3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(1):147-155. doi:10.7150/jca.28534 This issue Cite
Research Paper
A Functional 5'-UTR Polymorphism of MYC Contributes to Nasopharyngeal Carcinoma Susceptibility and Chemoradiotherapy Induced Toxicities
1. Department of Clinical Pharmacology, Xiangya Hospital, Central South University and Institute of Clinical Pharmacology, Central South University; Hunan Key Laboratory of Pharmacogenetics, Changsha, 410008, P.R. China;
2. National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, 410008, P.R. China;
3. Key Laboratory of Translational Radiation Oncology, Hunan Province; Department of Radiation Oncology, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, 410013, P.R. China;
4. Department of Laboratory Medicine, National Key Laboratory of Biotherapy/Collaborative Innovation Center of Biotherapy and Cancer Center, West China Hospital, Sichuan University, Chengdu, 610000, P.R. China.
Received 2018-7-15; Accepted 2018-10-24; Published 2019-1-1
Abstract
MYC is a transcription factor acting as a pivotal regulator of genes involved in cell cycle progression, apoptosis, differentiation and metabolism. In this study, we evaluated the association of MYC polymorphisms with nasopharyngeal carcinoma (NPC) risk and chemoradiotherapy induced toxicities among Chinese population. By using bioinformatic tools, five potential functional single nucleotide polymorphisms of MYC were genotyped in a case-control study with 684 NPC patients and 823 healthy controls. We found two SNPs rs4645948 (C>T) and rs2071346 (G>T) were significantly associated with increased risk of developing NPC (TT+CT vs CC, OR=1.557, P=3.34×10-4; TT+GT vs GG, OR=1.361, P=0.007, respectively). In addition, rs4645948 (C>T) was conferred with increased risk of anemia (CT vs CC, OR=2.152, P=0.001) and severe leukopenia (CT vs CC, OR=1.893, P=0.034) for NPC patients receiving chemoradiotherapy. We also found rs2071346 (G>T) variant genotype carriers were subjected to higher risk of anemia (GT vs GG, OR=1.665, P=0.022) and thrombocytopenia (GT vs GG, OR=1.685, P=0.035). Our results demonstrated that the relative expression of MYC was dramatically higher in NPC tissues compared to rhinitis tissues. Over-expression of MYC was positively correlated with advanced T stage, N stage, and late clinical stage. Notably, the expression of MYC in rs4645948 CT and TT genotypes carriers were significantly higher than CC genotype carriers. Luciferase reporter assay indicated that the T allele of rs4645948 led to significantly higher transcription activity of MYC compared to the C allele. These findings suggested that individual carrying the rs4645948 T allele may be at greater risk for NPC due to an increase of MYC transcriptional activity and an augment of MYC expression.
Keywords: nasopharyngeal carcinoma, susceptibility, chemoradiotherapy, MYC, polymorphisms
Introduction
Nasopharyngeal carcinoma (NPC) is a malignant tumor arising from the epithelium of the nasopharynx. Although relatively rare in the western world (<1 case per 100,000 people), NPC is highly prevalent in southern China, Southeast Asia and Northern Africa, with Epstein-Barr virus (EBV) infection, environmental factors and genetic susceptibility implicated in its aetiology [1,2]. Currently, radiation therapy is a key modality in the treatment of NPC, and the treatment efficiency is improved when combined with chemotherapy [3,4]. The overall five-year survival rate of NPC patients remains around 72-90% for stages I and II NPC and 30%-55% for stages III and IV NPC, suggesting that early diagnosis is a key for better prognosis [5]. Unfortunately, the majority of NPC is diagnosed at an advanced stage due to the non-specific symptoms at early stage and the lack of a thorough nasopharyngeal examination. A clearer understanding of the molecular signature and underlying mechanisms would be beneficial in providing effective biomarkers for early diagnosis and therapeutic intervention [6-8].
With the evolution of DNA sequencing and bioinformatics, evidence of genetic polymorphisms underlying NPC pathogenesis is rapidly increasing [9,10]. In the past decades, several large-scale case-control studies have present a genome-wide view of the changes in NPC by genome wide association study (GWAS), exome sequencing, targeted sequencing, and SNP array analysis. Many susceptibility loci of NPC in different populations have been identified, including ITGA9 at chromosome 3p22.2, HLA-A and GABBR1 at 6p22.1, HLA-B/C and MICA at 6p21.33, HLA-DQ/DR at 6p21.32, MECOM at 3q26.2, CDKN2A/2B at 9q21.3, TNFRSF19 at 13q12.12, and MST1R at 3p21.3 [11-14]. Despite these notable achievements, additional predisposing genetic variants remain to be discovered as the identified risk variants only explain a small proportion of nasopharyngeal carcinoma heritability.
MYC is a proto-oncogene encoding a transcription factor that regulates the expression of approximately 15% of human genes directly and indirectly [15]. MYC integrates multiple signaling pathways and function as a master protein in the regulation of cellular proliferation, differentiation, cellular metabolism, apoptosis, migration/invasion, angiogenesis, microenvironment remodeling and immune responses [16-20]. Mechanistically, MYC interacts with MAX and activates gene transcription by binding to E-box DNA elements in the regulatory regions of target genes [21]. In addition, MYC participates in the regulation of global chromatin structure by recruiting histone acetyltransferases and regulating histone acetylation [22]. Moreover, MYC can indirectly regulate gene expression at the translational level since it controls multiple components of ribosome biogenesis. These critical functions of MYC imply its pivotal role in tumorigenesis.
