysis
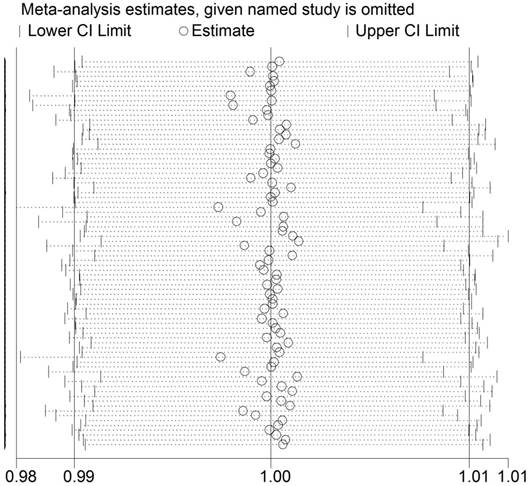
Between-study heterogeneity was first calculated by using Q test and I2 statistics. We used the random-effect model as significant heterogeneity was observed among all three genetic models (P<0.001) in the pooled analysis (TT vs. CC: P<0.001, I2 = 59.1%; TT vs. CT: P<0.001, I2 = 49.4%; T vs. C: P<0.001, I2 = 61.0%). In addition, sequential leave-one-out sensitivity analysis was adopted to evaluate the stability of the results. After removing each study, no substantial changes in pooled results were found (Figure 3).
Publication bias
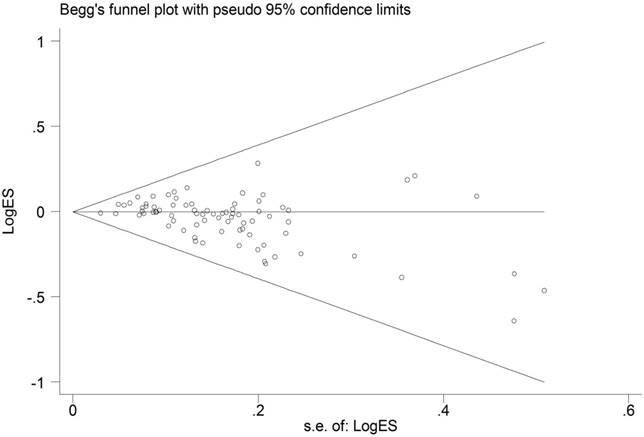
The shape of Begg's funnel plots was quite symmetry (Figure 4). Moreover, statistical evidence of Egger's test also provided the none-existence of publication bias among the studies (data not shown).
Discussion
In this meta-analysis, we comprehensively evaluated the association between ESR1 PvuII (rs2234693 T>C) polymorphism with cancer susceptibility. The obtained results suggested ESR1 PvuII (rs2234693 T>C) polymorphism may influence overall cancer risk in a low impact effect manner. So far, this meta-analysis represents the most powerful investigation in elucidating the role of ESR1 PvuII (rs2234693 T>C) in cancer risk.
The polymorphism of ESR1, PvuII (rs2234693 T>C), can affect ESR1 transcription activity and further contribute to the carcinogenesis. A myriad of studies has investigated the role of ESR1 PvuII (rs2234693 T>C) polymorphisms in cancer risk. In 2001, Massart et al. claimed that the PvuII and XbaI polymorphisms in the ESR1 gene do not produce different risks of developing uterine leiomyomas [52]. In another study performed in urban Shanghai with 1069 breast cancer patients and 1166 controls, Cai et al. found that ESR1 PvuII (rs2234693 T>C) polymorphism conferred to an enhanced risk of breast cancer among subjects carrying Pp (CT) and pp (TT) genotypes [21]. Yet, AI-Hendy et al. claimed that the ESR1PvuII PP (CC) genotype contributed to a significantly increased risk of uterine leiomyomas in black and white women, but not in Hispanic women [15]. Many meta-analyses have been conducted aiming to obtain a clear association between ESR1 PvuII (rs2234693 T>C) and cancer risk. In 2010, Li et al. performed a meta-analysis regarding the association of several potentially functional SNPs in ESR1 with breast cancer risk. This analysis on 10,300 breast cancer cases and 16,620 controls in PvuII (rs2234693 T>C) polymorphism revealed a borderline significant decreased breast cancer risk for CC and CC/CT carriers (CC vs. TT: OR=0.92, 95% CI=0.86-0.99; CC/CT vs. TT: OR=0.95, 95% CI=0.89-1.00) [84]. In a meta-analysis updated to April 2014, 41 studies were included to analyze the relationship between ESR1 PvuII (rs2234693 T>C) and cancer risk. Results of the pooled analysis suggested a null relationship between PvuII (rs2234693 T>C) polymorphism and overall cancer risk. Subgroup analysis indicated that PvuII (rs2234693 T>C) polymorphism was associated with a decreased risk of gallbladder cancer, in contrast with the increased risk of prostate cancer and hepatocellular carcinoma (HCC). They also failed to observe significant association in Asian and Caucasian populations [85].
