3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(22):4150-4155. doi:10.7150/jca.26948 This issue Cite
Research Paper
Niclosamide Inhibits Cell Growth and Enhances Drug Sensitivity of Hepatocellular Carcinoma Cells via STAT3 Signaling Pathway
1. Key Laboratory of Nanobiological Technology of Chinese Ministry of Health, Xiangya Hospital, Central South University, Changsha, Hunan, 410008, P.R. China.
2. Department of Neurology, Xiangya Hospital, Central South University, Changsha, Hunan, 410008, P.R. China
3. Center for Experimental Medical Research, Third Xiangya Hospital, Central South University, Changsha, Hunan, 410013, P.R. China
4. Department of Nephrology, Xiangya Hospital,Central South University, Changsha, Hunan, 410008, P.R. China
Received 2018-4-28; Accepted 2018-9-7; Published 2018-10-18
Abstract
Aims: Hepatocellular carcinoma (HCC) is the sixth most common malignant tumor worldwide, with a high mortality rate at advanced stages. In this study, we investigated the effect of niclosamide on cell growth and drug sensitivity in human HCC and elucidated the underlying mechanism.
Methods: Three human HCC cell lines (HepG2, QGY-7703 and SMMC-7721) were used to evaluate the effect of niclosamide. Cell proliferation was measured by MTT assay and colony formation assay. Assessment of apoptosis was evaluated by flow cytometry and Hoechst staining. The mRNA and protein levels were analyzed by real-time PCR and western blot, respectively.
Results: Niclosamide suppressed cell viability, inhibited clone formation, and induced cell apoptosis in HCC cells dose- and time-dependently. Furthermore, niclosamide synergized with cisplatin to promote the apoptosis of HCC cells. With niclosamide treatment, phospho-STAT3 (Y705) was inactivated and the downstream antiapoptotic proteins Mcl-1 and survivin were downregulated at both mRNA and protein levels in HCC cells.
Conclusion: Niclosamide has effective function in anti-HCC and may be a single or combined drug treatment for HCC and acts via the STAT3 signaling pathway.
Keywords: Niclosamide, Cell growth, Drug sensitivity, STAT3 signaling, Mcl-1, Hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common malignant tumor and causes the second most common cancer-related deaths in the world[1]. The dominant risk factor of HCC is chronic infection with hepatitis B virus or hepatitis C virus, together with exposure to aflatoxin B1 and alcohol use[2,3]. Patients with advanced-stage HCC, which cannot be resected completely, have few treatment options available. Liver transplantation is an effective therapy for liver cancer but with serious shortage of liver donors[2]. Chemotherapy and radiotherapy are the main choice, with many side effects[4]. Sorafenib, a multi-kinase inhibitor, is the only US Food and Drug Administration (FDA)-approved drug with proven effective molecular drug for treatment of advanced HCC, which can significantly prolong the survival of patients[5]. Unfortunately, many patients may develop acquired resistance to sorafenib[6]. Thus, developing other novel molecular agents to prevent and treat advanced HCC would have potential clinical application value.
Niclosamide is an FDA-approved drug that has been used for anti-tapeworm infections for approximately 50 years[7]. In recent years, niclosamide was found a potential anticancer drug against various cancers including human lung cancer[8,9], ovarian cancer[10], prostate cancer[11,12], head and neck cancer[13], osteosarcoma[14] and breast cancer[15,16] both in vivo and in vitro. Niclosamide is a multi-targeted anticancer drug that may target Wnt/beta-catenin, NF-κB, Notch and signal transducer and activator of transcription 3 (STAT3) signaling pathways to induce cell proliferation inhibition, cell cycle arrest and apoptosis[17]. Furthermore, niclosamide synergizes with frontline chemotherapeutic drugs (erlotinib, Ara-C, VP-16, DNR and dasatinib) against head and neck cancer [13], acute myelogenous leukemia[18] and blast phase chronic myeloid leukemia[19]. However, the precise molecular mechanism underlying the effect of niclosamide on HCC treatment is not fully understood.
In this study, we investigated the effects of niclosamide and its possible molecular mechanisms in HCC cell lines. Niclosamide inhibited cell proliferation and induced apoptosis of HCC cells by inactivating STAT3 signaling. Furthermore, niclosamide enhanced drug sensitivity of HCC to cisplatin. These findings suggest that niclosamide may be a single or combined drug treatment for HCC.
