3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(3):611-619. doi:10.7150/jca.28780 This issue Cite
Research Paper
Long Noncoding RNAs Serve as Potential Diagnostic Biomarkers for Colorectal Cancer
1. Department of Oncology, The First Affiliated Hospital, Yijishan Hospital of Wannan Medical College, Wuhu 241000, China
2. Department of Gastrointestinal Surgery, The First Affiliated Hospital, Yijishan Hospital of Wannan Medical College, Wuhu 241000, China
3. Hepatobiliary Center, The First Affiliated Hospital of Nanjing Medical University, Key Laboratory of Liver Transplantation, Chinese Academy of Medical Sciences, Nanjing 210029, China
* These authors contributed equally to this work.
Received 2018-7-26; Accepted 2018-12-8; Published 2019-1-1
Abstract
Background: Mounting evidence has indicated that long noncoding RNAs (lncRNAs) are promising candidates for tumor diagnosis and prognosis. Nonetheless, the significance of lncRNAs in colorectal cancer (CRC) diagnosis remains to be clarified. Here, we performed a comprehensive meta-analysis to evaluate the utility of lncRNAs as diagnostic indicators for CRC.
Materials and Methods: Pertinent studies were searched using PubMed, PMC, Web of Science, Cochrane, and EMBASE database up to September 2018. Study quality was assessed with the Quality Assessment for Studies of Diagnostic Accuracy-2. Subgroup analyses by sample size and publication year were conducted. Threshold effect and meta-regression were performed to find the origin of heterogeneity. Statistical analyses were conducted using Stata and Meta-Disc.
Results: A total of 19 studies with 3,114 individuals were enrolled in the current analysis. The overall sensitivity and specificity of lncRNAs in the diagnosis of CRC were 0.83 [95% confidence interval (CI): 0.76-0.87] and 0.84 (95% CI: 0.77-0.89), respectively. The pooled positive likelihood ratio was 5.11 (95% CI: 3.57-7.31), and the pooled negative likelihood ratio was 0.21 (95% CI: 0.15-0.28). The overall area under the curve was 0.90 (95% CI: 0.87-0.92), with a diagnostic odds ratio of 24.57 (95% CI: 14.67-41.17).
Conclusions: The accuracy of lncRNAs for CRC diagnosis is high, and lncRNAs could be functioned as promising candidates for CRC diagnosis.
Keywords: long noncoding RNA, colorectal cancer, meta-analysis, diagnosis, biomarker
Introduction
Colorectal cancer (CRC) is the third most common malignancy with the occurrence of 1.3 million new cases and 0.7 million cancer-related deaths per year around the world [1]. In China, the mortality of CRC ranks fourth among various types of cancers [2]. Although substantial advances in multidisciplinary treatment for CRC have contributed to great improvements in survival outcome, early diagnosis of CRC is still a major issue to be solved [3]. Colonoscopy examination can provide a high diagnostic accuracy, but its invasiveness makes this procedure intolerant to many patients, limiting its use for large-scale screening. Fecal occult blood testing is a widely used test, whereas it has relatively unsatisfactory accuracy. The measurement of blood biomarkers such as carcinoembryonic antigen (CEA) has a limited utility due to low sensitivity for CRC, particularly in early stage of cancer [4]. An ideal approach for CRC screening is supposed to possess a very high degree of sensitivity and specificity for early cancer detection. Over the past decade, substantial endeavors were made by researchers to seek for more effective and reliable screening tests based on a systems biology method, using easily accessible human specimens, such as serum, urine, and feces. Therefore, it is essential to determine novel biomarkers for early diagnosis and targeted therapy for CRC patients.
Long non-coding RNAs (lncRNAs) participate in gene expression regulation at different levels, exerting a key role in various biological processes [5, 6]. Dysregulated lncRNA expression has been demonstrated in malignant transformation and tumor progression [7, 8]. A number of lncRNAs have been associated with clinical diagnosis and survival outcomes in cancer patients, and can be used as a predictor for tumor prognosis [9-13]. Recently, lncRNAs have been considered as novel markers for cancer diagnosis, but with varying diagnostic accuracy [14].
LncRNAs have been suggested as a promising marker for CRC diagnosis. Ye et al. examined the diagnostic efficiency of lnc-GNAT1-1 in CRC, and revealed that the area under the curve (AUC) of the receiver operator characteristic (ROC) curve is 0.72, indicating that serum lnc-GNAT1-1 level exhibits a moderate to strong diagnostic efficiency for CRC patients [15]. Based on a study performed by Svoboda et al., HOTAIR is dramatically up-regulated in plasma of CRC patients. ROC analysis presented an AUC of 0.87, with 67% sensitivity and 92.5% specificity of CRC detection, respectively [16]. With increasing evidence suggesting the diagnostic use of various lncRNAs in CRC, the utility of lncRNAs in CRC diagnosis, however, has not been comprehensively investigated yet.