Meta-analysis of the association between ESR1 PvuII polymorphism and cancer risk
| Variables | No. of studies | Homozygous | Heterozygous | Allele | |||||
|---|---|---|---|---|---|---|---|---|---|
| TT vs. CC | TT vs. CT | T vs. C | |||||||
| OR (95% CI) | P het | OR (95% CI) | P het | OR (95% CI) | P het | ||||
| All | 80 | 0.92 (0.84-1.01) | <0.001 | 0.94 (0.88-1.001) | <0.001 | 0.95 (0.91-0.99) | <0.001 | ||
| Cancer type | |||||||||
| Breast | 28 | 1.08 (0.98-1.19) | 0.001 | 1.01 (0.94-1.08) | 0.015 | 1.03 (0.99-1.08) | 0.004 | ||
| Prostate | 26 | 0.79 (0.66-0.94) | <0.001 | 0.89 (0.78-1.01) | 0.006 | 0.89 (0.82-0.98) | <0.001 | ||
| Leiomyoma | 11 | 0.72 (0.49-1.06) | 0.016 | 0.83 (0.61-1.12) | 0.003 | 0.82 (0.68-0.98) | 0.006 | ||
| HCC | 3 | 0.45 (0.28-0.71) | 0.353 | 0.63 (0.39-1.04) | 0.191 | 0.67 (0.47-0.95) | 0.145 | ||
| Endometrial | 3 | 0.73 (0.43-1.24) | 0.067 | 0.73 (0.40-1.35) | 0.005 | 0.84 (0.63-1.11) | 0.046 | ||
| Others | 9 | 1.26 (0.85-1.90) | 0.070 | 1.06 (0.88-1.40) | 0.203 | 1.06 (0.88-1.28) | 0.042 | ||
| Ethnicity | |||||||||
| Asian | 38 | 0.94 (0.80-1.10) | <0.001 | 0.93 (0.84-1.04) | <0.001 | 0.96 (0.89-1.03) | <0.001 | ||
| Caucasian | 36 | 0.93 (0.83-1.04) | <0.001 | 0.95 (0.88-1.04) | 0.003 | 0.96 (0.90-1.01) | <0.001 | ||
| African | 3 | 0.54 (0.30-0.98) | 0.292 | 0.83 (0.52-1.32) | 0.870 | 0.70 (0.49-1.001) | 0.185 | ||
| Hispanics | 1 | 0.39 (0.11-1.34) | - | 0.41 (0.17-0.99) | - | 0.55 (0.30-0.99) | - | ||
| Non-Hispanic Caucasian | 1 | 1.09 (0.81-1.47) | - | 1.04 (0.81-1.34) | - | 1.04 (0.90-1.21) | - | ||
| Hispanic Caucasian | 1 | 1.18 (0.71-1.96) | - | 1.08 (0.75-1.55) | - | 1.08 (0.85-1.38) | - | ||
| Control source | |||||||||
| HB | 44 | 1.02 (0.91-1.13) | <0.001 | 0.99 (0.92-1.08) | 0.009 | 0.89 (0.83-0.96) | <0.001 | ||
| PB | 36 | 0.81 (0.70-0.94) | <0.001 | 0.86 (0.78-0.96) | <0.001 | 0.99 (0.95-1.05) | <0.001 | ||
| HWE | |||||||||
| >0.05 | 74 | 0.94 (0.86-1.02) | <0.001 | 0.94 (0.88-1.00) | <0.001 | 0.96 (0.92-1.001) | <0.001 | ||
| ≤0.05 | 6 | 0.74 (0.33-1.67) | <0.001 | 0.98 (0.80-1.21) | 0.672 | 0.90 (0.70-1.14) | 0.009 | ||
| Quality score | |||||||||
| >9 | 17 | 1.07 (0.92-1.23) | 0.386 | 1.04 (0.98-1.11) | 0.327 | 1.03 (0.96-1.10) | <0.001 | ||
| ≤9 | 63 | 0.86 (0.77-0.96) | 0.008 | 0.88 (0.81-0.96) | <0.001 | 0.92 (0.87-0.97) | <0.001 | ||
Abbreviations: Het, heterogeneity; HB, hospital based; PB, population based.
Forest plot for the overall cancer susceptibility associated with the ESR1 PvuII (T>C) polymorphism under allele comparison model. Notes: The horizontal lines represent the study-specific ORs and 95% CIs, respectively. The diamond represents the pooled results of OR and 95% CI.
Sensitivity analysis of the association between ESR1 PvuII (T>C) polymorphism and cancer susceptibility. Each point represents the recalculated OR after deleting a separate study.
Funnel plot analysis to detect publication bias for ESR1 PvuII (T>C) polymorphism under allele comparison model. Notes: Each point represents a separate study for the indicated association.
