3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(2):488-493. doi:10.7150/jca.26967 This issue Cite
Research Paper
Potential Mutations in Uveal Melanoma Identified Using Targeted Next-Generation Sequencing
Peking University Cancer Hospital & Institute, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Renal Cancer and Melanoma, No. 52 Fucheng Road, Haidian District, Beijing, China, 100142.
#Contributed equally to this work.
Received 2018-4-30; Accepted 2018-11-18; Published 2019-1-1
Abstract
Background/objective: Uveal melanoma (UM) is the most common intraocular malignancy and has a high tendency to metastasize to the liver. Although primary tumours can be successfully treated, there is currently no effective treatment for metastatic UM. To gain insight into the genetics of UM, we performed the targeted next-generation sequencing (NGS) of UM samples from a non-Caucasian population.
Methods: This study included tumour samples and blood samples from 107 UM patients at Peking University Cancer Hospital & Institute. Clinical data were collected. DNA was extracted from formalin-fixed, paraffin-embedded (FFPE) specimens. Using the HaloPlex Target Enrichment System (Agilent Technologies), NGS was performed to investigate mutations in a 35-gene panel composed of cancer-related genes.
Results: Recurrent coding mutations were found in the known UM drivers GNAQ and GNA11. FOXO1, PIK3R1 and HIF1A were also found to harbour somatic mutations in more than 20% of patients, a result that may indicate previously undescribed associations between these genes and UM pathogenesis. Patients with HIF1A and FOXO1 mutations exhibited worse overall survival (OS). In multivariate analysis, FOXO1 mutation was an independent prognostic factor for OS (P<0.05) that was associated with an increase in the risk ratio by a factor of 1.35. Notably, we found that HIF1A and FOXO1 mutations were associated with metastatic transformation of UM (P<0.05 and P<0.001, respectively).
Conclusion: Our findings from analyses of targeted NGS data shed new light on the molecular genetics of UM and facilitate the exploration of mutations associated with metastatic potential.
Keywords: uveal melanoma, metastasis, FOXO1, HIF1A.
Introduction
Uveal melanoma (UM) is the most common intraocular malignancy and can arise at three sites: the iris, the choroid, and the ciliary body. [1]UM tumours are highly aggressive and exhibit a high tendency to metastasize to the liver. [2]Due to a lack of effective systemic therapy, the median survival time of patients with metastatic UM is 12.5 months. [3]
Dramatic breakthroughs in the treatment of cutaneous melanoma (CM) have been achieved over the past decade. Targeted therapy such as the use of BRAF and MEK mutation inhibitors can improve overall survival (OS) for CM patients who harbour specific mutations. [4] However, the development of targeted therapy for UM is hampered by a lack of understanding of genetic changes associated with UM. Therefore, investigating potential high-frequency mutations in UM is crucial and could provide benefits for direct clinical applications.
Based on genomic, transcriptomic, and proteomic high-throughput screening of UM samples, several driver genes for UM have been detected. The molecular alterations associated with UM differ from those associated with CM. Driver mutations related to the pathogenesis of UM, such as mutations in GNAQ, GNA11, and BAP1, have also been revealed; these mutations are distinct from those related to CM. [5-7] GNAQ or GNA11 mutations lead to constitutive activation of the mitogen-activated protein kinase (MAPK) pathway and represent early events in tumourigenesis in UM. [8]BAP1 mutations are strongly associated with the progression and metastasis of UM. [7]Most research on the molecular landscape of UM has focused on Caucasians, and little data are available for other races. Asian CM patients significantly vary from Caucasian CM patients with respect to genetic profiles and clinical characteristics; [9-11] however, the genetic mutations associated with UM in non-Caucasian populations remain unclear.
To gain insight into the genetics of UM, we performed the targeted NGS of UM samples from a non-Caucasian population. We evaluated 35 mutated genes in the MAPK and PI3K/AKT pathways. We then analysed correlations between mutation status and clinicopathological features, with a particular focus on metastatic transformation. Our findings can facilitate the selection of more appropriate therapeutic treatments for UM in clinical practice, especially for patients with a high risk of metastasis.