Materials and Methods
Materials
Materials and chemicals. Niclosamide was purchased from Sellck (TX, USA) and stored at 20 mM in dimethylformamide (DMF) at -20℃. 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) was procured from Sigma-Aldrich (MO, USA) and dissolved in ddH2O at 5 mg/mL. Giemsa staining was from Solarbio Life Sciences (Beijing). The Hoechst staining kit was from Beyotime Biotechnology (Shanghai). Primary antibodies against survivin were from Abcam (MA, USA), and primary antibodies against Mcl-1, phospho-STAT3 (Y705), STAT3, GAPDH and secondary antibodies were from Cell Signaling Technology (Cell Signaling Technology, MA, USA).
Cell lines and cell culture
The human HCC cell lines (HepG2, QGY-7703 and SMMC-7721) were obtained from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai) and maintained in Dulbecco's modified Eagle's medium (DMEM, Gibco, CA, USA) with 10% fetal bovine serum (FBS, Gibco, CA, USA) and 1% penicillin streptomycin (PS, Gibco, CA, USA) in a humidified cell incubator with 5% CO2 at 37℃.
Cell viability assay
Cell viability was measured by MTT assay. Cells were seeded in 96-well plates (1x104 per well, triplicate) and cultured overnight, then treated with different concentrations of niclosamide (0, 1, 5, 10 or 20 μM) for 24, 48, or 72 h. To each well was added 10 μL 0.5 mg/mL MTT solution for 4 h, then the medium was removed and 150 μL DMSO was added, and the absorbance was measured at 490 nm by using a microplate reader (BioTek, VT, USA). All experiments were repeated at least three times.
Cell colony formation assay
Cells (100-200 cells per well) were plated in 6-well plates and cultured overnight. Niclosamide was added and incubated for 2 weeks to form colonies. Cells were washed twice with phosphate buffered saline (PBS) and fixed with methanol for 15 min, then stained with 1 mL Giemsa for 20 min. Surviving colonies were counted under a light microscope and the surviving fraction (SF) was calculated by using the formula SF=treatment colony numbers/control colony numbers. All experiments were performed independently at least three times.
Cell apoptosis assay
Cells (1x106 per well) were plated in 6-well plates and cultured overnight. After treatment with niclosamide and cisplatin, cells were harvested 48 h later and resuspended in binding buffer. Apoptotic cells were detected by using an Annexin V-FITC kit according to the manufacturer's instructions (BD Pharmingen, CA, USA) and analyzed by flow cytometry (BD Biosciences, USA). As for Hoechst staining, cells (1x106 per well) were seeded in cover glass in 6-well plates and cultured overnight. Niclosamide was added and incubated for 48 h. The culture medium was removed and 0.5 mL Hoechst 33342 was added (Beyotime Biotechnology, Shanghai). After incubation for 10 min in the dark, cells were analyzed by fluorescence microscopy (Carl Zeiss, Thuringia, Germany).
Western blot analysis
After treatment with 5 μM niclosamide for 2 h, cells were washed with cold PBS twice and generated in lysis buffer containing RIPA (Thermo Scientific, IL, USA) and protease inhibitors for 30 min. Total protein extracts were collected and centrifuged at 13,000 x g at 4℃ for 10 min. The supernatants were obtained and stored at -80℃. Protein concentration was quantified by BCA assay. An equivalent amount of proteins was separated by SDS-PAGE gel and transferred onto PVDF membranes. The membranes were blocked in 5% bovine serum albumin in PBS and 0.1% Tween20 for 1 h, followed by incubation with primary antibodies overnight at 4℃ and secondary antibody for 2 h at room temperature. The immunoblots were developed and visualized with an enhanced chemiluminescence detection system and the FluorChem FC3 system (ProteinSimple, CA, USA).