The goal of our study is to determine the diagnostic significance of lncRNAs in CRC and to explore the potential of lncRNAs as biomarkers for CRC diagnosis. We comprehensively identified and enrolled pertinent studies and evaluated the overall value of these lncRNAs for CRC diagnosis.
Materials and Methods
Literature search
The current meta-analysis conformed with the protocol of the preferred reporting items for systematic reviews and meta-analyses (PRISMA) guidelines [17]. PubMed, PMC, Web of Science, Cochrane Library, and EMBASE were searched up to September 2018 for pertinent articles using the keywords as follows: (lncRNA or long noncoding RNA) and (CRC or colorectal cancer) and (sensitivity or specificity or diagnosis or receiver operating characteristic curve). The titles and abstracts were screened, and the relevant full-text manuscripts were acquired for perusal.
Inclusion and exclusion criteria
The inclusion criteria were as follows: (a) studies assessing the diagnostic value of lncRNAs in CRC; (b) studies should contain true negative, true positive, false negative, and false positive values to reconstruct the 2 × 2 contingency tables; and (c) CRC diagnosis was verified by two independent pathologists. The exclusion criteria were: (a) letters, case reports, and reviews; and (b) duplicate studies. The literature review process was accomplished by C.J. and Z.X.L. Group discussion was carried out to settle any disagreement and achieve a consensus.
Quality assessment
C.J. and C.Z.Q. independently evaluated the quality of the enrolled studies according to the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) [18]. Study quality was examined based on four dimensions: selection of subjects, index test, reference standard, and flow and timing, with each item scored as yes, no, or unclear.
Data extraction and analysis
The following data were extracted from the included studies: the first author, lncRNA expression pattern, year of publication, sample source, diagnostic indexes (AUC, sensitivity, and specificity), and sample size. True positive, false positive, false negative, and true negative values were directly acquired from the manuscript or calculated from the reported data in the text.
Meta-DiSc 1.4 (http://www.hrc.es/investigacion/metadisc_en.htm; Universidad Complutense, Madrid, Spain) and STATA 12.0 (Stata Corporation, College Station, TX, USA) were used for statistical analysis. We calculated sensitivity, specificity, positive likelihood ratio (LR), and negative LR of each enrolled study. An overall assessment of the diagnostic value of the lncRNA reported was acquired using diagnostic odds ratio (DOR) as well as AUC of the summary ROC (sROC) curve. The threshold effect was examined by Spearman's correlation analysis. The heterogeneity was evaluated with χ2 test and I2 statistics. Heterogeneity was concluded at P < 0.10 or I2 > 50%. Further subgroup analysis was carried out based on the size and source of sample. Meta-regression was also employed to elucidate the potential origins of heterogeneity. Moreover, we used Fagan's nomogram to evaluate the post-test probability. Deeks' funnel plot was employed to examine publication bias. A significant result was determined as P < 0.05.
Results
Study characteristics
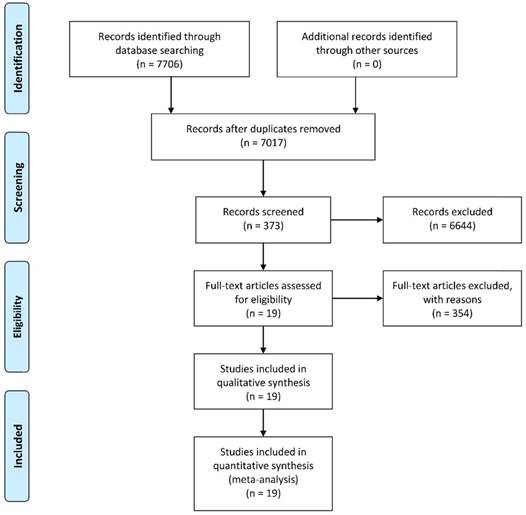
A total of 7,706 potentially relevant records were identified using the PubMed, PMC, Web of Science, and the Cochrane Library database according to the predetermined keywords. After excluding 689 duplicates, we screened the titles and abstracts of the remaining 7,017 studies. Subsequently, 373 articles were included for full-text screening and data extraction after exclusion of 6,644 unrelated studies, reviews, or letters. We further removed 354 articles due to inadequate data to construct 2 × 2 tables, and the remaining 19 studies with a total of 3,114 subjects were enrolled in the current meta-analysis [15, 16, 19-35]. A flowchart illustrating the process of study selection is shown in Figure 1.