Patients and Methods
Patient samples
Matched Formalin-fixed, paraffin-embedded (FFPE) tumor tissues and blood samples were obtained from 107 Chinese UM patients who visited Peking University Cancer Hospital. All of these samples were collected between June 2009 and December 2017. Clinical data, including gender, age, tumour side, tumour thickness, metastasis status, and survival (with follow-up continuing until September 2018, loss to follow-up or patient death), were collected. Diagnoses of UM were confirmed histopathologically for all patients. This study was approved by the medical ethics committee of Peking University Cancer Hospital & Institute and was conducted according to the Declaration of Helsinki Principles.
DNA preparation and NGS
Genomic DNA was extracted from FFPE sections using a QIAamp DNA FFPE Tissue Kit (Qiagen) and from blood lymphocytes using a QIAamp DNA Mini kit (Qiagen). Human Genomic DNA (G3041, Promega) as normal control. Using the HaloPlex Target Enrichment System (Agilent Technologies), NGS was performed to investigate mutations in a panel of 35 cancer-related genes. The panel was designed contain common genes relevant to melanoma pathogenesis, include MAPK, PI3K/AKT and cell cycle pathway. In accordance with the manufacturer's instructions, we utilized a custom-designed HaloPlex Target Enrichment Kit (Agilent) to capture target regions. In brief, genomic DNA was digested via eight different restriction reactions. The restricted segments were hybridized to a probe with ends complementary to those of the target fragment. During hybridization, the fragment was cyclized and integrated with the sequencing motif, which consisted of index sequences. The attachment of the cyclized target DNA fragment was captured using streptavidin magnetic beads. Finally, polymerase chain reaction (PCR) was performed to amplify the captured target libraries. Paired-end sequencing (with 100 bp reads) of all samples was performed on a HiSeq2500 instrument (Illumina). Clusters were generated using the TruSeq PE Cluster Kit V3 (Illumina), and the TruSeq SBS Kit V3 (Illumina) was used for sequencing. Image analysis and base calling were performed using Illumina RTA software. Sequence reads were trimmed to remove Illumina adapter sequences and aligned to the human reference genome (version hg19). Variants were invoked using Agilent SureCall software. Variants were further filtered using the dbSNP database and 1000 Genomes Project. The results were filtered by dbSNP database (https://www.ncbi.nlm.nih.gov/projects/SNP/) and 1000 Genomes projects (https://ftp.ncbi.nih.gov/). Shanghai Biotechnology Corporation (Shanghai, China) completed the targeted enrichment, sequencing, and data analysis for NGS.
Statistical analysis
IBM SPSS statistical software (version 20.0) was used for statistical assessments. Mean values were analysed using t tests for normally distributed continuous variables and estimated using the Mann-Whitney U test for abnormally distributed continuous variables. OS curves were evaluated using the Kaplan-Meier method. Cox univariate and multivariate analyses were performed to assess associations between prognostic indicators and OS. For all statistical analyses, P<0.05 (for two-tailed tests) was considered to be statistically significant.
Results
Patient characteristics and gene mutation rates
The records of 107 patients with UM were reviewed in this study. Among them, 40 patients were classified as training cohort, 67 patients were classified as validation cohort. Clinicopathological characteristics of these patients are shown in Table 1.
In training cohort, the gene mutation rates of GNAQ, GNA11, BRAF, NRAS, CCND1, HIF1A, AKT1, FOXO1, PIK3R1, PTEN, TSC1, TSC2, FGFR3, KIT and MET were 45% (18/40), 20% (8/40), 10% (4/40), 10% (4/40), 5% (2/40), 30% (12/40), 10% (4/40), 40% (16/40), 50% (20/40), 10% (4/40), 17.5% (7/40), 15% (6/40), 7.5% (3/40), 5% (2/40) and 5% (2/40), respectively.