From then on, several new case-control studies with larger samples were available. In addition, the former meta-analysis conducted only included studies published in English. To provide a robust clarification, we performed the updated meta-analysis by involving all the eligible studies published. Overall, statistically significant relationship was only observed for PvuII in allele model T vs. C (OR=0.95, 95% CI=0.91-0.99). However, we did not detect any significant relationship between ESR1 PvuII (rs2234693 T>C) polymorphism and cancer risk in the pooled analysis under homozygous and heterozygous model. Cancer type by subgroup analysis indicated that T genotype significantly decreased prostate cancer risk, Leiomyoma risk, and HCC risk. Yet no association was observed in other types of cancers. These data suggested that the PvuII (rs2234693 T>C) polymorphism on ESR1 may function in a wide manner regarding the different cancer types. When stratified by population, no significant association between ESR1 PvuII genotype and cancer risk among African, and Hispanics was detected. Such association was observed for the Africans. The limited statistical power caused by relatively small number of studies in Africans should be considered. In this meta-analysis, several measurements were performed to enhance the credibility of our conclusion. First, we adopted every effort to expand the numbers of included studies, such as incorporating all publications written both in Chinese and in English. The relatively large number of including studies was one of the important merits of the current study. We also performed publication bias and the sensitivity analysis under the guidance of Cochrane protocol. The sensitivity analysis and publication bias analysis revealed the strength of our conclusions. Although this meta-analysis has its own merits, limitations still exist. First, we only used unadjusted estimates to determine whether there is a relationship between ESR1 PvuII (rs2234693 T>C) polymorphism and cancer risk. Adjustment analysis was absence due to the lack of patient's clinical data such as life habit, smoking and drinking status, exposing factors, and gene-environment interactions, which restrains our further analysis for confounding factors. Second, the validity of conclusion was impaired as significant between-study heterogeneity was detected in some comparisons. Such heterogeneity might result from different quality of studies, and might impair the strength of the conclusion. Third, selection bias and language bias were inevitable, as only published studies and papers written in English or Chinese were analyzed, respectively. Moreover, selection bias might also generate as most of the studies included in this meta-analysis were from candidate gene based, but not from GWAS. Fourth, the sample size of subgroup analysis was relatively small in some strata, impaired the statistical power to estimate the real association. Last, the analyzed case-control studies were mostly performed using Caucasians and Asians populations. Therefore, more trials using different population background, especially Africans, are essential to further confirm such conclusion, due to the genetic and geographical differences.
Conclusion
In conclusion, the current meta-analysis suggests that ESR1 PvuII (rs2234693 T>C) polymorphism may not be strong enough to impact the risk of cancer, based on the pooled results of the published articles. Such relationship further helps to explain the etiology of cancer. Yet, further epidemiological studies with larger sample sizes, standardized unbiased design are warranted to confirm this conclusion.
Acknowledgements
This study was supported by grants from Science and Technology Program of Guangzhou (No. 201509010012).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7-30 doi: 10.3322/caac.21332
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30 doi: 10.3322/caac.21442
3. Foulkes WD. Inherited susceptibility to common cancers. N Engl J Med. 2008;359:2143-53 doi: 10.1056/NEJMra0802968
4. Pharoah PD, Dunning AM, Ponder BA, Easton DF. Association studies for finding cancer-susceptibility genetic variants. Nat Rev Cancer. 2004;4:850-60 doi: 10.1038/nrc1476
