3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(22):4166-4171. doi:10.7150/jca.27110 This issue Cite
Research Paper
CASP8 rs3834129 (-652 6N insertion/deletion) Polymorphism and Colorectal Cancer Susceptibility: An Updated Meta-Analysis
1. Department of Pharmacy, Tongde Hospital of Zhejiang Province, Hangzhou 310012, Zhejiang, China.
2. Department of ENT, Tongde Hospital of Zhejiang Province, Hangzhou 310012, Zhejiang, China.
3. Department of Pharmacy, Zhejiang Cancer Hospital, Hangzhou 310022, Zhejiang, China.
4. Department of Pathology, Tongde Hospital of Zhejiang Province, Hangzhou, Zhejiang, China.
Received 2018-5-6; Accepted 2018-8-15; Published 2018-10-18
Abstract
CASP8 rs3834129 polymorphism (-652 6N insertion/deletion) is a genetic alteration which might affect the apoptosis pathway caspase enzyme. The impaired caspase enzyme would lead to the change of cancer risk. By now, the role of CASP8 rs3834129 polymorphism has been widely investigated. However, the relationship of this genetic variant on colorectal cancer (CRC) susceptibility still remains inconsistent. Therefore, we further investigated the role of rs3834129 polymorphism on CRC risk. Eligible published studies were retrieved from EMBASE, PubMed, CNKI and WANFANG database updates to March 2018. Odds ratios (ORs) and 95% confidence intervals (CIs) were used to assess the relationship strengths. In general, we successfully retrieved 13 studies (8 publications) involving 13058 cases and 14418 controls. The meta-analysis results demonstrated that rs3834129 polymorphism was associated with a decreased CRC risk in heterozygous model (ID vs. II: OR = 0.94, 95% CI = 0.88-0.99), but not the homozygous and allele models. Furthermore, significantly decreased risk was also found among Asian (ID vs. II: OR = 0.86, 95% CI = 0.76-0.98), and high quality score group (ID vs. II: OR = 0.90, 95% CI = 0.81-1.00) in the stratified analyses. Taken together, we showed that CASP8 rs3834129 polymorphism influences CRC susceptibility in a weak impact manner. More case-control studies are warranted to validate such relationship.
Keywords: colorectal cancer, CASP8, polymorphism, susceptibility, meta-analysis
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and the fourth most cause of cancer death in the world [1]. In China, CRC ranks the top five cancers both in new diagnoses and in the cancer-related cause death [2]. The definitive etiology of CRC remains unknown. Evidence suggests that CRC derives from combinations of genetic and environmental factors [3]. Previous epidemiological studies have elucidated that inherited susceptibility is a major component of CRC predisposition.
Apoptosis, also called as programmed cell death, is a pivotal mechanism to maintain the normal cellular growth [4]. Aberrant functions of apoptosis pathway are associated with the development of CRC [5, 6]. Caspases are the main regulative and executive enzymes in apoptosis pathway [7]. Among them, caspase 8 was one of the most important components of the caspases family proteins [5, 8]. Caspase 8 plays a critical role in mediating the extrinsic apoptosis pathway [9]. The human gene CASP8 is located on chromosome 2q33‑q34 with 11 exons. Several SNPs of CASP8 gene have been identified to be associated with cancer risk [10-13]. Among them, CASP8 rs3834129 polymorphism (-652 6N ins/del), a six six-nucleotide insertion/deletion variant, leads to the decreased expression of CASP8 mRNA [14].
CASP8 SNPs are reported to predispose to the susceptibility of several cancers, including neuroblastoma [12], bladder cancer [15], breast cancer [13]. Among them, CASP8 rs3834129, namely -652 6N ins/del polymorphism, is one of the most investigated SNP. Extensively epidemiological studies have assessed the association between CASP8 rs3834129 (-652 6N ins/del) polymorphism and CRC risk, yet with discrepant results. Therefore, we conducted this meta-analysis to provide a precise evaluation of the association of interest.
Materials and Methods
Publication search
We first searched the following key words: 'Caspase 8' or 'CASP8' or 'rs3834129' and 'SNP' or 'polymorphism' or 'polymorphisms' or 'single nucleotide polymorphism' or 'variant' and 'colorectal cancer' or 'colorectal tumor' or 'colorectal carcinoma' or 'colorectal neoplasm' or 'CRC' in database of PubMed and EMBASE. We also searched the Chinese database CNKI and WANFANG to include more eligible studies. Further, additional studies were also manually extracted from the references of the above obtained publications. The latest search was done in March 2018 without any language restriction. The article will be considered as different studies if it contains more than two ethnicities. Among overlapping reports, only the largest one will be retained.