Deregulation of MYC has been frequently observed in the pathogenesis of numerous human cancers, including diffuse large B-cell lymphoma, breast cancer, cervical cancer, hepatocellular carcinaoma, esophageal squamous cell carcinoma, and lung cancer, and was strongly correlated with poor prognosis [23-29]. Several studies have indicated that ectopic expression of MYC was a frequent genetic abnormality in NPC [30-32]. Konckdown of MYC inhibits cell growth and cell cycle progression in NPC cells [33]. In addition, MYC regulates radioresistance in a stem cell-like population of NPC cells through transcriptional activation of CHEK1 and CHEK2 checkpoint kinases [34]. However, the underling mechanisms of MYC dysregulation in NPC remains unclear.
Since gene mutation and DNA amplification are two major causes for oncogene desregulation, we wondered if there exists single nucleotide polymorphism (SNP) that contributes to MYC over-expression. To date, few studies have directly investigated the genetic polymorphisms of MYC in NPC. This study fills in this knowledge gap by conducting a case-control study with 684 NPC cases and 823 healthy controls from China with bioinformatic tools for functional SNP selection of MYC, and attempted to understand their impact on MYC function and clinical outcomes.
Materials and Methods
Study population
684 newly diagnosed, histopathologically confirmed and untreated NPC patients were consecutively recruited in Hunan Provincial Cancer Hospital between October 2014 and September 2015. Patients who had other concomitant malignant diseases or a second malignancy were excluded. Moreover, 94 newly diagnosed NPC tissue biopsies and 8 rhinitis tissue biopsies were collected from patients receiving pharyngorhinoscopy inspection. At the same time, 823 cancer-free healthy control subjects were enrolled in Xiangya Hospital who were seeking health care. All participants were genetically unrelated, ethnic Han Chinese population. The patients that received induction chemotherapy (IC) plus concurrent chemoradiotherapy (CCRT) regimen were rigorously picked out for toxicity association study, as we described previously [35]. The toxic reactions were evaluated by the Common Terminology Criteria for Adverse Events (CTCAE 3.0) during the CCRT treatment period. This study was performed with the approval of the Independent Ethical Committee of Institute of Clinical Pharmacology, Central South University (CTXY-140007-2). At recruitment, written informed consent was obtained from all participants involved in this study.
Bioinformatic selection for functional SNPs
We applied three bioinformatic databases, including ENCODE (http://genome.ucsc.edu/ENCODE/), ENSEMBL (http://asia.ensembl.org/index.html?redirect=no) and dbSNP (http://www.ncbi.nlm.nih.gov/snp/) to select SNPs lying within putative regulatory regions. The potential functional SNPs were picked out based on the following criteria: 1) SNPs localize to DNaseI hypersensitivity regions (a feature of active cis-regulatory sequences); 2) SNPs within regions of H3K27 acetylation marks (near active regulatory element); 3) SNPs localize to dense transcription factor binding sites; 4) SNPs localize to active chromatin state segmentation predicted by ChIP-seq data from the Broad Histone track; 5) SNPs exhibit eQTL trait. Finally, five potential functional SNPs (rs4645948, rs2071346, rs3891248, rs3824120, rs10110283) of MYC were identified for genotyping. As shown in Figure S1 and Table S1, there was no significant LD (R2<0.8) between each SNP pair. The detailed characteristics of the candidate SNPs were listed in Table 1.
DNA extraction and genotyping
3 ml of peripheral venous blood of each participant was collected and stored at -80°C. Tissue samples were preserved in RNAlater and stored in liquid nitrogen. Genomic DNA was extracted using the QIAamp DNA Mini and Blood Mini Kit (Qiagen Inc., Valencia, CA) in accordance with the manufacturer's instructions. The Sequenom MassARRAY iPLEX platform (Sequenom, Inc., San Diego, CA) was applied for candidate SNPs genotyping. The SNPs with call rate > 95 % were allowed for further analysis.
RNA isolation and real-time PCR
Total RNA was extracted and purified from tissues with RNAiso reagent (Takara Bio Inc., Japan). Complementary DNA (cDNA) was synthesized using a PrimeScript TM RT reagent Kit (Takara Bio Inc., Japan). Real-time PCR was performed in Roche LightCycler 480 system (Roche, Basel, Switzerland). 2-ΔΔCt method was used to calculate the relative expression of MYC. The primer sequences were listed as follows: GAPDH forward: 5'-ACAACTTTGGTATCGTGGAAGG-3' and reverse: 5'-GCCATCACGCCACAGTTTC-3'; MYC forward: 5'-GTCAAGAGGCGAACACACAAC-3' and reverse: 5'-TTGGACGGACAGGATGTATGC-3'.
Dual luciferase reporter assay
The human nasopharyngeal carcinoma cell lines HONE1 and HNE1 were obtained from the Advanced Research Center of Central South University, cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA), penicillin (100 units/ml), and streptomycin (100 units/ml) at 37 °C in humidified 5% CO2 incubator. The 5'-UTR region of MYC containing putative functional polymorphism site rs4645948 was directly synthesized by Genechem Company (Genechem, Shanghai, China) and cloned into Hind III site of pGL3 vector to get the wild-type vector (pGL3-MYC-WT). The mutant-type vector (pGL3-MYC-MUT) of rs4645948 was generated by site-directed mutagenesis. Nucleotide sequence of the constructs were confirmed by DNA sequencing. For the luciferase reporter assays, HONE1 and HNE1 cells were cultured in 24-well plates and transfected with pGL3-MYC-WT or pGL3-MYC-MUT constructs using Lipofectamine 3000 (Invitrogen, Carlsbad, CA). 48h after transfection, cells were harvested and assayed for Firefly and Renilla luciferase activities using the Dual Luciferase Reporter Assay System (Promega, Madison, WI). Firefly luciferase activity was normalized to Renilla luciferase activity in order to correct the transfection efficiency.