Quantitative PCR (qPCR)
Cells (1x 106 per well) were plated in 6-well plates and cultured overnight, then treated with or without 5 μM niclosamide for 2 h. Total RNA was extracted with TRIzol according to the manufacturer's instructions (Invitrogen, NY, USA). The extracted total RNA was reverse transcribed into complementary DNA (cDNA) by using the cDNA Synthesis Kit (TOYOBO, Osaka, Japan). qPCR was performed using SYBR Green Master Mix (Bimake, TX, USA) and a QuantStudio 6 (Life Technologies, NY, USA). Primer sequence used for real-time PCR (Sangon Biotech, Shanghai) were (5'->3') Mcl-1-F: CAC TTC CGC TTC CTT CCA GT, Mcl-1-R: GGT GGC CAA AAG TCG CCC; survivin-F: AGG ACC ACC GCA TCT CTA CA, survivin-R: TTT CCT TTG CAT GGG GTC GT; GAPDH-F: GAA AGC CTG CCG GTG ACT AA, GAPDH-R: GCC CAA TAC GAC CAA ATC AGA GA. The melting curve analysis was used to monitor each amplification reaction for the absence of non-specific PCR products. The threshold cycle numbers obtained from qPCR were compared to generate the relative copy number. Data were normalized by co-amplification of GAPDH.
Statistical analysis
All data are presented as mean ± SD. Statistical analyses were performed with SPSS v18.0. Student's t-test and univariate analysis were used to analyze the statistical differences. P<0.05 was considered statistically significant.
Results
Niclosamide inhibits cell proliferation and colony formation of HCC cells
Niclosamide is a potential anticancer drug against various cancers[17]. To determine the inhibitory effect of niclosamide on HCC cells (HepG2, QGY-7703 and SMMC-7721), MTT assay was performed. Niclosamide inhibited the proliferation of three HCC cell lines (Fig. 1A-C) dose- and time-dependently. The 48-h IC50 values of HepG2, QGY-7703 and SMMC-7721 were 31.91, 10.24, 13.46 μM, respectively. We also tested the clone-forming ability of HCC cells after niclosamide treatment. Niclosamide at 1 μM had little effect on clone-forming ability of HCC cells; however, 5 μM niclosamide completely inhibited the clone-forming ability of HCC cells (Fig.1D-E). Thus, niclosamide could inhibit cell growth in HCC.
Niclosamide inhibits cell proliferation and colony formation of HCC cells. (A), (B) and (C) HepG2, QGY-7703 and SMMC-7721 cells were treated with niclosamide for 24, 48 and 72 h. Cell viability was determined by MTT assay. (D) and (E) Colony-forming ability of HepG2, QGY-7703 and SMMC-7721 cells analyzed by clone formation assay. Data are mean ± SD of three separate determinations.
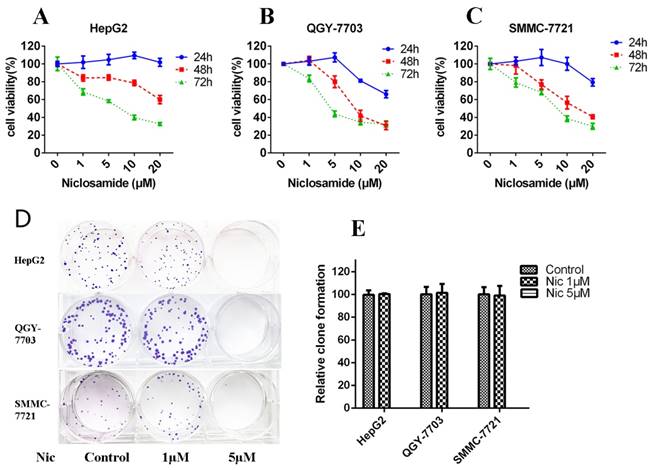
Niclosamide induced apoptosis of HCC cells. (A) and (B) HepG2 cells were treated with 5 μM niclosamide for 48 h; apoptosis was determined by analyzing Annexin V-FITC PI. (C) Hoechst 33342 staining of HepG2, QGY-7703 and SMMC-7721 cells after treatment with 5 μM niclosamide for 48 h; cell nuclei dyed bright blue or white were considered apoptotic cells. Data are mean ±SD of three separate determinations.
Niclosamide inhibits the expression of anti-apoptotic proteins associated with increased cell apoptosis
Mcl-1 is a major anti-apoptotic member of the Bcl2 family. Survivin is an anti-apoptotic protein more widely expressed in HCC cells. It plays an important role in cell survival and drug resistance. We tested the expression of Mcl-1 and survivin after niclosamide treatment. The protein (Fig. 4A) and mRNA (Fig. 4D) levels of anti-apoptosis protein Mcl-1 and survivin were downregulated by niclosamide in HepG2 and QGY-7703 cells. To assess cell apoptosis after niclosamide treatment, Annexin V-FITC/PI and Hoechst staining were performed. Niclosamide induced apoptosis of HCC cells (Fig. 2A-C). These findings suggest that the survival pathway is involved in niclosamide-induced anticancer therapy of HCC.