Among the included studies, the diagnostic value of 18 different lncRNAs and a 4-lncRNA panel was assessed. Most of the examined lncRNAs were increased in CRC. The sample source of thirteen studies was blood specimen, and eight studies used tissue samples to evaluate the diagnostic significance of lncRNAs in CRC. The sample size ranged between 38 and 331, with 200 as the median value. The study characteristics are listed in Table 1.
Assessment of study quality
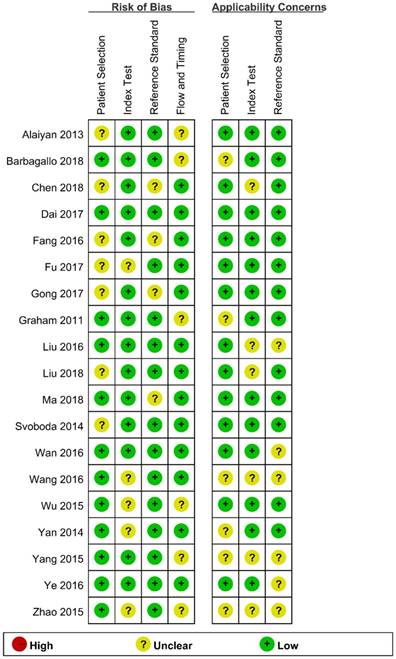
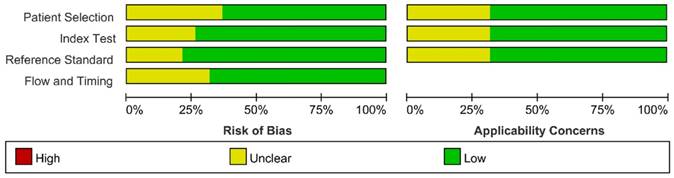
We assessed the study quality using QUADAS-2, and the results are indicated in Figure 2 and 3. The study quality of the enrolled manuscripts were mostly moderate and high.
Pooled diagnostic accuracy indexes
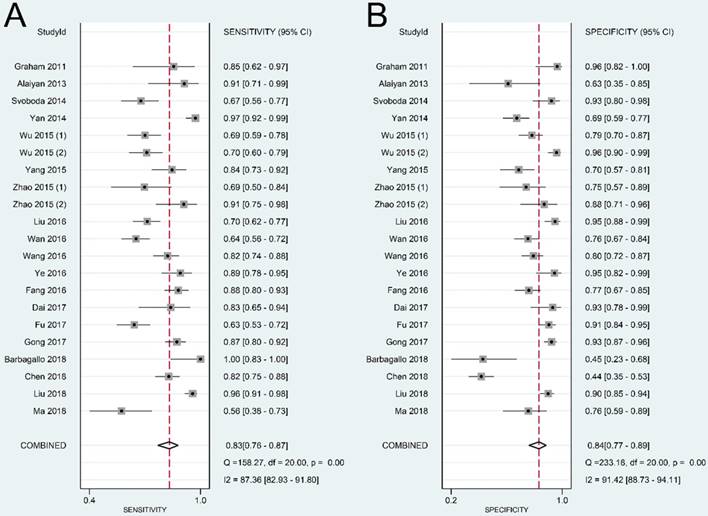
The summary sensitivity, specificity, positive LR, negative LR, DOR, and sROC curve were 0.83 [95% confidence interval (CI): 0.76-0.87], 0.84 (95% CI: 0.77-0.89), 5.11 (95% CI: 3.57-7.31), 0.21 (95% CI: 0.15-0.28), 24.57 (95% CI: 14.67-41.17), and 0.90 (95% CI: 0.87-0.92), respectively (Figure 4-6). The I2 value of DOR was used to detect the heterogeneity across the studies (Table 2). In order to locate the source of heterogeneity, we conducted subsequent analysis on threshold effect, stratified analysis, and meta-regression to elucidate the potential source of the heterogeneity.
Source of heterogeneity
To determine the origin of the heterogeneity, we first calculated Spearman's correlation coefficient. Data showed that the Spearman correlation coefficient was 0.187 with a P value of 0.417 across the included studies, suggesting that the threshold effect was not the primary source of the heterogeneity.
A flow diagram demonstrating the study selection process.