Characteristics of the candidate SNPs.
| SNPs | Chromosome | Localization | MAFa (Global/ EUR/ AMR/ EAS/ SAS/ CHS) | CpG island | H3K27 acetylation marks | DNaseI hypersensitivity clusters | Dense transcription factor binding region | Active chromatin state segmentation by HMM | eQTL trait |
|---|---|---|---|---|---|---|---|---|---|
| rs4645948 (C>T) | 8:127736252 | 5'-UTR | 7%/2%/6%/14%/ 11%/17% | √ | √ | √ | √ | Promoter | √ |
| rs2071346 (G>T) | 8:127736777 | Intron | 13%/2%/8%/20%/ 13%25% | √ | √ | √ | √ | Promoter | √ |
| rs3891248 (A>T) | 8:127737893 | Intron | 40%/17%/26%/40%/33%/39% | √ | √ | √ | √ | Promoter | √ |
| rs3824120 (G>T) | 8:127735707 | 5'-UTR | 12%/12%/19%/15%/18%/16% | √ | √ | √ | √ | Promoter | √ |
| rs10110283 (G>A) | 8:127739932 | Intron | 9%/3%/2%/7%/6%/5% | × | √ | √ | √ | Enhancer | √ |
Abbreviations: MAF, minor allele frequency; EUR, European; AMR, American; EAS, East Asian; SAS, South Asian; CHS, Southern Han Chinese, China.
aData from 1000 Genomes Project Phase 3 allele frequencies.
Statistical analysis
The distribution of demographic characteristics between cases and controls were examined by the χ2 test. Genotype frequencies of candidate SNPs were tested for the Hardy-Weinberg equilibrium (HWE). The association between SNPs and the risk of NPC susceptibility as well as chemoradiotherapy induced toxicities were demonstrated as odd ratios (ORs) and their 95% confidence intervals (CIs) computed by multivariate logistic regression analyses. All experiments were performed in triplicate, and the data was shown as the mean± standard deviation caculated by Student t test. All tests were two-sided, and a P value < 0.05 was considered as statistical significant. Haplotype analysis was conducted by the Web tool SHEsis (http://analysis.bio-x.cn/myAnalysis.php). Statistical analyses were carried out using the SPSS 19.0 (SPSS Inc, Chicago, IL).
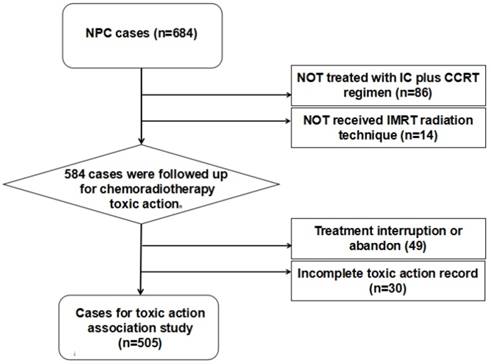
Patient selection diagram. The patients that received intensity-modulated radiotherapy (IMRT) and treated with induction chemotherapy (IC) plus concurrent chemoradiotherapy (CCRT) regimen with complete follow-ups data were rigorously picked out for toxicity association study.
Results
Population Characteristics
The demographics characteristics of patients with NPC and control subjects were shown in Table 2. Overall, the controls were comparable with patients with regard to age and BMI. However, a significantly higher male-to-female ratio (case vs control: 3.10 vs 0.85) was observed in cases, suggesting the cases representation paralleled the demographics of NPC in the general population. After excluding the cases with non-uniform treatment regimen, 505 NPC patients were included in the following chemoradiotherapy toxicity association analysis (Figure 1). The detailed clinical and therapy information for NPC patients were listed in Table S2. Most of the patients (90.1%) were diagnosed at late stages (III and IV) according to the 2002 American Joint Committee on Cancer (AJCC) staging system. All the patients received platinum-based induction chemotherapy plus concurrent radiochemotherapy with an average pGTVnx irradiation dose of 71.34 Gy. According to the CTCAE, there were 51 (10.1%) patients developed severe dermatitis, 129 (25.5%) patients developed severe oral mucositis, 121 (24.0%) patients developed severe myelosuppression, 94 (18.6%) patients developed severe neutropenia, 73 (14.5%) patients developed severe leukopenia, 217 (43%) patients developed anemia, and 98 (19.4%) patients developed thrombocytopenia during the CCRT treatment period.