Baseline Characteristics of Uveal Melanoma Patients.
| Characteristics | Training Cohort, n=40 | Validation Cohort, n=67 | ||
|---|---|---|---|---|
| N | % | N | % | |
| Gender | ||||
| Male | 24 | 60 | 36 | 53.7 |
| Female | 16 | 40 | 31 | 46.3 |
| Age/year | ||||
| ≥60 | 15 | 37.5 | 28 | 41.8 |
| <60 | 25 | 62.5 | 39 | 58.2 |
| Side | ||||
| Left eye | 18 | 45 | 32 | 47.8 |
| Right eye | 22 | 55 | 35 | 52.2 |
| Thickness | ||||
| ≤2mm | 8 | 20 | 16 | 23.9 |
| 2-4mm | 12 | 30 | 27 | 40.3 |
| >4 mm | 20 | 50 | 24 | 35.8 |
| Ciliary body involvement | ||||
| yes | 22 | 55 | 31 | 46.3 |
| no | 18 | 45 | 36 | 53.7 |
| Cell type | ||||
| Spindle | 20 | 50 | 34 | 50.7 |
| Mixed or epithelioid | 20 | 50 | 33 | 49.3 |
| Extraocular extension | ||||
| None or superficial | 24 | 60 | 42 | 62.7 |
| Deep, total, or episcleral | 16 | 40 | 25 | 37.3 |
| with metastasis | ||||
| without | 24 | 60 | 38 | 56.7 |
| with | 16 | 40 | 29 | 43.3 |
| GNAQ Mutation | ||||
| yes | 18 | 45 | 32 | 47.8 |
| no | 22 | 55 | 35 | 52.2 |
| GNA11 Mutation | ||||
| yes | 8 | 20 | 15 | 22.4 |
| no | 32 | 80 | 52 | 77.6 |
| BRAF Mutation | ||||
| yes | 4 | 10 | 9 | 13.4 |
| no | 36 | 90 | 58 | 86.6 |
| NRAS Mutation | ||||
| yes | 4 | 10 | 6 | 9 |
| no | 36 | 90 | 61 | 91 |
| HIF1A Mutation | ||||
| yes | 12 | 30 | 18 | 26.9 |
| no | 28 | 70 | 49 | 73.1 |
| FOXO1 Mutation | ||||
| yes | 16 | 40 | 29 | 43.3 |
| no | 24 | 60 | 38 | 56.7 |
| PIK3R1 Mutation | ||||
| yes | 20 | 50 | 27 | 40.3 |
| no | 20 | 50 | 40 | 59.7 |
| TSC1 Mutation | ||||
| yes | 7 | 17.5 | 12 | 17.9 |
| no | 33 | 82.5 | 55 | 82.1 |
| TSC2 Mutation | ||||
| yes | 6 | 15 | 7 | 10.4 |
| no | 34 | 85 | 60 | 89.6 |
In validation cohort, the gene mutation rates of GNAQ, GNA11, BRAF, NRAS, CCND1, HIF1A, AKT1, FOXO1, PIK3R1, PTEN, TSC1, TSC2, FGFR3, KIT and MET were 47.8% (32/67), 22.4% (15/67), 13.4% (9/67), 9% (6/67), 6% (4/67), 26.9% (18/67), 7.5% (5/67), 43.3% (29/67), 40.3% (27/67), 7.5% (5/67), 17.9% (12/67), 10.4% (7/40), 7.5% (5/67), 9% (6/67) and 6% (4/67), respectively.
MAPK pathway gene mutations
Associations between mutations in MAPK pathway genes and patients' clinicopathological features were analysed. We found that HIF1A mutation was significantly correlated with tumour metastasis (Table 2, P<0.05).
Associations between mutation status of genes in the MAPK and PI3K/AKT pathways and OS for uveal melanoma patients in univariate Cox regression analyses.