Eligibility criteria
The final including studies in this meta-analysis should fulfill all the following requirements: 1) unrelated case-control studies; 2) original epidemiological studies; 3) analyzing the relationship between CASP8 rs3834129 polymorphism and CRC risk; 4) Sufficient genotype data were presented to obtain odds ratios (ORs) and 95% confidence intervals (CIs); 5) articles written in English or in Chinese.
Data extraction
We arranged two authors to identify all eligible studies independently. The following items were recorded from each study: first author's name, year of publication, Hardy-Weinberg equilibrium (HWE), quality score, country, ethnicity, source of controls, genotyping method, and genotype distributions of cases and controls. All the disagreed information was settle down after fully discussed by the two authors.
Statistical analysis
Fisher's exact test was applied to check whether the genotype frequency distribution of rs3834129 in controls was deviated from Hardy-Weinberg equilibrium. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated from multivariate logistic regression, and then used to estimate the associations between rs3834129 and CRC risk. Subgroup analyses were performed by ethnicity, source of control, and quality score. Between-study heterogeneity was determined by a chi-square-based Q-Test. The random-effects model (the DerSimonian and Laird method) would be performed in the presence of heterogeneity, whereas the fixed-effects model (the Mantel-Haenszel method) would be performed. Publication bias was assessed by visual inspection of funnel plots and the Egger's linear regression test. The asymmetric plot and P value less than 0.5 was considered as the existence of publication bias. In addition, sensitivity analysis was also applied to assess the robustness of the results. Quality assessment for each study was performed using the quality assessment criteria (Table 1). The meta-analysis was conducted using STATA version 11.0 (Stata Corporation, College Station, TX, USA). All p values were two-sided.
Score of quality assessment
| Criteria | Score |
|---|---|
| Representativeness of cases | |
| Selected from population cancer registry | 2 |
| Selected from hospital | 1 |
| No method of selection described | 0 |
| Representativeness of controls | |
| Population-based | 3 |
| Blood donors | 2 |
| Hospital-based | 1 |
| Not described | 0 |
| Ascertainment of cancer cases | |
| Histopathologic confirmation | 2 |
| Patient medical record | 1 |
| Not described | 0 |
| Control selection | |
| Controls matched with cases by age and sex | 2 |
| Controls matched with cases only by age or by sex | 1 |
| Not matched or not descried | 0 |
| Genotyping examination | |
| Genotyping done blindly and quality control | 2 |
| Only genotyping done blindly or quality control | 1 |
| Unblinded and without quality control | 0 |
| Total sample size for both cases and controls | |
| Larger than 1000 | 3 |
| Larger than 500, but less than 1000 | 2 |
| Larger than 200, but less than 500 | 1 |
| Less than 200 | 0 |
Results
Study characteristics
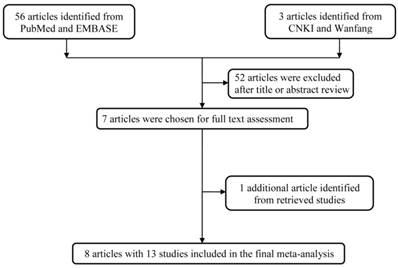
Using the above mentioned database, we firstly identified 59 potentially relevant published records. After literature screening and abstracts reading, we kept 7 publications in the analysis [16-22]. We also extracted 1 article from the references of the retrieval articles [23]. The general work flow of the selection process was graphically shown in Figure 1. In all, 13 studies (8 publications) with 13058 cases and 14418 controls were used in the pooled analysis (Table 2). Among them, 4 studies focused on Asians and 9 on Caucasians. 4 studies were hospital-based design, 9 were population-based design. 6 studies with a quality score >9, and 7 studies with a quality score ≤9.