Distribution of clinical characteristics of NPC cases and cancer-free controls.
| Variables | Controls N (%) | Cases N (%) | P a |
|---|---|---|---|
| Gender | |||
| Male | 378 (45.9) | 517 (75.6) | 1.91×10-14` |
| Female | 445 (54.1) | 167 (24.4) | |
| Age, years | |||
| ≥ 47 | 349 (42.4) | 305 (44.6) | 0.394 |
| < 47 | 474 (57.6) | 379 (55.4) | |
| BMI | |||
| ≥ 24 | 312 (37.9) | 275 (40.2) | 0.542 |
| 18.5 ~ 24 | 466 (56.6) | 368 (53.8) | |
| ≤ 18.5 | 45 (5.5) | 41 (6.0) | |
| Smoking status | |||
| Smoker | 192 (23.3) | 326 (47.7) | 2.37×10-14` |
| Nonsmoker | 631 (76.7) | 358 (52.3) | |
| Drinking status | |||
| Drinker | 121 (14.7) | 128 (18.7) | 0.043 |
| Nondrinker | 702 (85.3) | 556 (81.3) |
aTwo-sided x2 test
MYC candidate SNPs and NPC susceptibility
All of the SNPs showed a call rate > 95% except for rs3891248, and the genotype distributions of other SNPs in controls were in accordance with HWE (p>0.05). Multivariate logistic regression adjusted for age, gender, BMI, smoking status, and drinking status was used to assess the association between candidate SNPs and the risk of NPC. We found two SNPs rs4645948 (C>T) and rs2071346 (G>T) were significantly associated with increased risk of developing NPC (TT+CT vs CC, OR=1.557, P=3.34×10-4; TT+GT vs GG, OR=1.361, P=0.007, respectively) (Table 3). The statistical power for the two SNPs were 0.869 and 0.672 calculated by PS software (version 3.0.43). We failed to found any association between the other candidate SNPs and NPC susceptibility in our study population.
Multivariate logistic regression analysis of MYC candidate SNPs and NPC susceptibility.
| Genotypes | Controls N (%) | Cases N (%) | ORa (95% CI) | P a |
|---|---|---|---|---|
| rs4645948 (C>T) | ||||
| CC | 631 (76.7) | 464 (67.8) | 1.00 (reference) | |
| CT | 174 (21.1) | 197 (28.8) | 1.557 (1.212-2.001) | 0.001 |
| TT | 17 (2.1) | 20 (2.9) | 1.556 (0.781-3.102) | 0.209 |
| TT+CT vs CC | 1.557 (1.223-1.984) | 3.34×10-4 | ||
| TT vs CT+CC | 1.402 (0.703-2.794) | 0.337 | ||
| rs2071346 (G>T) | ||||
| GG | 555 (67.4) | 408 (59.6) | 1.00 (reference) | |
| GT | 234 (28.4) | 240 (35.1) | 1.361 (1.078-1.720) | 0.010 |
| TT | 32 (3.9) | 35 (5.1) | 1.360 (0.809-2.288) | 0.246 |
| TT+GT vs GG | 1.361 (1.088-1.702) | 0.007 | ||
| TT vs GT+GG | 1.210 (0.722-2.028) | 0.468 |
a Adjusted for gender, age, BMI, drinking status and smoking status.
Interaction analyses further revealed a significant multiplicative interaction between selected SNPs and gender on NPC risk. As shown in Table 4, compared to females with rs4645948 CC genotype, significantly increased risk of NPC was observed for males with CC (OR=2.754, P=9.07×10-11), CT (OR=4.678, P=2.75×10-15) and TT genotypes (OR=4.998, P=2.34×10-4). Likewise, the risk of developing NPC increased to 2.782 (P=3.20×10-10), 3.818 (P=4.69×10-13), 4.608 (P=4.89×10-6) fold for males with rs2071346 GG, GT, TT genotype carriers, compared to females carrying rs2071346 GG genotype.
The frequency of inferred haplotypes of MYC gene based on observed genotypes and their association with the risk of NPC were shown in Table S3. The result demonstrated that the TTGG haplotype was associated with increased risk of NPC (OR=1.327, P=0.009).
MYC candidate SNPs and chemoradiotherapy induced toxicities
Furthermore, we tried to reveal the relationship between MYC polymorphisms and chemoradiotherapy induced toxicities for NPC patients. By applying multivariate logistic regression analysis adjusting for patient clinical and treatment factors, our result demonstrated that rs4645948 (C>T) was associated with increased risk of anemia (CT vs CC, OR=2.152, P=0.001) and severe leukopenia (CT vs CC, OR=1.893, P=0.034) for NPC patients receiving chemoradiotherapy. We also found rs2071346 (G>T) variant genotype carriers were conferred with increased risk of anemia (GT vs GG, OR=1.665, P=0.022) and thrombocytopenia (GT vs GG, OR=1.685, P=0.035) (Table 5). Our result indicated that neither of the candidate SNPs have a linkage with chemoradiotherapy induced dermatitis, oral mucositis and neutropenia for NPC patients.
Genotype-specific expression effect of rs4645948
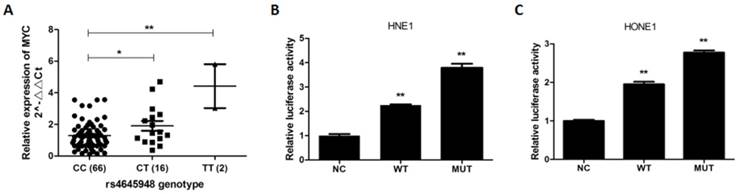
Since rs4645948 was predicted to have regulatory feature in the active promoter region of MYC, we wondered if rs4645948 could affect MYC transcription activity and hence MYC expression. Therefore, we examined the expression of MYC in NPC tissues with different rs4645948 genotypes. As shown in Figure 2A, the relative expression level of MYC was significantly higher in samples carrying CT and TT genotypes compared to samples with CC genotype.