Niclosamide synergizes with cisplatin against hepatocellular carcinoma
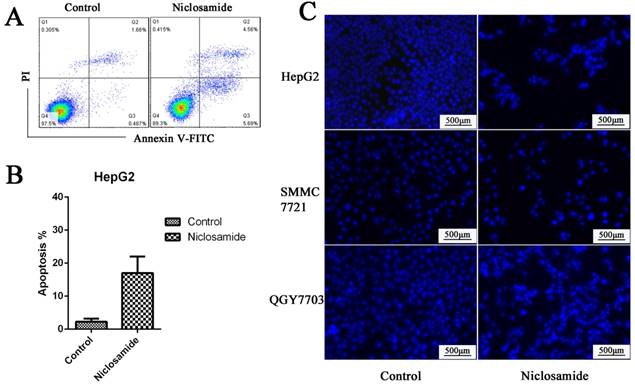
We have identified that niclsoamide can inhibit cell proliferation and induce apoptosis of hepotama carcinoma cells. To test whether niclosamide is a combination drug for treating HCC, HCC cells were treated with cisplatin, a common clinical chemotherapeutic drug used in the treatment of cancers, in the absence or presence of niclosamide. Niclosamide enhanced cisplatin-induced cell apoptosis (P<0.05) The results (Fig. 3A and 3B), which indicates that niclosamide has synergistic effects with cisplatin to increase the drug sensitivity of HCC cells.
Niclosamide downregulates anti-apoptotic proteins by blocking activation of the STAT3 signal pathway
The anti-apoptosis protein Mcl-1 and survivin play an important role in cell survival and drug resistance. Mcl-1 and survivin are downstream proteins of the STAT3 signaling pathway. To test this, we analyzed the protein expression and phosphorylation of STAT3 in various HCC cell lines by western blot analysis in the absence or presence of niclosamide. Niclosamide could suppress the phosphorylation of STAT3 at Y705 but not inhibit its expression (Fig. 4A-C). This may be a potential mechanism by which niclosamide-induced STAT3 inactivation increases the drug sensitivity of HCC cells and downregulates the expression of anti-apoptotic proteins.
Discussion
Niclosamide, an FDA-approved anthelmintic drug, has been used for treating tapeworm infections for approximately 50 years[7]. Recently, niclosamide received more attention for its anticancer and nontoxic roles in humans. Both in vitro and in vivo research show the anticancer role of niclosamide by regulating cell proliferation, apoptosis and metastasis in various cancers[11,14,15]. STAT3[8,11,13], Wnt/ beta-catenin[20], and NF-κB[18] signaling pathways associated with cell proliferation have been found involved in the anticancer actions of niclosamide. However, its effects on HCC are unknown. Thus, the aim of this study was to detect whether niclosamide works against HCC cell lines and further to investigate the possible mechanisms. Our data showed that niclosamide inhibits STAT3 phosphorylation at Tyr705 in association with decreased Mcl-1 and survivin levels at both mRNA and protein levels in various HCC cells.
Niclosamide synergizes with cisplatin against HCC. (A) and (B) HepG2 and QGY-7703 cells were treated with 5 μM niclosamide, 20 μg/mL cisplatin or both for 72 h. Apoptosis was detected by flow cytometry by FITC Annexin V-FITC PI staining. Data are mean ±SD of three separate determinations. *P<0.05.