Meta-analysis results
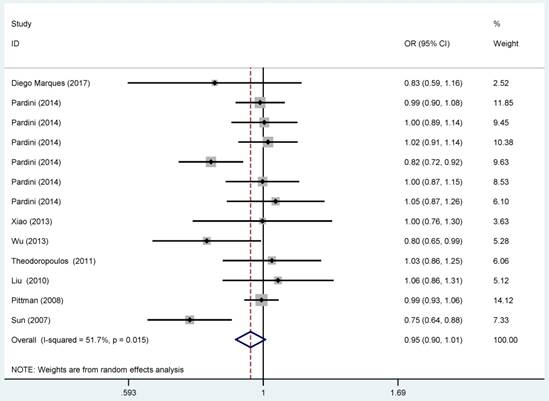
Overall meta-analysis information was shown in Table 3 and Figure 2. In the pooled analysis, statistically significant protection role of CASP8 rs3834129 polymorphism in CRC was observed among heterozygous models (ID vs. II: OR=0.94, 95% CI=0.88-0.99). Statistically significant relationship was not observed in homozygous and allele model. When stratified by population, significant association between CASP8 rs3834129 polymorphism and CRC risk was detected among African (ID vs. II: OR=0.86, 95% CI=0.76-0.98). Such association was not observed for the Caucasians. In terms of source of controls, we failed to detect any significant relationship in hospital-based group and in population-based group. Further subgroup analysis by quality score yielded a significant association for allele model (D vs. I: OR=0.90, 95% CI=0.81-1.00).
The work flow of the current process of handling selection.
Characteristics of studies included in the current meta-analysis
| Surname | Year | Country | Ethnicity | Control Source | Genotype method | Case | Control | MAF | HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| II | ID | DD | All | II | ID | DD | All | ||||||||
| Sun | 2007 | China | Asian | PB | PCR-RFLP | 605 | 280 | 33 | 918 | 528 | 304 | 58 | 890 | 0.24 | 0.116 |
| Pittman | 2008 | England | Caucasian | PB | AS-PCR | 995 | 1897 | 987 | 3879 | 892 | 1872 | 897 | 3661 | 0.50 | 0.170 |
| Liu | 2010 | China | Asian | PB | PCR-RFLP | 233 | 116 | 21 | 370 | 528 | 278 | 32 | 838 | 0.20 | 0.538 |
| Theodoropoulos | 2011 | Greece | Caucasian | HB | RFLP-PCR | 103 | 201 | 98 | 402 | 120 | 254 | 106 | 480 | 0.49 | 0.194 |
| Xiao | 2013 | China | Asian | HB | PCR-PAGE | 187 | 107 | 11 | 305 | 212 | 115 | 15 | 342 | 0.21 | 0.905 |
| Wu | 2013 | China | Asian | HB | PCR-SSCP | 284 | 152 | 15 | 451 | 358 | 244 | 29 | 631 | 0.24 | 0.119 |
| Pardini | 2014 | Spain | Caucasian | PB | Taqman | 500 | 996 | 482 | 1978 | 425 | 802 | 420 | 1647 | 0.50 | 0.290 |
| Pardini | 2014 | Italy | Caucasian | PB | Taqman | 195 | 285 | 137 | 617 | 783 | 1230 | 538 | 2551 | 0.45 | 0.178 |
| Pardini | 2014 | USA | Caucasian | PB | Taqman | 237 | 514 | 259 | 1010 | 383 | 794 | 403 | 1580 | 0.51 | 0.835 |
| Pardini | 2014 | England | Caucasian | PB | Taqman | 410 | 825 | 341 | 1576 | 165 | 393 | 209 | 767 | 0.53 | 0.436 |
| Pardini | 2014 | Czech | Caucasian | PB | Taqman | 239 | 479 | 249 | 967 | 169 | 326 | 177 | 672 | 0.51 | 0.443 |
| Pardini | 2014 | Netherlands | Caucasian | PB | Taqman | 169 | 282 | 134 | 585 | 106 | 177 | 76 | 359 | 0.46 | 0.895 |
| Diego Marques | 2017 | Brazil | Caucasian | HB | PCR | 49 | 64 | 27 | 140 | 42 | 65 | 33 | 140 | 0.47 | 0.424 |
MAF, minor allele frequency; HWE, Hardy-Weinberg equilibrium; PB, population based; HB, hospital based; PCR-PAGE, polymerase chain reaction-polyacrylamide gel electrophoresis; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; AS-PCR, allele-specific polymerase chain reaction.