The interaction between rs4645948, rs2071346 and gender on NPC risk.
| Genotypes | Gender | Controls N (%) | Cases N (%) | ORa (95% CI) | P a |
|---|---|---|---|---|---|
| rs4645948 (C>T) | |||||
| CC | female | 337 (40.9) | 116 (17.0) | 1.00 (reference) | |
| CT | female | 99 (12.0) | 46 (6.7) | 1.353 (0.899-2.036) | 0.147 |
| TT | female | 8 (1.0) | 3 (0.4) | 1.081 (0.282-4.145) | 0.910 |
| CC | male | 294 (35.7) | 348 (50.9) | 2.754 (2.027-3.740) | 9.07×10-11 |
| CT | male | 75 (9.1) | 151 (22.1) | 4.678 (3.190-6.858) | 2.75×10-15 |
| TT | male | 9 (1.1) | 17 (2.5) | 4.998 (2.121-11.777) | 2.34×10-4 |
| rs2071346 (G>T) | |||||
| GG | female | 303 (36.8) | 105 (15.4) | 1.00 (reference) | |
| GT | female | 125 (15.2) | 58 (8.5) | 1.342 (0.916-1.968) | 0.131 |
| TT | female | 15 (1.8) | 4 (0.6) | 0.767 (0.249-2.363) | 0.644 |
| GG | male | 252 (30.6) | 303 (44.3) | 2.782 (2.023-3.828) | 3.20×10-10 |
| GT | male | 109 (13.2) | 182 (26.6) | 3.818 (2.656-5.488) | 4.69×10-13 |
| TT | male | 17 (2.1) | 31 (4.5) | 4.608 (2.393-8.872) | 4.89×10-6 |
a Adjusted for age, BMI, drinking status and smoking status.
Multivariate logistic analysis of candidate SNPs and concurrent chemoradiotherapy induced hematotoxicities in NPC patients.
| Genotypes | Leukopenia | Thrombocytopenia | Anemia | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grade ≤2 N (%) | Grade > 2 N (%) | ORa (95% CI) | P a | Grade 0 N (%) | Grade > 0 N (%) | ORa (95% CI) | P a | Grade 0 N (%) | Grade > 0 N (%) | ORa (95% CI) | P a | |
| rs4645948 (C>T) | ||||||||||||
| CC | 302 (69.9) | 46 (63.0) | 1.00 (reference) | 284 (69.8) | 64 (65.3) | 1.00 (reference) | 213 (74.0) | 135 (62.2) | 1.00 (reference) | |||
| CT | 116 (26.9) | 25 (34.2) | 1.893 (1.048-3.419) | 0.034 | 109 (26.8) | 32 (32.7) | 1.305 (0.780-2.184) | 0.311 | 66 (22.9) | 75 (34.6) | 2.152 (1.372-3.375) | 0.001 |
| TT | 12 (2.8) | 2 (2.7) | 0.778 (0.129-4.694) | 0.784 | 12 (2.9) | 2 (2.0) | 0.369 (0.065-2.094) | 0.261 | 7 (2.4) | 7 (3.2) | 1.613 (0.484-5.370) | 0.436 |
| TT+CT vs CC | 1.747 (0.985-3.099) | 0.056 | 1.176 (0.712-1.941) | 0.527 | 2.095 (1.356-3.235) | 0.001 | ||||||
| TT vs CT+CC | 0.667 (0.113-3.927) | 0.654 | 0.345 (0.062-1.934) | 0.226 | 1.378 (0.406-4.684) | 0.607 | ||||||
| rs2071346 (G>T) | ||||||||||||
| GG | 264 (61.1) | 41 (56.2) | 1.00 (reference) | 253 (62.2) | 52 (53.1) | 1.00 (reference) | 184 (63.9) | 121 (55.8) | 1.00 (reference) | |||
| GT | 143 (33.1) | 28 (38.4) | 1.593 (0.903-2.812) | 0.108 | 131 (32.2) | 40 (40.8) | 1.685 (1.037-2.738) | 0.035 | 91 (31.6) | 80 (36.9) | 1.665 (1.077-2.575) | 0.022 |
| TT | 24 (5.6) | 4 (5.5) | 1.098 (0.312-3.861) | 0.884 | 23 (5.7) | 5 (5.1) | 0.800 (0.270-2.366) | 0.686 | 13 (4.5) | 15 (6.9) | 2.403 (1.001-5.773) | 0.050 |
| TT+GT vs GG | 1.517 (0.879-2.618) | 0.134 | 1.518 (0.953-2.419) | 0.079 | 1.755 (1.157-2.662) | 0.008 | ||||||
| TT vs GT+GG | 0.934 (0.272-3.212) | 0.914 | 0.604 (0.203-1.792) | 0.363 | 1.981 (0.841-4.665) | 0.118 | ||||||
a Adjusted for gender, age, BMI, smoking status, drinking status, histological type, clinical stage, induced chemotherapy regimen, concurrent chemoradiotherapy regimen, and irradiation dose.
The effect of rs4645948 on MYC expression and transcription activity. (A) The expression level of MYC in NPC tissues with different rs4645948 genotypes. (B) The TT genotype of rs4645948 increases MYC transcription activity in HNE1 cell. (C) The TT genotype of rs4645948 increases MYC transcription activity in HONE1 cell.