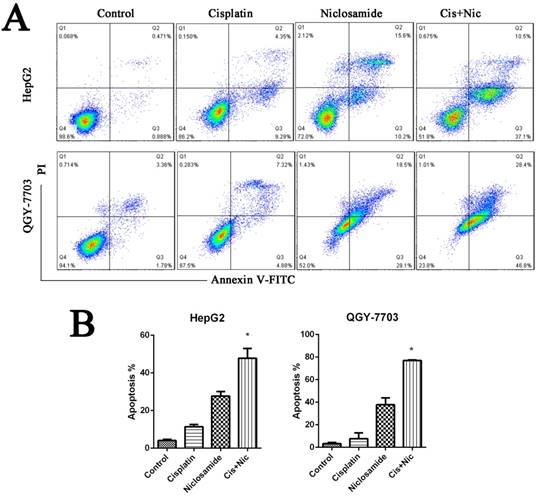
Niclosamide downregulates anti-apoptotic proteins by blocking activation of the STAT3 signal pathway. (A), (B) and (C) HepG2 and QGY-7703 cells were treated with 5 μM niclosamide for 2 h. P-Stat3, Stat3, Mcl-1 and Survivin protein levels were examined by western blot analysis. Signal intensity was quantified with ImageJ and normalized to GAPDH. (D) HepG2 cells were treated with 5 μM niclosamide for 2 h, Mcl-1 and survivin mRNA levels were analyzed by qRT-PCR. Each mRNA level was normalized to that of the GAPDH housekeeping gene. Data are mean ± SD of three separate determinations. *P<0.05
The main problem with developing new chemotherapy reagents is safety and efficacy. Many new drugs fail to enter the clinical trials due to lack of safety. Niclosamide has been used for about 50 years. It has been verified as with very low toxicity to mammals at an oral median lethal dose of >5000 mg/kg in rats[21]. Furthermore, niclosamide induces acute and chronic myelogenous leukemia cell apoptosis but not normal bone marrow[18,19]. The same effects were verified in prostate cancer[12]. Niclosamide treatment has been reported to be well tolerated without weight loss and does not influence vital organ functions such as liver, kidney, and bone marrow[13]. In this study, we determined the efficacy of niclosamide as a candidate for advanced HCC treatment by evaluating its effects on HCC cell lines. Niclosamide inhibited cell viability and colony formation remarkably in HCC cell lines dose- and time-dependently (Fig. 1). Furthermore, niclosamide induced cell apoptosis significantly (Fig. 2) and was more effective when combined with cisplatin (Fig. 3). Our findings together with previous reports demonstrate that niclosamide may be a single and combined anticancer drug against HCC.
STAT3 is a member of the STAT transcription factor family and has been validated as an attractive therapeutic in cancer therapy[22]. STAT3 is activated constitutively in many human cancers[23] including 50% in HCC[24]. STAT3 activation can support tumor cell survival by upregulating the downstream target genes, including apoptosis inhibitors (Bcl-2, Mcl-1 and survivin) and subsequently suppressing pro-apoptotic molecules (Bax, Bad and Bid)[23,25]. Bcl-2 and Mcl-1 are anti-apoptotic genes of the Bcl-2 family that play vital roles in regulating intrinsic mitochondrial pathways. Survivin is a member of the family of inhibitor of apoptosis proteins expressed at high levels in HCC. Survivin inhibits apoptosis and also promotes aberrant proliferation and cell cycle progression of HCC cells[26]. Niclosamide has been reported to be a new potent STAT3 inhibitor. Our results showed that niclosamide suppresses activation of STAT3 and downregulates the mRNA and protein levels of STAT3-targeted downstream genes Mcl-1 and Survivin in HCC cells (Fig. 4). This study indicates that targeting STAT3 is an ideal strategy for developing novel anticancer agents for the treatment of HCC.
In summary, our results show that niclosamide inhibited cell growth and enhanced drug sensitivity of HCC cells by suppressing the STAT3 signaling pathway and blocking the anti-apoptotic function of Mcl-1 and survivin in HCC cells. Niclosamide may be developed into a single and combined anticancer drug against HCC.