| Characteristics | HR | 95%CI | P value |
|---|---|---|---|
| Gender (female/male) | 0.85 | 0.47-1.27 | 0.245 |
| Age(>60 years/ 60 years) | 0.61 | 0.19-1.76 | 0.284 |
| Side(left/right) | 0.83 | 0.35-1.79 | 0.392 |
| Thickness (2 mm/>2 mm) | 0.77 | 0.14-1.84 | 0.289 |
| Ciliary body involvement(with/without) | 1.14 | 0.46-2.98 | 0.121 |
| Cell type(spindle/mixed or epithelioid) | 0.91 | 0.07-1.48 | 0.348 |
| Mixed or epithelioid(extraocular extension/none or superficial) | 0.97 | 0.27-1.36 | 0.249 |
| with metastasis(with/without) | 2.34 | 1.13-4.14 | 0.008 |
| GNA11 mutation(yes/no) | 0.87 | 0.69-1.23 | 0.489 |
| GNAQ mutation(yes/no) | 1.13 | 0.31-3.02 | 0.941 |
| BRAF mutation(yes/no) | 1.07 | 0.09-2.38 | 0.125 |
| NRAS mutation(yes/no) | 1.32 | 0.37-3.12 | 0.637 |
| HIF1A mutation(yes/no) | 1.39 | 0.27-2.99 | 0.022 |
| FOXO1 mutation(yes/no) | 1.47 | 0.09-3.87 | 0.013 |
| PIK3R1 mutation(yes/no) | 0.52 | 0.03-1.69 | 0.524 |
| TSC1 mutation(yes/no) | 1.03 | 0.09-2.63 | 0.134 |
| TSC2 mutation(yes/no) | 0.39 | 0.11-2.98 | 0.537 |
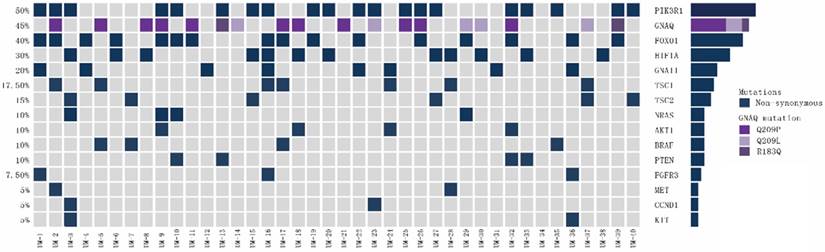
We further analysed the types of gene mutations detected in BRAF, NRAS, GNA11, GNAQ, HIF1A and PTEN in training cohort, as shown in Figure 1. For the 4 patients harbouring BRAF mutations, the mutation, V600E, involved the well-known V600 amino acid residue. All 4 patients with NRAS mutations had hot-spot mutations, including mutations at Q61 (n=3, 75%) and G12 (n=1, 25%). The 8 patients carrying GNA11 mutations shared a Q209 mutation. Among the 18 patients carrying GNAQ mutations, 11 (61%) patients shared the Q209P mutation, 5 (28%) patients shared the Q209L mutation, and 2 (11%) patients shared the R183Q mutation. Among the 12 patients with HIF1A mutations, 5 (42%) patients had the P606S mutation, 3 (25%) patients had the I136T mutation, and 4 (33%) patients had both the D373N and P606S mutations. All 4 patients harbouring PTEN mutations shared the M270I mutation.
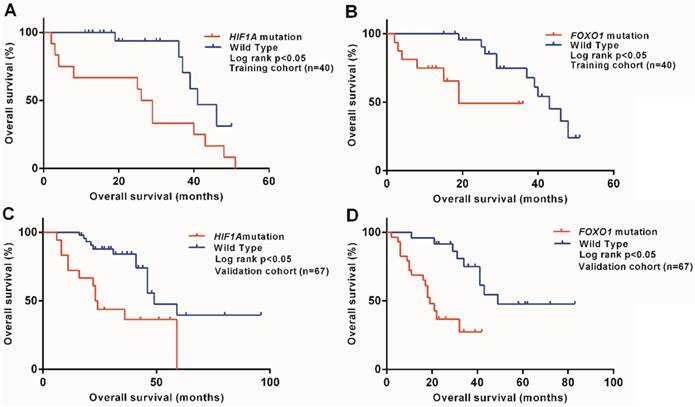
We also investigated associations between MAPK pathway gene mutations and prognosis for UM patients and demonstrated that HIF1A mutation was associated with a worse prognosis in training cohort (P<0.05), as shown in Figure 2A. The result were further verified in validation cohort (P<0.05), as shown in Figure 2C.
In univariate analyses, HIF1A mutation status was significantly associated with OS for UM patients (P<0.05), as shown in Table 2. However, in a multivariate Cox analysis, no gene mutation in the MAPK pathway was significantly correlated with OS.
Mutation patterns for MAPK and PI3K/AKT pathway genes in uveal melanoma patients of training cohort (n=40).