5. Gong J, Tian J, Lou J, Wang X, Ke J, Li J, Yang Y, Gong Y, Zhu Y, Zou D, Peng X, Yang N, Mei S. et al. A polymorphic MYC response element in KBTBD11 influences colorectal cancer risk, especially in interaction with an MYC-regulated SNP rs6983267. Ann Oncol. 2018;29:632-9 doi: 10.1093/annonc/mdx789
6. Li J, Zou L, Zhou Y, Li L, Zhu Y, Yang Y, Gong Y, Lou J, Ke J, Zhang Y, Tian J, Zou D, Peng X. et al. A low-frequency variant in SMAD7 modulates TGF-beta signaling and confers risk for colorectal cancer in Chinese population. Mol Carcinog. 2017;56:1798-807 doi: 10.1002/mc.22637
7. Lou J, Gong J, Ke J, Tian J, Zhang Y, Li J, Yang Y, Zhu Y, Gong Y, Li L, Chang J, Zhong R, Miao X. A functional polymorphism located at transcription factor binding sites, rs6695837 near LAMC1 gene, confers risk of colorectal cancer in Chinese populations. Carcinogenesis. 2017;38:177-83 doi: 10.1093/carcin/bgw204
8. Holt SK, Kwon EM, Fu R, Kolb S, Feng Z, Ostrander EA, Stanford JL. Association of variants in estrogen-related pathway genes with prostate cancer risk. Prostate. 2013;73:1-10 doi: 10.1002/pros.22534
9. Cowley SM, Hoare S, Mosselman S, Parker MG. Estrogen receptors alpha and beta form heterodimers on DNA. J Biol Chem. 1997;272:19858-62 doi
10. Katzenellenbogen BS, Katzenellenbogen JA. Estrogen receptor transcription and transactivation: Estrogen receptor alpha and estrogen receptor beta: regulation by selective estrogen receptor modulators and importance in breast cancer. Breast Cancer Res. 2000;2:335-44 doi
11. Li L, Zhang X, Xia Q, Ma H, Chen L, Hou W. Association between estrogen receptor alpha PvuII polymorphism and prostate cancer risk. Tumour Biol. 2014;35:4629-35 doi: 10.1007/s13277-014-1606-9
12. Dai ZM, Zhang TS, Lin S, Zhang WG, Liu J, Cao XM, Li HB, Wang M, Liu XH, Liu K, Li SL, Dai ZJ. Role of IL-17A rs2275913 and IL-17F rs763780 polymorphisms in risk of cancer development: an updated meta-analysis. Sci Rep. 2016;6:20439. doi: 10.1038/srep20439
13. Dai ZJ, Wang XJ, Kang AJ, Ma XB, Min WL, Lin S, Zhao Y, Yang PT, Wang M, Kang HF. Association between APE1 Single Nucleotide Polymorphism (rs1760944) and Cancer Risk: a Meta-Analysis Based on 6,419 Cancer Cases and 6,781 Case-free Controls. J Cancer. 2014;5:253-9 doi: 10.7150/jca.8085
14. He J, Liao XY, Zhu JH, Xue WQ, Shen GP, Huang SY, Chen W, Jia WH. Association of MTHFR C677T and A1298C polymorphisms with non-Hodgkin lymphoma susceptibility: evidence from a meta-analysis. Sci Rep. 2014;4:6159. doi: 10.1038/srep06159
15. Al-Hendy A, Salama SA. Ethnic distribution of estrogen receptor-alpha polymorphism is associated with a higher prevalence of uterine leiomyomas in black Americans. Fertil Steril. 2006;86:686-93 doi: 10.1016/j.fertnstert.2006.01.052
16. Anghel A, Narita D, Seclaman E, Popovici E, Anghel M, Tamas L. Estrogen receptor alpha polymorphisms and the risk of malignancies. Pathol Oncol Res. 2010;16:485-96 doi: 10.1007/s12253-010-9263-9
17. Ashton KA, Proietto A, Otton G, Symonds I, McEvoy M, Attia J, Gilbert M, Hamann U, Scott RJ. Estrogen receptor polymorphisms and the risk of endometrial cancer. BJOG. 2009;116:1053-61 doi: 10.1111/j.1471-0528.2009.02185.x
18. Balistreri CR, Caruso C, Carruba G, Miceli V, Candore G. Genotyping of sex hormone-related pathways in benign and malignant human prostate tissues: data of a preliminary study. OMICS. 2011;15:369-74 doi: 10.1089/omi.2010.0128
19. Berndt SI, Chatterjee N, Huang WY, Chanock SJ, Welch R, Crawford ED, Hayes RB. Variant in sex hormone-binding globulin gene and the risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:165-8 doi: 10.1158/1055-9965.EPI-06-0689
20. Beuten J, Gelfond JA, Franke JL, Weldon KS, Crandall AC, Johnson-Pais TL, Thompson IM, Leach RJ. Single and multigenic analysis of the association between variants in 12 steroid hormone metabolism genes and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:1869-80 doi: 10.1158/1055-9965.EPI-09-0076
21. Cai Q, Shu XO, Jin F, Dai Q, Wen W, Cheng JR, Gao YT, Zheng W. Genetic polymorphisms in the estrogen receptor alpha gene and risk of breast cancer: results from the Shanghai Breast Cancer Study. Cancer Epidemiol Biomarkers Prev. 2003;12:853-9 doi
22. Cao L, Li H, Liu L. ERα Gene Polymorphism and Breast Cancer Risk among Females in Sichuan Province: A Case-control Study. Cancer intervention and therapy. 2014;27:171-5 doi