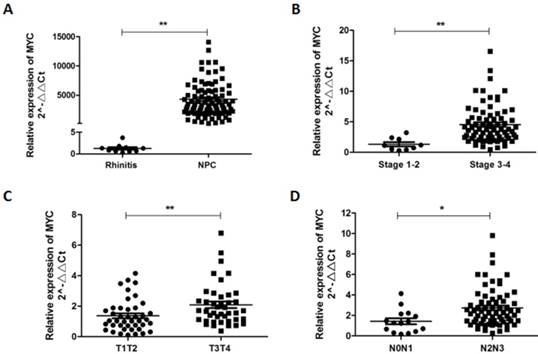
The relative expression of MYC in NPC tissues and its clinical significance. (A) MYC was significantly higher expressed in NPC tissues compared to rhinitis tissues. MYC over expression was correlated with late clinical stage (B), advanced T stage (C) and N stage (D).
rs4645948 increases MYC transcriptional activity
Due to the expression variation of MYC with different rs4645948 genotypes in NPC tissues, we want to know if this 5'-UTR polymorphism exerts its role through transcription regulation. We applied luciferase reporter assay to evalute the effects of rs4645948 on the MYC transcriptional activity. We found that the luciferase activity of the cells transfected with pGL3-MYC-MUT increased to about 1.5 times compared with the cells transfected with pGL3-MYC-WT (Figure 2B, 2C). These findings indicated that individual carrying the MYC T allele may be at greater risk for NPC due to an increase of MYC transcriptional activity and an augment of MYC expression.
Up regulation of MYC in NPC tissues and its clinical significance
To assess the significance of MYC in NPC pathogenesis, we examined and quantified the expression of MYC by qRT-PCR in 94 NPC tissues and 10 rhinitis tissues. As presented in Figure 3, MYC was prominently up-regulated (5792 fold change) in clinical NPC tissues compared with rhinitis tissues (P < 0.01). Moreover, we evaluated the correlation of MYC expression with patients' clinicopathological parameters to assess its clinical significance. As shown, over-expression of MYC was positively correlated with advanced T stage (P<0.05), N stage (P<0.05), and late clinical stage (P<0.01), suggesting that MYC could be a potential progressive factor for NPC patients.
Discussion
The remarkable ethnic and geographical distributions indicate genetic factors may play a leading role in the pathogenesis of NPC. Nevertheless, genomic background of this disease remains largely obscure, and no targeted therapy has been established. A number of studies have indicated that deregulation of MYC contributed to the development of most human malignancies both in vitro and in vivo since it had an effect on all basic cellular processes [16]. In this study, we wanted to know if SNPs of MYC contribute to aberrant expression of MYC in NPC, and eventually lead to NPC tumorigenesis.
In the past decade, a number of large scale GWAS have identified nearly 6,500 loci associated with diseases. However, only 7% of such loci are located in protein-coding regions, whereas 93% are located in noncoding regions [36-38]. Such results imply that many genetic variants predisposing to the risk of human diseases are likely to exert their role by affecing the regulation of genes rather than directly altering protein function. In light of this, we took full advantage of bioinformatic databases for selection of potentially functional SNPs lies in non-coding regions of MYC, as mentioned in the method. Herein, for the first time we identified MYC rs4645948 and rs2071346 as new susceptibility locus for NPC among a Chinese population. Notably, we found the risk of NPC increased dramatically to 5.488 fold and 5.266 fold for male mutant homozygote carriers of rs4645948 and rs2071346, compared to female wild homozygote carriers. Moreover, rs4645948 and rs2071346 mutant genotype carriers were exposed to increased risk of developing anemia, thrombocytopenia, and severe leukopenia in the concurrent chemoradiotherapy period.
Since rs4645948 was located in 5'-UTR of MYC, we conducted a dual luciferase reporter assay to estimate its role on MYC transcription activity. As predicted, the luciferase activity of pGL3-MYC-MUT was significantly higher than pGL3-MYC-WT. This was in accordance with the finding in NPC tissues, as the relative expression of MYC was significantly higher in CT and TT genotypes in comparison with CC genotype. These may explain our clinical association study result to a certain extent. Given the fact that MYC was a oncogene, the mutant genotype of rs4645948 increased transcriptional activity of MYC, leading to high level expression of MYC, and consequently increased risk of NPC. By the way, the mutation frequency of rs4645948 was particularly higher in East Asian (14%) and South Asian (11%) in contrast to American (6%) and European (2%) (data from 1000 Genomes Project Phase 3). This was interesting since NPC was endemic to Southeast Asia, the higher MAF of rs4645948 in southeast Asia indicated its pathogenic role in NPC. To the best of our knowledge, this is the first study that investigates the function of rs4645948 on MYC transcriptional activity and its mRNA expression, which might provide new insight for the genetic basis of NPC.
We further demonstrated that MYC was over expressed in NPC tissues, and was correlated with advanced T stage, N stage, and late clinical stage, indicating MYC was involved in NPC progression. Actually, previous studies have also reported that MYC gene amplification was associated with poor clinical stage and prognosis in many kinds of cancers [23-28]. Considering the pleiotropic effect of MYC on tumorigenesis transformation, MYC is an ideal target for pharmacological intervention, and targeting MYC could have broad-range anti-tumor effect [39]. Indeed, a mouse model expressing a switchable dominant negative form of MYC, called OmoMyc has demonstrated that MYC can be inactivated in cancer cells without causing detrimental tissue homeostasis [40]. Several small-molecule MYC inhibitors, such as KJ-Pyr-9, ME47, 10058-F4, have been proved to antagonize tumour cell growth in vitro and in vivo through disrupting the MYC:MAX:E-box interaction/ binding and blocking transcriptional MYC activity [41-43].