Meta-analysis of the association between CASP8 rs3834129 (-652 6N Ins/Del) polymorphism and CRC risk
| Variables | No. of | Homozygous | Heterozygous | Allele | |||||
|---|---|---|---|---|---|---|---|---|---|
| studies | DD vs. II | ID vs. II | D vs. I | ||||||
| OR (95% CI) | P het | OR (95% CI) | P het | OR (95% CI) | P het | ||||
| All | 13 | 0.92 (0.82-1.04) | 0.022 | 0.94 (0.88-0.99) | 0.602 | 0.95 (0.90-1.01) | 0.015 | ||
| Ethnicity | |||||||||
| Asian | 4 | 0.78 (0.47-1.32) | 0.028 | 0.86 (0.76-0.98) | 0.734 | 0.88 (0.74-1.04) | 0.046 | ||
| Caucasian | 9 | 0.96 (0.87-1.06) | 0.166 | 0.96 (0.90-1.02) | 0.408 | 0.98 (0.93-1.03) | 0.191 | ||
| Source of controls | |||||||||
| PB | 9 | 0.93 (0.81-1.07) | 0.007 | 0.94 (0.89-1.00) | 0.566 | 0.96 (0.90-1.02) | 0.009 | ||
| HB | 4 | 0.88 (0.67-1.16) | 0.493 | 0.89 (0.75-1.04) | 0.469 | 0.02 (0.81-1.05) | 0.279 | ||
| Quality score | |||||||||
| >9 | 6 | 0.82 (0.64-1.06) | 0.004 | 0.91 (0.82-1.02) | 0.174 | 0.90 (0.81-1.00) | 0.006 | ||
| ≤9 | 7 | 1.00 (0.91-1.10) | 0.925 | 0.95 (0.87-1.02) | 0.891 | 1.00 (0.96-1.05) | 0.942 | ||
Het, heterogeneity; HB, hospital based; PB, population based.
Heterogeneity and sensitivity analysis
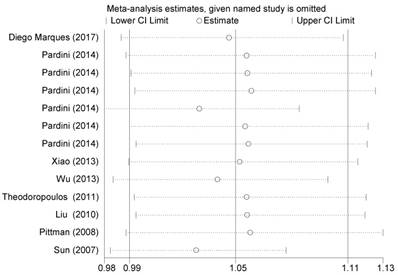
We first used I2 statistics and Q test to calculate between-study heterogeneity. Significant heterogeneity was detected among all three genetic models (P<0.001) in the pooled analysis. Therefore, we adopted the random-effect model to generate wider CIs. We also conducted sequential leave-one-out sensitivity analysis to assess the stability of the results. The results showed that no substantial changes in pooled results, after removing each study (Figure 3).
Forest plot for the CRC susceptibility associated with the CASP8 rs3834129 polymorphism under allele comparison model. The horizontal lines represent the study-specific ORs and 95% CIs, respectively. The diamond represents the pooled results of OR and 95% CI.
Publication bias
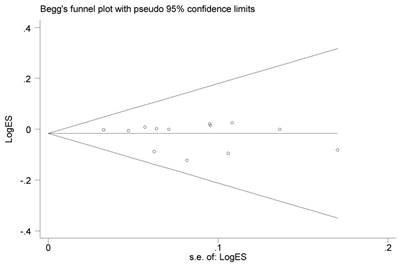
The Begg's funnel plots of the included studies showed no evidence of obvious asymmetry (Figure 4). Moreover, none-existence of publication bias among the studies was also approved by statistical evidence of Egger's test (data not shown).
Discussion
In this present meta-analysis, we comprehendsively assessed the relationship between CASP8 rs3834129 polymorphism with CRC susceptibility. The obtained results suggested CASP8 rs3834129 polymorphism may influence CRC risk in a low impact effect manner. To date, this meta-analysis represents the most powerful investigation in elucidating the role of CASP8 rs3834129 polymorphism in CRC risk.
The role of CASP8 rs3834129 polymorphism in CRC risk has attracted intensive attentions. The first case-control study with 4995 cases and 4972 controls was conducted by Sun et al. in 2007 [16]. They identified that -652 6N deletion allele would decrease the susceptibility of lung, colorectal, esophageal, breast, cervical and gastric cancer. Further biochemical assays illustrated that this variant might lead to decreased apoptotic reactivity of T lymphocytes upon cancer cells stimulation. A multi-centric study conducted by Pardini et al. indicated that rs3834129 was not associated with CRC risk in the full data set [21]. This study recruited 6,733 CRC cases and 7,576 controls by six different centers located in Spain, Italy, USA, England, Czech Republic and the Netherlands collaborating to the international consortium COGENT (Colorectal cancer GENeTics). Such null associations were also presented in a study conducted by Xiao MS et al. in Chinese population using 305 CRC patients and 342 healthy individuals [20].