Kaplan-Meier survival curves for OS according to gene mutation status for patients with uveal melanoma. (A) In training cohort, Patients with HIF1A mutation had a shorter median OS than patients with wild-type HIF1A (P<0.05). (B) In training cohort, Patients with FOXO1 mutation had a shorter median OS than patients with wild-type FOXO1 (P<0.05). (C) In validation cohort, Patients with HIF1A mutation had a shorter median OS than patients with wild-type HIF1A (P<0.05). (D) In validation cohort, Patients with FOXO1 mutation had a shorter median OS than patients with wild-type FOXO1 (P<0.05).
PI3K/AKT pathway gene mutations
We evaluated associations between mutations in PI3K/AKT pathway genes and patients' clinicopathological features and found that FOXO1 mutation was associated with metastatic transformation (P<0.001).
Details regarding the types of gene mutations detected in FOXO1, PIK3R1, TSC1, TSC2 and AKT1 in training cohort are shown in Figure 1. Among the 16 patients carrying FOXO1 mutations, 10 (63%) patients had the D82N mutation, and 6 (37%) patients had the A511V mutation. The 20 patients harbouring PIK3R1 mutations had the M56I mutation. The 4 patients with NRAS mutations had hot-spot mutations, including mutations at Q61 (n=3, 75%) and G12 (n=1, 25%). Among the 7 patients with TSC1 mutations, 2 (29%) patients had the E894Q mutation, 2 (29%) patients had the M271T mutation, 2 (29%) patients had the Q603E mutation, and 1 (13%) patient had the M374V mutation. Six patients harboured TSC2 mutations, including 3 (50%) patients with the R1115W mutation and 3 (50%) patients with the V591I mutation. The 4 patients carrying AKT mutations shared the V416A mutation.
We evaluated the effects of individual gene mutations in the PI3K/AKT pathway on UM patient survival and showed that FOXO1 mutation was correlated with a worse prognosis in training cohort (P<0.05), as shown in Figure 2B. The result were further verified in validation cohort (P<0.05), as shown in Figure 2D.
Cox analyses were performed to investigate the prognostic value of gene mutations in the PI3K/AKT pathway. In univariate analyses, FOXO1 mutation status was significantly correlated with worse OS for UM patients (P<0.05), as shown in Table 2. Parameters that were significant in univariate analyses were assessed in a multivariate Cox analysis. In this multivariate analysis, FOXO1 mutation was an independent prognostic factor for OS (P<0.05) that was associated with an increase in the risk ratio by a factor of 1.35, as shown in Table 3.
Association between FOXO1 gene mutation status and OS for uveal melanoma patients in multivariate Cox regression analyses.
| Characteristics | HR | 95%CI | P value |
|---|---|---|---|
| with metastasis(with/without) | 2.83 | 1.03-4.98 | 0.009 |
| FOXO1 mutation(yes/no) | 1.35 | 0.45-3.13 | 0.044 |
Discussion
Comprehensive genetic profiling of UM has been performed in Caucasian populations; [12] however, our research is the investigation involving genetic profiling of UM in a non-Caucasian population. Using NGS, we identified potential targeted therapy parameters and clinical biomarkers of disease metastasis and prognosis for UM.
With respect to genetic mutation profiles, UM is different from CM in that BRAF and NRAS mutations are not predominant. Instead, 83% of UM patients harbour mutations in GNAQ or GNA11.[6] The reported frequencies of GNAQ mutation range from 42-49% in Caucasian populations. [5 8 13] In the cohort examined in this investigation, the gene mutation rate for GNAQ was 46.7%, which was similar to previously reported rates in Caucasian populations. Although the GNA11 mutation rate was reported to be 32.6% in a Caucasian population, [13] this rate was only 21.5% in the current study, a much lower frequency than that observed in the Caucasian cohort. It remains unclear whether this discrepancy in the frequency of GNA11 mutation was attributable to ethnic differences.
UM is extremely deadly due to its high metastatic potential. Several biomarkers for predicting UM metastasis have been studied, such as BAP1, EIF1AX and SF3B1. [14-16] In our cohort, we found that HIF1A and FOXO1 mutations were associated with metastatic transformation of UM (P<0.05 and P<0.001, respectively).