In conclusion, our study demonstrated that MYC rs4645948 increased transcriptional activity of MYC, leading to high level expression of MYC, and consequently increased risk of developing NPC and concurrent radiochemotherapy induced toxicities in southern Chinese populations. The result of our study may provide new insight in the molecular pathogenesis of NPC and have certain implications for NPC diagnostic and therapeutic strategies.
Abbreviations
NPC: nasopharyngeal carcinoma; SNPs: single nucleotide polymorphisms; EBV: Epstein-Barr virus; GWAS: genome wide association study; IMRT: intensity-modulated radiotherapy; IC: induction chemotherapy; CCRT: concurrent chemoradiotherapy; CTCAE: Common Terminology Criteria for Adverse Events; cDNA: Complementary DNA; HWE: Hardy-Weinberg equilibrium; OR: odd ratio; CI: confidence interval; ROC: receiver operating characteristic curve; AJCC: American Joint Committee on Cancer; AUC: area under the curve; ENCODE: Encyclopedia of DNA Elements; DHSs: Deoxyribonuclease I hypersensitive sites.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by the National Key Research and Development Program (No.2016YFC0905000 and No.2016YFC0905001), National Natural Science Foundation of China (No.81522048, No.81573511 and No.81472802); Innovation Driven Project of Central South University (No.2016CX024); Graduate Students Independent Exploration and Innovation Project (No. 2017zzts226); Open Research Fund Program of Key Laboratory of Translational Radiation Oncology (2015TP1009).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Tang LL, Chen WQ, Xue WQ. et al. Global trends in incidence and mortality of nasopharyngeal carcinoma. Cancer Lett. 2016;374:22-30
2. Bruce JP, Yip K, Bratman SV. et al. Nasopharyngeal Cancer: Molecular Landscape. J Clin Oncol. 2015;33:3346-55
3. Lee AW, Tung SY, Chua DT. et al. Randomized trial of radiotherapy plus concurrent-adjuvant chemotherapy vs radiotherapy alone for regionally advanced nasopharyngeal carcinoma. J Natl Cancer Inst. 2010;102:1188-98
4. Sun Y, Li WF, Chen NY. et al. Induction chemotherapy plus concurrent chemoradiotherapy versus concurrent chemoradiotherapy alone in locoregionally advanced nasopharyngeal carcinoma: a phase 3, multicentre, randomised controlled trial. Lancet Oncol. 2016;17:1509-20
5. Chua MLK, Wee JTS, Hui EP. et al. Nasopharyngeal carcinoma. Lancet. 2016;387:1012-24
6. Xiao L, Xiao T, Wang ZM. et al. Biomarker discovery of nasopharyngeal carcinoma by proteomics. Expert Rev Proteomics. 2014;11:215-25
7. Feng XP, Yi H, Li MY. et al. Identification of biomarkers for predicting nasopharyngeal carcinoma response to radiotherapy by proteomics. Cancer Res. 2010;70:3450-62
8. Guo Z, Shu Y, Zhou H. et al. Radiogenomics helps to achieve personalized therapy by evaluating patient responses to radiation treatment. Carcinogenesis. 2015;36:307-17
9. Bei JX, Jia WH, Zeng YX. Familial and large-scale case-control studies identify genes associated with nasopharyngeal carcinoma. Semin Cancer Biol. 2012;22:96-106
10. Lin DC, Meng X, Hazawa M. et al. The genomic landscape of nasopharyngeal carcinoma. Nat Genet. 2014;46:866-71
11. Ng CC, Yew PY, Puah SM. et al. A genome-wide association study identifies ITGA9 conferring risk of nasopharyngeal carcinoma. J Hum Genet. 2009;54:392-7
12. Tse KP, Su WH, Chang KP. et al. Genome-wide association study reveals multiple nasopharyngeal carcinoma-associated loci within the HLA region at chromosome 6p21.3. Am J Hum Genet. 2009;85:194-203
13. Bei JX, Li Y, Jia WH. et al. A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat Genet. 2010;42:599-603
14. Dai W, Zheng H, Cheung AK. et al. Whole-exome sequencing identifies MST1R as a genetic susceptibility gene in nasopharyngeal carcinoma. Proc Natl Acad Sci U S A. 2016;113:3317-22
15. Gearhart J, Pashos EE, Prasad MK. Pluripotency redux-advances in stem-cell research. N Engl J Med. 2007;357:1469-72
16. Cotterman R, Jin VX, Krig SR. et al. N-Myc regulates a widespread euchromatic program in the human genome partially independent of its role as a classical transcription factor. Cancer Res. 2008;68:9654-62
17. Deb-Basu D, Karlsson A, Li Q. et al. MYC can enforce cell cycle transit from G1 to S and G2 to S, but not mitotic cellular division, independent of p27-mediated inhibition of cyclin E/CDK2. Cell Cycle. 2006;5:1348-55
18. Kim J, Woo AJ, Chu J. et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313-24
19. Dejure FR, Eilers M. MYC and tumor metabolism: chicken and egg. EMBO J. 2017;36:3409-20
20. Casey SC, Baylot V, Felsher DW. MYC: Master Regulator of Immune Privilege. Trends Immunol. 2017;38:298-305
21. Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635-45
22. Conacci-Sorrell M, Ngouenet C, Eisenman RN. Myc-nick: a cytoplasmic cleavage product of Myc that promotes alpha-tubulin acetylation and cell differentiation. Cell. 2010;142:480-93