To further clarify the role of CASP8 rs3834129 polymorphism on the risk of CRC, we performed this meta-analysis. CASP8 rs3834129 polymorphism was not associated with CRC risk, in some genetic models. This phenomenon may be due to the relatively small sample or the weak impact of single polymorphism in singe gene. Stratified analysis by ethnicity showed that significant association was observed between CASP8 rs3834129 polymorphism and CRC risk among Asian, but not Caucasian. Allelic distributions of CASP8 rs3834129 polymorphisms could vary geographically and ethnically.
When interpreting this meta-analysis, several limitations should be noted. First, we only used unadjusted estimates to assess the strength of association between CASP8 rs3834129 polymorphism and CRC risk. We failed to conduct adjustment analysis as we could not obtain original data such as life habitat, environmental exposes, and gene-environment interactions, which restrains our further analysis for confounding factors. Second, language bias and selection bias could not be ruled out, as only published studies and papers written in English or Chinese were analyzed. Third, significant between-study heterogeneity was detected, which would impair the validity of conclusion. Fourth, in some subgroup analysis, the sample size was relatively small. Thus, the statistical power is to be impaired to estimate the real association. Last, nearly all the eligible case-control studies included were conducted in Asians and Caucasians. The studies of other ethnicities, such as Africans, were absence. Therefore, more studies from other ethnicities, especially Africans, are necessary to further confirm such conclusion, due to the geographical and genetic differences.
Sensitivity analysis of the summary OR coefficients on the association between CASP8 rs3834129 polymorphism and CRC risk under allele comparison model.
Funnel plot analysis to detect publication bias for CASP8 rs3834129 polymorphism under allele comparison model. Each point represents a separate study for the indicated association.
Conclusion
In conclusion, the current meta-analysis provides strong evidence that CASP8 rs3834129 polymorphism may not be strong enough to impact the risk of CRC, from the perspective of the formed case-control studies. Such relationship further helps to explain the etiology of CRC. Yet, further case-control studies with larger sample sizes, standardized unbiased design are warranted to confirm our findings.
Acknowledgements
Financial support was provided by Zhejiang Natural Science Foundation Project (LYY18H280005) and Public Service Technology Research Project of Zhejiang Province of China (2016C33127).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177-193 doi: 10.3322/caac.21395
2. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108 doi: 10.3322/caac.21262
3. Obuch JC, Ahnen DJ. Colorectal Cancer: Genetics is Changing Everything. Gastroenterol Clin North Am. 2016;45:459-76 doi: 10.1016/j.gtc.2016.04.005
4. Chen M, Wang J. Initiator caspases in apoptosis signaling pathways. Apoptosis. 2002;7:313-9 doi
5. Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell. 2001;7:673-82 doi
6. Yang SY, Sales KM, Fuller B, Seifalian AM, Winslet MC. Apoptosis and colorectal cancer: implications for therapy. Trends Mol Med. 2009;15:225-33 doi: 10.1016/j.molmed.2009.03.003
7. Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491-501 doi
8. Kuwana T, Smith JJ, Muzio M, Dixit V, Newmeyer DD, Kornbluth S. Apoptosis induction by caspase-8 is amplified through the mitochondrial release of cytochrome c. J Biol Chem. 1998;273:16589-94 doi
9. Juo P, Kuo CJ, Yuan J, Blenis J. Essential requirement for caspase-8/FLICE in the initiation of the Fas-induced apoptotic cascade. Curr Biol. 1998;8:1001-8 doi
10. Li C, Zhao H, Hu Z, Liu Z, Wang LE, Gershenwald JE, Prieto VG, Lee JE, Duvic M, Grimm EA, Wei Q. Genetic variants and haplotypes of the caspase-8 and caspase-10 genes contribute to susceptibility to cutaneous melanoma. Hum Mutat. 2008;29:1443-51 doi: 10.1002/humu.20803