Acknowledgements
This work was supported by The National Natural Science Foundation of China (grant nos. 81302075, 81272193, 81402001 and 81770743) and The Fundamental Research Funds for Central South University (grant no. 2017zzts391).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL. et al. Global cancer statistics, 2012. CA: A Cancer Journal for Clinicians. 2015;65:87-108
2. Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. LANCET. 2012;379:1245-55
3. de Lope CR, Tremosini S, Forner A, Reig M, Bruix J. Management of HCC. J HEPATOL. 2012;56:S75-87
4. Raza A. Hepatocellular carcinoma review: Current treatment, and evidence-based medicine. WORLD J GASTROENTERO. 2014;20:4115
5. Colagrande S, Inghilesi AL, Aburas S. et al. Challenges of advanced hepatocellular carcinoma. WORLD J GASTROENTERO. 2016;22:7645
6. Gauthier A, Ho M. Role of sorafenib in the treatment of advanced hepatocellular carcinoma: An update. HEPATOL RES. 2013;43:147-54
7. Andrews P, Thyssen J, Lorke D. The biology and toxicology of molluscicides, Bayluscide. Pharmacol Ther. 1982;19:245-95
8. You S, Li R, Park D. et al. Disruption of STAT3 by Niclosamide Reverses Radioresistance of Human Lung Cancer. MOL CANCER THER. 2014;13:606-16
9. Li R, Hu Z, Sun SY. et al. Niclosamide Overcomes Acquired Resistance to Erlotinib through Suppression of STAT3 in Non-Small Cell Lung Cancer. MOL CANCER THER. 2013;12:2200-12
10. Walters Haygood CL, Arend RC, Gangrade A. et al. Niclosamide Analogs for Treatment of Ovarian Cancer. INT J GYNECOL CANCER. 2015;25:1377-85
11. Liu C, Lou W, Armstrong C. et al. Niclosamide suppresses cell migration and invasion in enzalutamide resistant prostate cancer cells via Stat3-AR axis inhibition. The Prostate. 2015;75:1341-53
12. Liu C, Lou W, Zhu Y. et al. Niclosamide Inhibits Androgen Receptor Variants Expression and Overcomes Enzalutamide Resistance in Castration-Resistant Prostate Cancer. CLIN CANCER RES. 2014;20:3198-210
13. Li R, You S, Hu Z. et al. Inhibition of STAT3 by niclosamide synergizes with erlotinib against head and neck cancer. PLOS ONE. 2013;8:e74670
14. Li Z, Yu Y, Sun S. et al. Niclosamide inhibits the proliferation of human osteosarcoma cell lines by inducing apoptosis and cell cycle arrest. ONCOL REP. 2015;33:1763
15. Ye T, Xiong Y, Yan Y. et al. The anthelmintic drug niclosamide induces apoptosis, impairs metastasis and reduces immunosuppressive cells in breast cancer model. PLOS ONE. 2014;9:e85887
16. Wang YC, Chao TK, Chang CC. et al. Drug screening identifies niclosamide as an inhibitor of breast cancer stem-like cells. PLOS ONE. 2013;8:e74538
17. Li Y, Li P, Roberts MJ. et al. Multi-targeted therapy of cancer by niclosamide: A new application for an old drug. CANCER LETT. 2014;349:8-14
18. Jin Y, Lu Z, Ding K. et al. Antineoplastic Mechanisms of Niclosamide in Acute Myelogenous Leukemia Stem Cells: Inactivation of the NF- B Pathway and Generation of Reactive Oxygen Species. CANCER RES. 2010;70:2516-27
19. Liu Z, Li Y, Lv C, Wang L, Song H. Anthelmintic drug niclosamide enhances the sensitivity of chronic myeloid leukemia cells to dasatinib through inhibiting Erk/Mnk1/eIF4E pathway. BIOCHEM BIOPH RES CO. 2016;478:893-9
20. King ML, Lindberg ME, Stodden GR. et al. WNT7A/beta-catenin signaling induces FGF1 and influences sensitivity to niclosamide in ovarian cancer. ONCOGENE. 2015;34:3452-62
21. Merschjohann K, Steverding D. In vitro trypanocidal activity of the anti-helminthic drug niclosamide. EXP PARASITOL. 2008;118:637-40
22. Johnson DE, O'Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. NAT REV CLIN ONCOL. 2018;15:234-48
23. Carpenter R, Lo H. STAT3 Target Genes Relevant to Human Cancers. Cancers. 2014;6:897-925
24. Subramaniam A, Shanmugam MK, Perumal E. et al. Potential role of signal transducer and activator of transcription (STAT)3 signaling pathway in inflammation, survival, proliferation and invasion of hepatocellular carcinoma. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 2013;1835:46-60
25. Wu N, Liu J, Zhao X. et al. Cardamonin induces apoptosis by suppressing STAT3 signaling pathway in glioblastoma stem cells. TUMOR BIOL. 2015;36:9667-76
26. Su C. Survivin in survival of hepatocellular carcinoma. CANCER LETT. 2016;379:184-90
Author contact
![]() Corresponding author: E-mail: rosnowcom
Corresponding author: E-mail: rosnowcom

 Global reach, higher impact
Global reach, higher impact