23. Chang HL, Cheng YJ, Su CK, Chen MC, Chang FH, Lin FG, Liu LF, Yuan SS, Chou MC, Huang CF, Yang CC. Association of estrogen receptor alpha gene PvuII and XbaI polymorphisms with non-small cell lung cancer. Oncol Lett. 2012;3:462-8 doi: 10.3892/ol.2011.482
24. Chattopadhyay S, Siddiqui S, Akhtar MS, Najm MZ, Deo SV, Shukla NK, Husain SA. Genetic polymorphisms of ESR1, ESR2, CYP17A1, and CYP19A1 and the risk of breast cancer: a case control study from North India. Tumour Biol. 2014;35:4517-27 doi: 10.1007/s13277-013-1594-1
25. Chen F, Zhu X, Qian X, Yang Y, Fan L. The study on the relationship between polymorphism of estrogen receptor-alpha gene and leiomyomata. J Clin Obstet Gynecol. 2006;7:290-1 doi
26. Cunningham JM, Hebbring SJ, McDonnell SK, Cicek MS, Christensen GB, Wang L, Jacobsen SJ, Cerhan JR, Blute ML, Schaid DJ, Thibodeau SN. Evaluation of genetic variations in the androgen and estrogen metabolic pathways as risk factors for sporadic and familial prostate cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:969-78 doi: 10.1158/1055-9965.EPI-06-0767
27. de Giorgi V, Sestini S, Gori A, Mazzotta C, Grazzini M, Rossari S, Mavilia C, Crocetti E, Brandi ML, Lotti T, Massi D. Polymorphisms of estrogen receptors: risk factors for invasive melanoma - a prospective study. Oncology. 2011;80:232-7 doi: 10.1159/000328321
28. Deng L, Lu Y. Research on polymorphism of estrogen α receptor sites Xba I and Pvu II in relation to breast cancer. Chin J of Oncol Prev and Treat. 2011;3:19-22 doi
29. Denschlag D, Bentz EK, Hefler L, Pietrowski D, Zeillinger R, Tempfer C, Tong D. Genotype distribution of estrogen receptor-alpha, catechol-O-methyltransferase, and cytochrome P450 17 gene polymorphisms in Caucasian women with uterine leiomyomas. Fertil Steril. 2006;85:462-7 doi: 10.1016/j.fertnstert.2005.07.1308
30. Dunning AM, Healey CS, Baynes C, Maia AT, Scollen S, Vega A, Rodriguez R, Barbosa-Morais NL, Ponder BA, SEARCH, Low YL, Bingham S, EPIC. et al. Association of ESR1 gene tagging SNPs with breast cancer risk. Hum Mol Genet. 2009;18:1131-9 doi: 10.1093/hmg/ddn429
31. Fukatsu T, Hirokawa Y, Araki T, Hioki T, Murata T, Suzuki H, Ichikawa T, Tsukino H, Qiu D, Katoh T, Sugimura Y, Yatani R, Shiraishi T. et al. Genetic polymorphisms of hormone-related genes and prostate cancer risk in the Japanese population. Anticancer Res. 2004;24:2431-7 doi
32. Gonzalez-Mancha R, Galan JJ, Crespo C, Iglesias Perez L, Gonzalez-Perez A, Moron FJ, Moreno Nogueira JA, Real LM, Pascual MH, Ruiz A, Royo JL. Analysis of the ERalpha germline PvuII marker in breast cancer risk. Med Sci Monit. 2008;14:CR136-43 doi
33. Gonzalez-Zuloeta Ladd AM, Vasquez AA, Rivadeneira F, Siemes C, Hofman A, Stricker BH, Pols HA, Uitterlinden AG, van Duijn CM. Estrogen receptor alpha polymorphisms and postmenopausal breast cancer risk. Breast Cancer Res Treat. 2008;107:415-9 doi: 10.1007/s10549-007-9562-3
34. Gupta L, Thakur H, Sobti RC, Seth A, Singh SK. Role of genetic polymorphism of estrogen receptor-alpha gene and risk of prostate cancer in north Indian population. Mol Cell Biochem. 2010;335:255-61 doi: 10.1007/s11010-009-0275-2
35. Han J, Jiang T, Bai H, Gu H, Dong J, Ma H, Hu Z, Shen H. Genetic variants of 6q25 and breast cancer susceptibility: a two-stage fine mapping study in a Chinese population. Breast Cancer Res Treat. 2011;129:901-7 doi: 10.1007/s10549-011-1527-x
36. Han Z, Zhang L, Zhu R, Luo L, Zhu M, Fan L, Wang G. Relationship of oestrogen receptor alpha gene polymorphisms with risk for benign prostatic hyperplasia and prostate cancer in Chinese men. Medicine (Baltimore). 2017;96:e6473. doi: 10.1097/MD.0000000000006473
37. Hernandez J, Balic I, Johnson-Pais TL, Higgins BA, Torkko KC, Thompson IM, Leach RJ. Association between an estrogen receptor alpha gene polymorphism and the risk of prostate cancer in black men. J Urol. 2006;175:523-7 doi: 10.1016/S0022-5347(05)00240-5
38. Hsieh YY, Wang YK, Chang CC, Lin CS. Estrogen receptor alpha-351 XbaI*G and -397 PvuII*C-related genotypes and alleles are associated with higher susceptibilities of endometriosis and leiomyoma. Mol Hum Reprod. 2007;13:117-22 doi: 10.1093/molehr/gal099
39. Hu Z, Song CG, Lu JS, Luo JM, Shen ZZ, Huang W, Shao ZM. A multigenic study on breast cancer risk associated with genetic polymorphisms of ER Alpha, COMT and CYP19 gene in BRCA1/BRCA2 negative Shanghai women with early onset breast cancer or affected relatives. J Cancer Res Clin Oncol. 2007;133:969-78 doi: 10.1007/s00432-007-0244-7