Characteristics of the studies included in the meta-analysis.
| Study | lncRNA | Expression level | Specimen source | AUC | Sensitivity | Specificity | TP | FP | FN | TN | No. of patients | No. of controls | Sample size |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Graham 2011 | CRNDE-b | U | Tissue | 0.939 | 0.85 | 0.96 | 17 | 1 | 3 | 27 | 20 | 28 | 48 |
| Alaiyan 2013 | CCAT1 | U | Tissue | NA | 0.91 | 0.63 | 20 | 6 | 2 | 10 | 22 | 16 | 38 |
| Svoboda 2014 | HOTAIR | U | Plasma | 0.87 | 0.670 | 0.925 | 56 | 3 | 28 | 37 | 84 | 40 | 124 |
| Yan 2014 | ncRuPAR | D | Tissue | 0.81 | 0.9714 | 0.6587 | 102 | 33 | 3 | 72 | 105 | 105 | 210 |
| Wu 2015 (1) | NEAT1_v1 | U | Whole blood | 0.787 | 0.69 | 0.79 | 69 | 21 | 31 | 79 | 100 | 100 | 200 |
| Wu 2015 (2) | NEAT1_v2 | U | Whole blood | 0.871 | 0.70 | 0.96 | 70 | 4 | 30 | 96 | 100 | 100 | 200 |
| Yang 2015 | PRNCR1 | U | Tissue | 0.799 | 0.841 | 0.698 | 53 | 19 | 10 | 44 | 63 | 63 | 126 |
| Zhao 2015 (1) | HOTAIR | U | Plasma | 0.777 | 0.688 | 0.750 | 22 | 8 | 10 | 24 | 32 | 32 | 64 |
| Zhao 2015 (2) | CCAT1 | U | Plasma | 0.836 | 0.906 | 0.875 | 29 | 4 | 3 | 28 | 32 | 32 | 64 |
| Liu 2016 | CRNDE-h | U | Serum | 0.892 | 0.703 | 0.950 | 104 | 4 | 44 | 76 | 148 | 80 | 228 |
| Wan 2016 | HOTAIRMI | D | Plasma | 0.780 | 0.640 | 0.765 | 96 | 24 | 54 | 77 | 150 | 101 | 251 |
| Wang 2016 | Four lncRNA panel | BANCR: U NR_026817: D NR_029373: D NR_034119: D | Serum | 0.881 | 0.82 | 0.80 | 98 | 24 | 22 | 96 | 120 | 120 | 240 |
| Ye 2016 | lnc-GNATH | D | Plasma | 0.720 | 0.8871 | 0.9459 | 55 | 2 | 7 | 35 | 62 | 37 | 99 |
| Fang 2016 | ZFAS1 | U | Plasma | 0.88 | 0.9238 | 0.7684 | 92 | 22 | 13 | 73 | 105 | 95 | 200 |
| Dai 2017 | BLACAT1 | U | Serum | 0.858 | 0.833 | 0.767 | 25 | 2 | 5 | 28 | 30 | 30 | 60 |
| Fu 2017 | ZEB1-AS1 | U | Tissue | 0.846 | 0.630 | 0.907 | 68 | 10 | 40 | 98 | 108 | 108 | 216 |
| Gong 2017 | HIF1A-AS1 | U | Serum | 0.960 | 0.868 | 0.925 | 131 | 12 | 20 | 148 | 151 | 160 | 311 |
| Chen 2018 | LINC00472 | D | Tissue | 0.680 | 0.823 | 0.439 | 107 | 73 | 23 | 57 | 130 | 130 | 260 |
| Liu 2018 | GAS5 | U | Tissue | 0.791 | 0.954 | 0.899 | 151 | 17 | 7 | 156 | 158 | 173 | 331 |
| Barbagallo 2018 | UCA1 | D | Serum | 0.719 | 1.00 | 0.43 | 20 | 11 | 0 | 9 | 20 | 20 | 40 |
| Ma 2018 | RP1-85F18.6 | U | Tissue | 0.651 | 0.559 | 0.765 | 19 | 8 | 15 | 26 | 34 | 34 | 68 |
lncRNA: long noncoding RNA; AUC: area under the curve; TP: true positive; FP: false positive; FN: false negative; TN: true negative; D: down-regulated; U: up-regulated; NA: not available.
Methodological quality graph.
We performed subgroup analyses based on sample size and source of specimen. The DOR value was higher in studies with sample size greater than 200 (DOR = 26.00, 95% CI: 10.57-63.97) compared to that in studies with sample size fewer than 200 (DOR = 22.71, 95% CI: 12.45-41.41). Studies using blood sample exhibited a higher DOR (25.26, 95% CI: 14.34-44.50) compared with studies using tissue as specimen source (DOR = 22.42, 95% CI: 8.20-61.28) (Table 2).
The subsequent meta-regression analysis showed no statistically significant correlation between sample size (P = 0.9582), source of specimen (P = 0.6203) and DOR in this analysis.
Publication bias
We adopted Deeks' funnel plot to evaluate the publication bias of the current meta-analysis (Figure 7). A P value of 0.828 showed the absence of statistically significant publication bias.