Tumour hypoxia is a prominent characteristic of the tumour microenvironment and could activate a series of genes that mediate metastasis.[17] Several studies have demonstrated the role of hypoxia in the metastatic behaviour of melanoma.[18] HIF1A (hypoxia-inducible factor 1 alpha) is an essential regulator of the hypoxic stress response.[19] Furthermore, the association between HIF1A and melanoma metastasis has already been studied.[20] Hanna found that inactivation of HIF1A led to decreased melanoma metastasis.[20] Chang also demonstrated that HIF1A was an accurate predictor of UM metastasis in a Caucasian population.[21] Consistent with these previous results, we found that HIF1A mutation was significantly associated with tumour metastasis in UM. Cox univariate analyses also indicated that HIF1A mutation was significantly associated with outcome for UM.
There are four FOXO (Forkhead box class O) proteins: FOXO1 (FKHR), FOXO3a (FKHRL1), FOXO4 (AFX) and FOXO6. These proteins play crucial roles in regulating series of tumour-related processes.[21] Nakamura found that FOXO1 is a critical effector of PTEN-mediated tumour suppression.[22] Zhang showed that in prostate cancer, inactivation of FOXO1 could drive the promiscuous expression of Runx2 target genes involved in cell migration and invasion and promote tumour progression.[23] Dong found that in hepatocellular carcinoma, FOXO1 could suppress the ZEB2-induced epithelial-to-mesenchymal transition (EMT) to inhibit tumour metastasis.[24] In our cohort, the FOXO1 mutation was significantly associated with metastatic transformation of UM. The FOXO1 mutation was also found to be an independent prognostic factor for UM in both univariate and multivariate analyses.
The prognosis of patients with UM remains dismal due to a lack of effective treatment. Overall, 80% of UM patients have mutations in the GNAQ and GNA11 genes, which constitutively activate the MAPK and PI3K/AKT pathways; therapies targeting downstream effectors of these pathways, such as MEK, AKT, and protein kinase C (PKC), are currently being investigated. Unfortunately, the results to date have been disappointing. Selumetinib, a highly selective inhibitor of MEK, improves progression-free survival (PFS) in UM, but no OS benefit has been observed. [25]Efforts to optimize the efficacy of targeted therapy remain ongoing.
In conclusion, in this study, we showed for the mutation patterns of UM in a non-Caucasian cohort, with a focus on gene mutations that affect key molecules in the MAPK and PI3K/AKT pathways. These findings illustrate the distinct biological mechanisms underlying UM pathogenesis and metastatic transformation and may provide evidence to improve targeted therapy for UM.
Abbreviations
UM: uveal melanoma; NGS: next-generation sequencing; FFPE: formalin-fixed, paraffin-embedded; OS: overall survival; CM: cutaneous melanoma; MAPK: mitogen-activated protein kinase; PCR: polymerase chain reaction; FOXO: Forkhead box class O; EMT: epithelial-to-mesenchymal transition; PKC: protein kinase C; PFS: progression-free survival.
Acknowledgements
We would like to thank Shanghai Biotechnology Corporation (Shanghai, China) for targeted enrichment, sequencing, and data analysis of NGS. We would like to thank American Journal Experts (AJE) for English language editing.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81672696; 81772912), Beijing Municipal Administration of Hospitals Clinical medicine Development of special funding support (ZYLX201603), Fostering Young Scholars of Peking University Health Science Center and Beijing Baiqianwan Talents Project.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Shields CL, Kaliki S, Furuta M. et al. Clinical spectrum and prognosis of uveal melanoma based on age at presentation in 8,033 cases. Retina (Philadelphia, Pa.). 2012;32:1363-72
2. Harbour JW, Chao DL. A molecular revolution in uveal melanoma: implications for patient care and targeted therapy. Ophthalmology. 2014;121:1281-8
3. Rietschel P, Panageas KS, Hanlon C. et al. Variates of survival in metastatic uveal melanoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005;23:8076-80