40. Iwamoto I, Fujino T, Douchi T, Nagata Y. Association of estrogen receptor alpha and beta3-adrenergic receptor polymorphisms with endometrial cancer. Obstet Gynecol. 2003;102:506-11 doi
41. Iwasaki M, Hamada GS, Nishimoto IN, Netto MM, Motola J Jr, Laginha FM, Kasuga Y, Yokoyama S, Onuma H, Nishimura H, Kusama R, Kobayashi M, Ishihara J. et al. Isoflavone, polymorphisms in estrogen receptor genes and breast cancer risk in case-control studies in Japanese, Japanese Brazilians and non-Japanese Brazilians. Cancer Sci. 2009;100:927-33 doi: 10.1111/j.1349-7006.2009.01118.x
42. Jurecekova J, Sivonova MK, Evinova A, Kliment J, Dobrota D. The association between estrogen receptor alpha polymorphisms and the risk of prostate cancer in Slovak population. Mol Cell Biochem. 2013;381:201-7 doi: 10.1007/s11010-013-1703-x
43. Kadiyska T, Yakulov T, Kaneva R, Nedin D, Alexandrova A, Gegova A, Savov A, Mitev V, Kremensky I. Vitamin D and estrogen receptor gene polymorphisms and the risk of colorectal cancer in Bulgaria. Int J Colorectal Dis. 2007;22:395-400 doi: 10.1007/s00384-006-0163-0
44. Kjaergaard AD, Ellervik C, Tybjaerg-Hansen A, Axelsson CK, Gronholdt ML, Grande P, Jensen GB, Nordestgaard BG. Estrogen receptor alpha polymorphism and risk of cardiovascular disease, cancer, and hip fracture: cross-sectional, cohort, and case-control studies and a meta-analysis. Circulation. 2007;115:861-71 doi: 10.1161/CIRCULATIONAHA.106.615567
45. Liu Y, Liu Y, Huang X, Sui J, Mo C, Wang J, Peng Q, Deng Y, Huang L, Li S, Qin X. Association of PvuII and XbaI polymorphisms in estrogen receptor alpha gene with the risk of hepatitis B virus infection in the Guangxi Zhuang population. Infect Genet Evol. 2014;27:69-76 doi: 10.1016/j.meegid.2014.07.002
46. Low YL, Taylor JI, Grace PB, Mulligan AA, Welch AA, Scollen S, Dunning AM, Luben RN, Khaw KT, Day NE, Wareham NJ, Bingham SA. Phytoestrogen exposure, polymorphisms in COMT, CYP19, ESR1, and SHBG genes, and their associations with prostate cancer risk. Nutr Cancer. 2006;56:31-9 doi: 10.1207/s15327914nc5601_5
47. Lu H, Chen D, Hu LP, Zhou LL, Xu HY, Bai YH, Lin XY. Estrogen receptor alpha gene polymorphisms and breast cancer risk: a case-control study with meta-analysis combined. Asian Pac J Cancer Prev. 2014;14:6743-9 doi
48. Lu X, Li B, Wei JM, Hua B. [The XbaI and PvuII gene polymorphisms of the estrogen receptor alpha gene in Chinese women with breast cancer]. Zhonghua Wai Ke Za Zhi. 2005;43:290-3 doi
49. Lu X, Yamano Y, Takahashi H, Koda M, Fujiwara Y, Hisada A, Miyazaki W, Katoh T. Associations between estrogen receptor genetic polymorphisms, smoking status, and prostate cancer risk: a case-control study in Japanese men. Environ Health Prev Med. 2015;20:332-7 doi: 10.1007/s12199-015-0471-5
50. Lundin E, Wirgin I, Lukanova A, Afanasyeva Y, Krogh V, Axelsson T, Hemminki K, Clendenen TV, Arslan AA, Ohlson N, Sieri S, Roy N, Koenig KL. et al. Selected polymorphisms in sex hormone-related genes, circulating sex hormones and risk of endometrial cancer. Cancer Epidemiol. 2012;36:445-52 doi: 10.1016/j.canep.2012.04.006
51. Madeira KP, Daltoe RD, Sirtoli GM, Carvalho AA, Rangel LB, Silva IV. Estrogen receptor alpha (ERS1) SNPs c454-397T>C (PvuII) and c454-351A>G (XbaI) are risk biomarkers for breast cancer development. Mol Biol Rep. 2014;41:5459-66 doi: 10.1007/s11033-014-3419-8
52. Massart F, Becherini L, Gennari L, Facchini V, Genazzani AR, Brandi ML. Genotype distribution of estrogen receptor-alpha gene polymorphisms in Italian women with surgical uterine leiomyomas. Fertil Steril. 2001;75:567-70 doi
53. Massart F, Becherini L, Marini F, Noci I, Piciocchi L, Del Monte F, Masi L, Falchetti A, Tanini A, Scarselli G, Brandi L. Analysis of estrogen receptor (ERalpha and ERbeta) and progesterone receptor (PR) polymorphisms in uterine leiomyomas. Med Sci Monit. 2003;9:BR25-30 doi
54. Modugno F, Weissfeld JL, Trump DL, Zmuda JM, Shea P, Cauley JA, Ferrell RE. Allelic variants of aromatase and the androgen and estrogen receptors: toward a multigenic model of prostate cancer risk. Clin Cancer Res. 2001;7:3092-6 doi