Clinical application of lncRNAs in CRC diagnosis
Fagan's nomogram is a useful tool to evaluate the post-test probability, and was assessed in our study. As indicated in Figure 8, we set the pre-test probability at 20% as previously reported [36]. A 56% post-test probability with a positive LR of 5 and a 5% post-test probability with a negative LR of 0.21 were achieved.
Methodological quality summary.
Forest plot of sensitivity (A) and specificity (B) of lncRNAs in colorectal cancer.
Assessment of diagnostic accuracy and heterogeneity in subgroup analysis.
| Subgroups | No. of studies | Pooled sensitivity (95% CI) | I2 (%) sensitivity | Pooled specificity (95% CI) | I2 (%) specificity | Pooled positive LR (95% CI) | I2 (%) positive LR | Pooled negative LR (95% CI) | I2 (%) negative LR | Pooled DOR | I2 (%) DOR | AUC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All studies | 21 | 0.83 (0.76-0.87) | 87.36 | 0.84 (0.77-0.89) | 91.42 | 5.11(3.57-7.31) | 89.72 | 0.21 (0.15-0.28) | 85.82 | 24.57 (14.67-41.17) | 100.00 | 0.90 (0.87-0.92) |
| Sample size | ||||||||||||
| ≤200 | 13 | 0.81 (0.74-0.87) | 76.12 | 0.84 (0.75-0.90) | 81.28 | 5.06 (3.24-7.93) | 70.55 | 0.22 (0.16-0.31) | 72.72 | 22.71 (12.45-41.41) | 99.62 | 0.89 (0.86-0.92) |
| >200 | 8 | 0.84 (0.72-0.91) | 93.49 | 0.83 (0.71-0.91) | 95.72 | 5.05 (2.84-8.98) | 94.69 | 0.19 (0.11-0.35) | 92.82 | 26.00 (10.57-63.97) | 100.00 | 0.90 (0.88-0.93) |
| Specimen source | ||||||||||||
| Tissue | 8 | 0.86 (0.74-0.93) | 92.51 | 0.78 (0.64-0.88) | 93.95 | 4.00 (2.30-6.95) | 92.12 | 0.18 (0.09-0.35) | 92.40 | 22.42 (8.20-61.28) | 100.00 | 0.89 (0.86-0.92) |
| Blood | 13 | 0.80 (0.73-0.85) | 81.05 | 0.87 (0.79-0.91) | 84.14 | 5.91 (3.82-9.14) | 78.40 | 0.23 (0.17-0.31) | 80.33 | 25.26 (14.34-44.50) | 100.00 | 0.89 (0.86-0.92) |
CI: confidence interval; LR: likelihood ratio; DOR: diagnostic odds ratio; AUC: area under the curve.
Forest plot of positive likelihood ratio (A) and negative likelihood ratio (B) of lncRNAs in colorectal cancer.
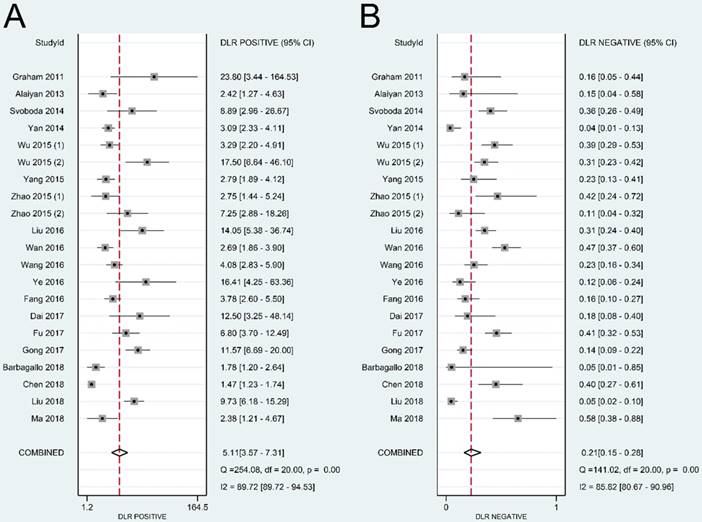
Overall performance of lncRNAs in diagnosis of colorectal cancer. (A) Diagnostic odds ratio of lncRNAs in diagnosis of colorectal cancer. (B) Summary receiver operator characteristic curve of lncRNAs in diagnosis of colorectal cancer.