11. Camp NJ, Parry M, Knight S, Abo R, Elliott G, Rigas SH, Balasubramanian SP, Reed MW, McBurney H, Latif A, Newman WG, Cannon-Albright LA, Evans DG. et al. Fine-mapping CASP8 risk variants in breast cancer. Cancer Epidemiol Biomarkers Prev. 2012;21:176-81 doi: 10.1158/1055-9965.EPI-11-0845
12. Rihani A, De Wilde B, Zeka F, Laureys G, Francotte N, Tonini GP, Coco S, Versteeg R, Noguera R, Schulte JH, Eggert A, Stallings RL, Speleman F. et al. CASP8 SNP D302H (rs1045485) is associated with worse survival in MYCN-amplified neuroblastoma patients. PLoS One. 2014;9:e114696. doi: 10.1371/journal.pone.0114696
13. Zhang Y, Li W, Hong Y, Wu G, He K, Liu D. A systematic analysis of the association studies between CASP8 D302H polymorphisms and breast cancer risk. J Genet. 2017;96:283-9 doi
14. Hashemi M, Eskandari-Nasab E, Fazaeli A, Rezaei H, Mashhadi MA, Arbabi F, Taheri M. Bi-directional PCR allele-specific amplification (bi-PASA) for detection of caspase-8 -652 6N ins/del promoter polymorphism (rs3834129) in breast cancer. Gene. 2012;505:176-9 doi: 10.1016/j.gene.2012.05.043
15. Srivastava K, Srivastava A, Mittal B. Caspase-8 polymorphisms and risk of gallbladder cancer in a northern Indian population. Mol Carcinog. 2010;49:684-92 doi: 10.1002/mc.20641
16. Sun T, Gao Y, Tan W, Ma S, Shi Y, Yao J, Guo Y, Yang M, Zhang X, Zhang Q, Zeng C, Lin D. A six-nucleotide insertion-deletion polymorphism in the CASP8 promoter is associated with susceptibility to multiple cancers. Nat Genet. 2007;39:605-13 doi: 10.1038/ng2030
17. Liu B, Zhang Y, Jin M, Ni Q, Liang X, Ma X, Yao K, Li Q, Chen K. Association of selected polymorphisms of CCND1, p21, and caspase8 with colorectal cancer risk. Mol Carcinog. 2010;49:75-84 doi: 10.1002/mc.20579
18. Theodoropoulos GE, Gazouli M, Vaiopoulou A, Leandrou M, Nikouli S, Vassou E, Kouraklis G, Nikiteas N. Polymorphisms of caspase 8 and caspase 9 gene and colorectal cancer susceptibility and prognosis. Int J Colorectal Dis. 2011;26:1113-8 doi: 10.1007/s00384-011-1217-5
19. Wu Z, Li Y, Li S, Zhu L, Li G, Yu Z, Zhao X, Ge J, Cui B, Dong X, Tian S, Hu F, Zhao Y. Association between main Caspase gene polymorphisms and the susceptibility and prognosis of colorectal cancer. Med Oncol. 2013;30:565. doi: 10.1007/s12032-013-0565-0
20. Xiao MS, Chang L, Li WL, Du YS, Pan Y, Zhang DF, Wen Y, Luo J, Li XY, Yao YG. Genetic polymorphisms of the CASP8 gene promoter may not be associated with colorectal cancer in Han Chinese from southwest China. PLoS One. 2013;8:e67577. doi: 10.1371/journal.pone.0067577
21. Pardini B, Verderio P, Pizzamiglio S, Nici C, Maiorana MV, Naccarati A, Vodickova L, Vymetalkova V, Veneroni S, Daidone MG, Ravagnani F, Bianchi T, Bujanda L. et al. Association between CASP8 -652 6N del polymorphism (rs3834129) and colorectal cancer risk: results from a multi-centric study. PLoS One. 2014;9:e85538. doi: 10.1371/journal.pone.0085538
22. Marques D, Ferreira-Costa LR, Ferreira-Costa LL, Correa RDS, Borges AMP, Ito FR, Ramos CCO, Bortolin RH, Luchessi AD, Ribeiro-Dos-Santos A, Santos S, Silbiger VN. Association of insertion-deletions polymorphisms with colorectal cancer risk and clinical features. World J Gastroenterol. 2017;23:6854-67 doi: 10.3748/wjg.v23.i37.6854
23. Pittman AM, Broderick P, Sullivan K, Fielding S, Webb E, Penegar S, Tomlinson I, Houlston RS. CASP8 variants D302H and -652 6N ins/del do not influence the risk of colorectal cancer in the United Kingdom population. Br J Cancer. 2008;98:1434-6 doi: 10.1038/sj.bjc.6604314
Author contact
![]() Corresponding author: Yue Yang, Department of Pathology, Tongde Hospital of Zhejiang Province, 234 Gucui Road, Hangzhou 310012, Zhejiang, China. E-mail: tdyangyuecom; Phone: 86-571-89972240
Corresponding author: Yue Yang, Department of Pathology, Tongde Hospital of Zhejiang Province, 234 Gucui Road, Hangzhou 310012, Zhejiang, China. E-mail: tdyangyuecom; Phone: 86-571-89972240

 Global reach, higher impact
Global reach, higher impact