55. Modugno F, Zmuda JM, Potter D, Cai C, Ziv E, Cummings SR, Stone KL, Morin PA, Greene D, Cauley JA. Association of estrogen receptor alpha polymorphisms with breast cancer risk in older Caucasian women. Int J Cancer. 2005;116:984-91 doi: 10.1002/ijc.21105
56. Nyante SJ, Gammon MD, Kaufman JS, Bensen JT, Lin DY, Barnholtz-Sloan JS, Hu Y, He Q, Luo J, Millikan RC. Genetic variation in estrogen and progesterone pathway genes and breast cancer risk: an exploration of tumor subtype-specific effects. Cancer Causes Control. 2015;26:121-31 doi: 10.1007/s10552-014-0491-2
57. Onland-Moret NC, van Gils CH, Roest M, Grobbee DE, Peeters PH. The estrogen receptor alpha gene and breast cancer risk (The Netherlands). Cancer Causes Control. 2005;16:1195-202 doi: 10.1007/s10552-005-0307-5
58. Onsory K, Sobti RC, Al-Badran AI, Watanabe M, Shiraishi T, Krishan A, Mohan H, Kaur P. Hormone receptor-related gene polymorphisms and prostate cancer risk in North Indian population. Mol Cell Biochem. 2008;314:25-35 doi: 10.1007/s11010-008-9761-1
59. Park SK, Andreotti G, Rashid A, Chen J, Rosenberg PS, Yu K, Olsen J, Gao YT, Deng J, Sakoda LC, Zhang M, Shen MC, Wang BS. et al. Polymorphisms of estrogen receptors and risk of biliary tract cancers and gallstones: a population-based study in Shanghai, China. Carcinogenesis. 2010;31:842-6 doi: 10.1093/carcin/bgq038
60. Pazarbasi A, Yilmaz MB, Alptekin D, Luleyap U, Tansug Z, Ozpak L, Izmirli M, Onatoglu-Arikan D, Kocaturk-Sel S, Erkoc MA, Turgut O, Bereketoglu C, Tunc E. et al. Genetic polymorphisms of estrogen receptor alpha and catechol-O-methyltransferase genes in Turkish patients with familial prostate carcinoma. Indian J Hum Genet. 2013;19:408-11 doi: 10.4103/0971-6866.124366
61. Ramalhinho AC, Marques J, Fonseca-Moutinho JA, Breitenfeld L. Genetic polymorphims of estrogen receptor alpha -397 PvuII (T>C) and -351 XbaI (A>G) in a portuguese population: prevalence and relation with breast cancer susceptibility. Mol Biol Rep. 2013;40:5093-103 doi: 10.1007/s11033-013-2611-6
62. Ruza E, Sotillo E, Sierrasesumaga L, Azcona C, Patino-Garcia A. Analysis of polymorphisms of the vitamin D receptor, estrogen receptor, and collagen Ialpha1 genes and their relationship with height in children with bone cancer. J Pediatr Hematol Oncol. 2003;25:780-6 doi
63. Safarinejad MR, Safarinejad S, Shafiei N, Safarinejad S. Estrogen receptors alpha (rs2234693 and rs9340799), and beta (rs4986938 and rs1256049) genes polymorphism in prostate cancer: evidence for association with risk and histopathological tumor characteristics in Iranian men. Mol Carcinog. 2012;51(Suppl 1):E104-17 doi: 10.1002/mc.21870
64. Sakoda LC, Blackston CR, Doherty JA, Ray RM, Lin MG, Gao DL, Stalsberg H, Feng Z, Thomas DB, Chen C. Selected estrogen receptor 1 and androgen receptor gene polymorphisms in relation to risk of breast cancer and fibrocystic breast conditions among Chinese women. Cancer Epidemiol. 2011;35:48-55 doi: 10.1016/j.canep.2010.08.005
65. Shen Y, Li DK, Wu J, Zhang Z, Gao E. Joint effects of the CYP1A1 MspI, ERalpha PvuII, and ERalpha XbaI polymorphisms on the risk of breast cancer: results from a population-based case-control study in Shanghai, China. Cancer Epidemiol Biomarkers Prev. 2006;15:342-7 doi: 10.1158/1055-9965.EPI-05-0485
66. Shin A, Kang D, Nishio H, Lee MJ, Park SK, Kim SU, Noh DY, Choe KJ, Ahn SH, Hirvonen A, Kim JH, Yoo KY. Estrogen receptor alpha gene polymorphisms and breast cancer risk. Breast Cancer Res Treat. 2003;80:127-31 doi
67. Sissung TM, Danesi R, Kirkland CT, Baum CE, Ockers SB, Stein EV, Venzon D, Price DK, Figg WD. Estrogen receptor alpha and aromatase polymorphisms affect risk, prognosis, and therapeutic outcome in men with castration-resistant prostate cancer treated with docetaxel-based therapy. J Clin Endocrinol Metab. 2011;96:E368-72 doi: 10.1210/jc.2010-2070
68. Sobczuk A, Smolarz B, Romanowicz-Makowska H, Pertynski T. Estrogen receptor alpha (ER-alpha) gene polymorphism in patients from the Lodz region of Poland with sporadic endometrial cancer. Eur J Gynaecol Oncol. 2009;30:431-4 doi
69. Sobti RC, Gupta L, Singh SK, Seth A, Kaur P, Thakur H. Role of hormonal genes and risk of prostate cancer: gene-gene interactions in a North Indian population. Cancer Genet Cytogenet. 2008;185:78-85 doi: 10.1016/j.cancergencyto.2008.04.022