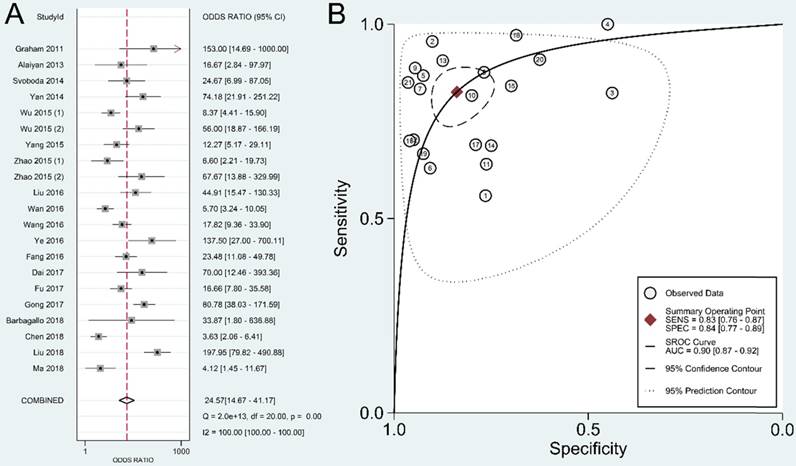
Deeks' funnel plot evaluating the potential publication bias of the included studies.
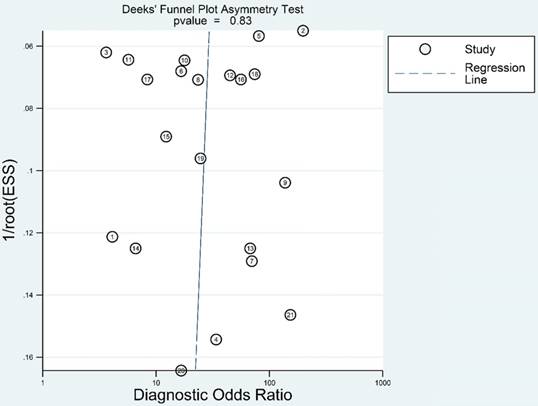
Fagan's nomogram evaluating the overall value of lncRNAs for diagnosis of colorectal cancer.
Discussion
Mounting evidence has implicated that lncRNAs play an indispensable role in the carcinogenesis, proliferation, and metastasis of various tumors [37-40]. LncRNAs have been found to be involved in tumorigenesis and associated with clinical outcomes of CRC [41]. For example, Ma et al. revealed that lncRNA SNHG17 is highly over-expressed in CRC tissues and associated with dismal prognosis, thereby potentially representing an unfavorable prognostic factor for CRC patients [42]. LncRNA CCAT2 is upregulated in microsatellite-stable CRC, and promotes cancer proliferation and metastasis. Increased lncRNA CCAT2 expression could be a potential diagnostic biomarker for CRC and an independent predictor of prognosis in patients with CRC [43, 44]. Besides, it has been reported that lncRNA is an independent predictive indicator for colon cancer recurrence [45]. Given their specific expression patterns in CRC, lncRNAs are potential candidates for detecting precancerous lesions and diagnosing tumors. Therefore, lncRNAs seem to possess great advantages as tumor diagnostic biomarkers.
The present study is the first meta-analysis that evaluated the significance of lncRNAs in CRC diagnosis. According to our data, the pooled sensitivity and specificity were 0.83 (95% CI: 0.76-0.87) and 0.84 (95% CI: 0.77-0.89), respectively. A high DOR value shows an excellent discriminatory performance [46]. A summary DOR of 24.57 indicated the diagnostic value of lncRNAs for CRC patients. Reportedly, an AUC with a value greater than 0.75 is acceptable for diagnostic test [47]. Our data revealed that lncRNAs with an AUC of 0.90 (95% CI: 0.87-0.92) have good diagnostic accuracy in CRC.
Currently, there are still no diagnostic signatures for CRC with satisfactory specificity and sensitivity. CEA is a classic colon cancer screening biomarker. As reported, the pooled sensitivity of CEA for CRC is 0.46, and the specificity is 0.89[48]. The specificity of CEA is high, but the sensitivity is unsatisfactory. LncRNAs, with an overall sensitivity of 0.83, could be served as a potential candidate for diagnosing CRC patients. AUC assesses the overall performance of diagnostic markers, and has been acknowledged as the most important parameter. According to our results, the AUC of lncRNA in CRC diagnosis was 0.90, which was much higher than the reported AUC of CEA in CRC (0.79) [49]. The comparation between CEA and lncRNAs showed that lncRNAs have an edge over CEA in CRC diagnosis.
According to the abovementioned results, it seems that we may have proposed an effective CRC screening approach using lncRNAs that could be used for clinical diagnosis, but we usually do not detect multiple lncRNAs at the same time in practical application. HIF1A-AS1, a lncRNA with the highest AUC (0.96) in CRC diagnosis, has the most potential diagnostic value and may be the most promising subject for population screening. In addition, we could combine two or more lncRNAs, thereby greatly increasing their application potential.