23. Quesada AE, Medeiros LJ, Desai PA. et al. Increased MYC copy number is an independent prognostic factor in patients with diffuse large B-cell lymphoma. Mod Pathol. 2017;30:1688-97
24. Schick M, Habringer S, Nilsson JA. et al. Pathogenesis and therapeutic targeting of aberrant MYC expression in haematological cancers. Br J Haematol. 2017;179:724-38
25. Kim EY, Kim A, Kim SK. et al. MYC expression correlates with PD-L1 expression in non-small cell lung cancer. Lung Cancer. 2017;110:63-7
26. Batistatou A, Kotoula V, Bobos M. et al. Correlation of MYC gene and protein status with breast cancer subtypes and outcome of patients treated with anthracycline-based adjuvant chemotherapy. Pooled analysis of 2 Hellenic Cooperative Group Phase III Trials. Clin Breast Cancer. 2018;18:53-62
27. Lian Y, Niu X, Cai H. et al. Clinicopathological significance of c-MYC in esophageal squamous cell carcinoma. Tumour Biol. 2017;39:1010428317715804
28. Liu J, Zhu M, Xia X. et al. Jumonji domain-containing protein 1A promotes cell growth and progression via transactivation of c-Myc expression and predicts a poor prognosis in cervical cancer. Oncotarget. 2016;7:85151-62
29. Koutb F, Abdel-Rahman S, Hassona E. et al. Association of c-myc and p53 gene expression and polymorphisms with hepatitis C (HCV) chronic infection, cirrhosis and hepatocellular carcinoma (HCC) stages in Egypt. Asian Pac J Cancer Prev. 2017;18:2049-57
30. Fan CS, Wong N, Leung SF. et al. Frequent c-myc and Int-2 overrepresentations in nasopharyngeal carcinoma. Hum Pathol. 2000;31:169-78
31. Porter MJ, Field JK, Leung SF. et al. The detection of the c-myc and ras oncogenes in nasopharyngeal carcinoma by immunohistochemistry. Acta Otolaryngol. 1994;114:105-9
32. Zhou W, Feng X, Ren C. et al. Over-expression of BCAT1, a c-Myc target gene, induces cell proliferation, migration and invasion in nasopharyngeal carcinoma. Mol Cancer. 2013;12:53
33. Niu Z, Liu H, Zhou M. et al. Knockdown of c-Myc inhibits cell proliferation by negatively regulating the Cdk/Rb/E2F pathway in nasopharyngeal carcinoma cells. Acta Biochim Biophys Sin (Shanghai). 2015;47:183-91
34. Wang WJ, Wu SP, Liu JB. et al. MYC regulation of CHK1 and CHK2 promotes radioresistance in a stem cell-like population of nasopharyngeal carcinoma cells. Cancer Res. 2013;73:1219-31
35. Guo Z, Wang Y, Zhao Y. et al. Genetic polymorphisms of long non-coding RNA GAS5 predict platinum-based concurrent chemoradiotherapy response in nasopharyngeal carcinoma patients. Oncotarget. 2017;8:62286-97
36. Maurano MT, Humbert R, Rynes E. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190-5
37. Zhang X, Bailey SD, Lupien M. Laying a solid foundation for Manhattan-'setting the functional basis for the post-GWAS era'. Trends Genet. 2014;30:140-9
38. Hindorff LA, Sethupathy P, Junkins HA. et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362-7
39. McKeown MR, Bradner JE. Therapeutic strategies to inhibit MYC. Cold Spring Harb Perspect Med. 2014:4 pii: a014266
40. Soucek L, Whitfield J, Martins CP. et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679-83
41. Hart JR, Garner AL, Yu J. et al. Inhibitor of MYC identified in a Kröhnke pyridine library. Proc Natl Acad Sci U S A. 2014;111:12556-61
42. Lustig LC, Dingar D, Tu WB. et al. Inhibiting MYC binding to the E-box DNA motif by ME47 decreases tumour xenograft growth. Oncogene. 2017;36:6830-7
43. Tan Y, Sementino E, Chernoff J. et al. Targeting MYC sensitizes malignant mesothelioma cells to PAK blockage-induced cytotoxicity. Am J Cancer Res. 2017;7:1724-37
Author contact
![]() Corresponding authors: Prof. Wei Zhang, Department of Clinical Pharmacology, Xiangya Hospital, Central South University and Institute of Clinical Pharmacology, Central South University; Hunan Key Laboratory of Pharmacogenetics, 110 Xiangya Road, Changsha, 410008, China. Tel: +86 13973168774; Fax: +86 731 8235 4476; Email: yjsd2003com, and Prof. Hui Wang, Key Laboratory of Translational Radiation Oncology, Hunan Province; Department of Radiation Oncology, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, 283 Tongzipo Road, Changsha, 410013, China. Tel: +86 13973135460; Fax: +86 731 8865 1999; Email: wanghui710327com
Corresponding authors: Prof. Wei Zhang, Department of Clinical Pharmacology, Xiangya Hospital, Central South University and Institute of Clinical Pharmacology, Central South University; Hunan Key Laboratory of Pharmacogenetics, 110 Xiangya Road, Changsha, 410008, China. Tel: +86 13973168774; Fax: +86 731 8235 4476; Email: yjsd2003com, and Prof. Hui Wang, Key Laboratory of Translational Radiation Oncology, Hunan Province; Department of Radiation Oncology, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, 283 Tongzipo Road, Changsha, 410013, China. Tel: +86 13973135460; Fax: +86 731 8865 1999; Email: wanghui710327com

 Global reach, higher impact
Global reach, higher impact