70. Sonestedt E, Ivarsson MI, Harlid S, Ericson U, Gullberg B, Carlson J, Olsson H, Adlercreutz H, Wirfalt E. The protective association of high plasma enterolactone with breast cancer is reasonably robust in women with polymorphisms in the estrogen receptor alpha and beta genes. J Nutr. 2009;139:993-1001 doi: 10.3945/jn.108.101691
71. Sonoda T, Suzuki H, Mori M, Tsukamoto T, Yokomizo A, Naito S, Fujimoto K, Hirao Y, Miyanaga N, Akaza H. Polymorphisms in estrogen related genes may modify the protective effect of isoflavones against prostate cancer risk in Japanese men. Eur J Cancer Prev. 2010;19:131-7 doi: 10.1097/CEJ.0b013e328333fbe2
72. Srivastava A, Sharma KL, Srivastava N, Misra S, Mittal B. Significant role of estrogen and progesterone receptor sequence variants in gallbladder cancer predisposition: a multi-analytical strategy. PLoS One. 2012;7:e40162. doi: 10.1371/journal.pone.0040162
73. Suzuki K, Nakazato H, Matsui H, Koike H, Okugi H, Kashiwagi B, Nishii M, Ohtake N, Nakata S, Ito K, Yamanaka H. Genetic polymorphisms of estrogen receptor alpha, CYP19, catechol-O-methyltransferase are associated with familial prostate carcinoma risk in a Japanese population. Cancer. 2003;98:1411-6 doi: 10.1002/cncr.11639
74. Szendroi A, Speer G, Tabak A, Kosa JP, Nyirady P, Majoros A, Romics I, Lakatos P. The role of vitamin D, estrogen, calcium sensing receptor genotypes and serum calcium in the pathogenesis of prostate cancer. Can J Urol. 2011;18:5710-6 doi
75. Taghizade Mortezaee F, Tabatabaiefar MA, Hashemzadeh Chaleshtori M, Miraj S. Lack of Association between ESR1 and CYP1A1 Gene Polymorphisms and Susceptibility to Uterine Leiomyoma in Female Patients of Iranian Descent. Cell J. 2014;16:225-30 doi
76. Tanaka Y, Sasaki M, Kaneuchi M, Shiina H, Igawa M, Dahiya R. Polymorphisms of estrogen receptor alpha in prostate cancer. Mol Carcinog. 2003;37:202-8 doi: 10.1002/mc.10138
77. Tang LY, Chen LJ, Qi ML, Su Y, Su FX, Lin Y, Wang KP, Jia WH, Zhuang ZX, Ren ZF. Effects of passive smoking on breast cancer risk in pre/post-menopausal women as modified by polymorphisms of PARP1 and ESR1. Gene. 2013;524:84-9 doi: 10.1016/j.gene.2013.04.064
78. Wang J, Higuchi R, Modugno F, Li J, Umblas N, Lee J, Lui LY, Ziv E, Tice JA, Cummings SR, Rhees B. Estrogen receptor alpha haplotypes and breast cancer risk in older Caucasian women. Breast Cancer Res Treat. 2007;106:273-80 doi: 10.1007/s10549-007-9497-8
79. Wang J, Wang Z, Ding L, Hao M, Shao S. Effects of estradiol and estrogen receptor alpha Xba I/Pvu II gene polymorphismon cervical cancer. CHIN J CANCE RPREVT REAT. 2011;18:746-9 doi
80. Wang J, Zhang H, Guan Y, Zhou Q. Estrogen receptor α gene polymorphisms in women with leiomyoma. Beijing Med J. 2010;32:880-2 doi
81. Wang X, Ceng J, Chen J, Hu T, Xiong W, Wang W. Association between estrogen receptor-alpha gene polymorphism and uterine leiomyomas. Sichuan Med J. 2010;31:1740-3 doi
82. Wedren S, Lovmar L, Humphreys K, Magnusson C, Melhus H, Syvanen AC, Kindmark A, Landegren U, Fermer ML, Stiger F, Persson I, Baron J, Weiderpass E. Oestrogen receptor alpha gene haplotype and postmenopausal breast cancer risk: a case control study. Breast Cancer Res. 2004;6:R437-49 doi: 10.1186/bcr811
83. Zhai Y, Zhou G, Deng G, Xie W, Dong X, Zhang X, Yu L, Yang H, Yuan X, Zhang H, Zhi L, Yao Z, Shen Y. et al. Estrogen receptor alpha polymorphisms associated with susceptibility to hepatocellular carcinoma in hepatitis B virus carriers. Gastroenterology. 2006;130:2001-9 doi: 10.1053/j.gastro.2006.02.030
84. Li N, Dong J, Hu Z, Shen H, Dai M. Potentially functional polymorphisms in ESR1 and breast cancer risk: a meta-analysis. Breast Cancer Res Treat. 2010;121:177-84 doi: 10.1007/s10549-009-0532-9
85. Sun H, Deng Q, Pan Y, He B, Ying H, Chen J, Liu X, Wang S. Association between estrogen receptor 1 (ESR1) genetic variations and cancer risk: a meta-analysis. J BUON. 2015;20:296-308 doi
Author contact
![]() Corresponding author: Haiyan Lao, Department of Pharmacy, Guangdong General Hospital, Guangdong Academy of Medical Sciences, Guangzhou, 510080, China, Tel./Fax: +86-020-83827812-36066, Email: laohaiyan2010com
Corresponding author: Haiyan Lao, Department of Pharmacy, Guangdong General Hospital, Guangdong Academy of Medical Sciences, Guangzhou, 510080, China, Tel./Fax: +86-020-83827812-36066, Email: laohaiyan2010com

 Global reach, higher impact
Global reach, higher impact