Significant heterogeneity was detected in the meta-analysis. A Spearman correlation coefficient of 0.187 (P = 0.417) showed that the threshold effect was not the main cause of heterogeneity. Subgroup analysis and meta-regression showed that size and source of the included individuals did not cause the heterogeneity. Due to limited data, we did not include other important potential cofounding variates, such as gender, age, study design, and socioeconomic conditions.
Despite our efforts to conduct a comprehensive meta-analysis, several limitations should be addressed. First of all, it is crucial for diagnostic biomarkers that they could distinguish CRC patients from not only healthy individuals but also patients with other digestive system diseases, especially with similar symptoms. However, the control groups of most included studies were healthy people, which might lead to an overestimate of the diagnostic value. Second, a considerate amount of across-study heterogeneity was detected. Data from subgroup analyses and meta-regression could not fully explain the detected heterogeneity. Due to limited clinical and demographical data, we could not further elucidate potential sources of heterogeneity. Third, the number of lncRNAs that can be effectively used is still to be determined. Fourth, a large proportion of the included subjects were from Asia. It remains unclear whether these findings could be applied to other areas. Thus, more studies are warranted to further clarify the diagnostic value of lncRNAs for CRC patients.
To conclude, the present meta-analysis suggested that lncRNAs could distinguish CRC patients from the control group. The overall sensitivity and specificity of lncRNAs in CRC diagnosis were 0.83 and 0.84, respectively. The overall AUC was 0.90, with a pooled DOR of 24.57. LncRNAs could be used as potential candidates for CRC diagnosis.
Abbreviations
CRC: colorectal cancer; lncRNA: long noncoding RNA; QUADAS-2: Quality Assessment for Studies of Diagnostic Accuracy-2; CI: confidence interval; CEA: carcinoembryonic antigen; CA19-9: carbohydrate antigen 19-9; AUC: area under the curve; ROC: receiver operator characteristic; PRISMA: preferred reporting items for systematic reviews and meta-analyses; DOR: diagnostic odds ratio; LR: likelihood ratio.
Acknowledgements
This study was funded by the Natural Science Research of Anhui Education Department Key Project (No. KJ2018A0246), the Key Scientific Research Project of Wannan Medical College (No. WK2018ZF01), and the Natural Science Foundation of Anhui Province (No. 1708085MH202).
Ethics approval and consent to participate
This work did not require any written patient consent. The ethics committee of The First Affiliated Hospital of Wannan Medical College approved this work.
Authors' contributions
Study concept, study design, and manuscript drafting: Cai J and Zuo XL. Data acquisition: Zhang Y and Wang JF. Data interpretation: Ye XB and Wang JG. Statistical analysis: Zuo XL and Chen ZQ. Manuscript editing and critical revision: Chen ZQ and Zhao WY.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
2. Chen W, Zheng R, Zeng H. et al. Annual report on status of cancer in China, 2011. Chin J Cancer Res. 2015;27:2-12
3. Sehdev A, Sherer EA, Hui SL. et al. Patterns of computed tomography surveillance in survivors of colorectal cancer at Veterans Health Administration facilities. Cancer. 2017;123:2338-51
4. Yang L, Wang J, Li J. et al. Identification of serum biomarkers for gastric cancer diagnosis using a human proteome microarray. Mol Cell Proteomics. 2016;15:614-23
5. Lekka E, Hall J. Noncoding RNAs in disease. FEBS Lett. 2018;592(17):2884-900
6. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393-407
7. Zhang L, Peng D, Sood AK. et al. Shedding light on the dark cancer genomes: long noncoding RNAs as novel biomarkers and potential therapeutic targets for cancer. Mol Cancer Ther. 2018;17:1816-23
8. Lin C, Yang L. Long noncoding RNA in cancer: wiring signaling circuitry. Trends Cell Biol. 2018;28:287-301
9. Chen C, He W, Huang J. et al. LNMAT1 promotes lymphatic metastasis of bladder cancer via CCL2 dependent macrophage recruitment. Nat Commun. 2018;9:3826
10. Wang X, Mo FM, Bo H. et al. Upregulated expression of long non-coding RNA, LINC00460, suppresses proliferation of colorectal cancer. J Cancer. 2018;9:2834-43