4. Iams WT, Sosman JA, Chandra S. Novel Targeted Therapies for Metastatic Melanoma. Cancer journal (Sudbury, Mass.). 2017;23:54-58
5. Van Raamsdonk CD, Bezrookove V, Green G. et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599-602
6. Van Raamsdonk CD, Griewank KG, Crosby MB. et al. Mutations in GNA11 in uveal melanoma. The New England journal of medicine. 2010;363:2191-9
7. Harbour JW, Onken MD, Roberson ED. et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science (New York, N.Y.). 2010;330:1410-3
8. Onken MD, Worley LA, Long MD. et al. Oncogenic mutations in GNAQ occur early in uveal melanoma. Investigative ophthalmology & visual science. 2008;49:5230-4
9. Curtin JA, Fridlyand J, Kageshita T. et al. Distinct sets of genetic alterations in melanoma. The New England journal of medicine. 2005;353:2135-47
10. Chi Z, Li S, Sheng X. et al. Clinical presentation, histology, and prognoses of malignant melanoma in ethnic Chinese: a study of 522 consecutive cases. BMC cancer. 2011;11:85
11. Bai X, Kong Y, Chi Z. et al. MAPK Pathway and TERT Promoter Gene Mutation Pattern and Its Prognostic Value in Melanoma Patients: A Retrospective Study of 2,793 Cases. Clinical cancer research: an official journal of the American Association for Cancer Research. 2017;23:6120-27
12. Royer-Bertrand B, Torsello M, Rimoldi D. et al. Comprehensive Genetic Landscape of Uveal Melanoma by Whole-Genome Sequencing. American journal of human genetics. 2016;99:1190-98
13. Dono M, Angelini G, Cecconi M. et al. Mutation frequencies of GNAQ, GNA11, BAP1, SF3B1, EIF1AX and TERT in uveal melanoma: detection of an activating mutation in the TERT gene promoter in a single case of uveal melanoma. British journal of cancer. 2014;110:1058-65
14. Kalirai H, Dodson A, Faqir S, Damato BE, Coupland SE. Lack of BAP1 protein expression in uveal melanoma is associated with increased metastatic risk and has utility in routine prognostic testing. British journal of cancer. 2014;111:1373-80
15. Ewens KG, Kanetsky PA, Richards-Yutz J. et al. Chromosome 3 status combined with BAP1 and EIF1AX mutation profiles are associated with metastasis in uveal melanoma. Investigative ophthalmology & visual science. 2014;55:5160-7
16. Yavuzyigitoglu S, Koopmans AE, Verdijk RM. et al. Uveal Melanomas with SF3B1 Mutations: A Distinct Subclass Associated with Late-Onset Metastases. Ophthalmology. 2016;123:1118-28
17. Semenza GL. Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends in pharmacological sciences. 2012;33:207-14
18. Cheli Y, Giuliano S, Fenouille N. et al. Hypoxia and MITF control metastatic behaviour in mouse and human melanoma cells. Oncogene. 2012;31:2461-70
19. Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Molecular cell. 2010;40:294-309
20. Hanna SC, Krishnan B, Bailey ST. et al. HIF1alpha and HIF2alpha independently activate SRC to promote melanoma metastases. The Journal of clinical investigation. 2013;123:2078-93
21. Chang SH, Worley LA, Onken MD, Harbour JW. Prognostic biomarkers in uveal melanoma: evidence for a stem cell-like phenotype associated with metastasis. Melanoma research. 2008;18:191-200
22. Nakamura N, Ramaswamy S, Vazquez F, Signoretti S, Loda M, Sellers WR. Forkhead transcription factors are critical effectors of cell death and cell cycle arrest downstream of PTEN. Molecular and cellular biology. 2000;20:8969-82
23. Zhang H, Pan Y, Zheng L. et al. FOXO1 inhibits Runx2 transcriptional activity and prostate cancer cell migration and invasion. Cancer research. 2011;71:3257-67
24. Dong T, Zhang Y, Chen Y. et al. FOXO1 inhibits the invasion and metastasis of hepatocellular carcinoma by reversing ZEB2-induced epithelial-mesenchymal transition. Oncotarget. 2017;8:1703-13
25. Carvajal RD, Sosman JA, Quevedo JF. et al. Effect of selumetinib vs chemotherapy on progression-free survival in uveal melanoma: a randomized clinical trial. Jama. 2014;311:2397-405
Author contact
![]() Corresponding authors: Jun Guo. E-mail: guoj307com; Telephone: 8601088196348, Yan Kong. E-mail: k-yan08com; Telephone: 8601088196707
Corresponding authors: Jun Guo. E-mail: guoj307com; Telephone: 8601088196348, Yan Kong. E-mail: k-yan08com; Telephone: 8601088196707

 Global reach, higher impact
Global reach, higher impact