11. Cai J, Zuo X, Chen Z. et al. Prognostic value and clinical significance of long noncoding RNA CASC2 in human malignancies: a meta-analysis. Cancer Manag Res. 2018;10:1403-12
12. Zuo X, Cai J, Chen Z. et al. The utility of long non-coding RNA ZEB1-AS1 as a prognostic biomarker in human solid tumors: A meta-analysis. Clin Chim Acta. 2018;485:14-20
13. Zhou Y, Chen S, Cheng S. et al. The prognostic value of high LncRNA AFAP1-AS1 expression in various cancers: A systematic review and meta-analysis containing 21 studies. Clin Chim Acta. 2018;481:147-53
14. Wang J, Du S, Wang J. et al. The prognostic value of abnormally expressed lncRNAs in colorectal cancer: A meta-analysis. PLoS One. 2017;12:e0179670
15. Ye C, Shen Z, Bo W. et al. A novel long non-coding RNA lnc-GNAT1-1 is low expressed in colorectal cancer and acts as a tumor suppressor through regulating RKIP-NF-κB-Snail circuit. J Exp Clin Cancer Res. 2016;35:187
16. Svoboda M, Slyskova J, Schneiderova M. et al. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis. 2014;35:1510
17. Moher D, Shamseer L, Clarke M. et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA Statement. Syst Rev. 2015;4:1
18. Whiting PF, Rutjes AW, Westwood ME. et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529-36
19. Alaiyan B, Ilyayev N, Stojadinovic A. et al. Differential expression of colon cancer associated transcript1 (CCAT1) along the colonic adenoma-carcinoma sequence. BMC Cancer. 2013;13:196
20. Dai M, Chen X, Mo S. et al. Meta-signature LncRNAs serve as novel biomarkers for colorectal cancer: integrated bioinformatics analysis, experimental validation and diagnostic evaluation. Sci Rep. 2017;7:46572
21. Gong W, Tian M, Qiu H. et al. Elevated serum level of lncRNA-HIF1A-AS1 as a novel diagnostic predictor for worse prognosis in colorectal carcinoma. Cancer Biomark. 2017;20:417-24
22. Graham LD, Pedersen SK, Brown GS. et al. Colorectal Neoplasia Differentially Expressed (CRNDE), a novel gene with elevated expression in colorectal adenomas and adenocarcinomas. Genes Cancer. 2011;2:829-40
23. Liu T, Zhang X, Gao S. et al. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget. 2016;7:85551
24. Wan L, Kong J, Tang J. et al. HOTAIRM1 as a potential biomarker for diagnosis of colorectal cancer functions the role in the tumour suppressor. J Cell Mol Med. 2016;20:2036-44
25. Wang R, Du L, Yang X. et al. Identification of long noncoding RNAs as potential novel diagnosis and prognosis biomarkers in colorectal cancer. J Cancer Res Clin Oncol. 2016;142:2291-301
26. Wu Y, Li Y, Jiang Z. et al. Nuclear-enriched abundant transcript 1 as a diagnostic and prognostic biomarker in colorectal cancer. Mol Cancer. 2015;14:191
27. Yan B, Gu W, Yang Z. et al. Downregulation of a long noncoding RNA-ncRuPAR contributes to tumor inhibition in colorectal cancer. Tumor Biol. 2014;35:11329
28. Yang L, Qiu M, Xu Y. et al. Upregulation of long non-coding RNA PRNCR1 in colorectal cancer promotes cell proliferation and cell cycle progression. Oncol Rep. 2016;35:318
29. Zhao W, Song M, Zhang J. et al. Combined identification of long non-coding RNA CCAT1 and HOTAIR in serum as an effective screening for colorectal carcinoma. Int J Clin Exp Pathol. 2015;8:14131-40
30. Barbagallo C1, Brex D1, Caponnetto A. et al. LncRNA UCA1, upregulated in CRC biopsies and downregulated in serum exosomes, controls mRNA expression by RNA-RNA interactions. Mol Ther Nucleic Acids. 2018;12:229-41
31. Ma Y, Chen Y, Lin C. et al. Biological functions and clinical significance of the newly identified long non-coding RNA RP1-85F18.6 in colorectal cancer. Oncol Rep. 2018;40:2648-58
32. Chen L, Zhang W, Li DY. et al. Regulatory network analysis of LINC00472, a long noncoding RNA downregulated by DNA hypermethylation in colorectal cancer. Clin Genet. 2018;93:1189-98
33. Fang C, Zan J, Yue B. et al. Long non-coding ribonucleic acid zinc finger antisense 1 promotes the progression of colonic cancer by modulating ZEB1 expression. J Gastroenterol Hepatol. 2017;32:1204-11
34. Fu J, Cui Y. Long noncoding RNA ZEB1-AS1 expression predicts progression and poor prognosis of colorectal cancer. Int J Biol Markers. 2017;32:e428-e433
35. Liu L, Meng T, Yang XH. et al. Prognostic and predictive value of long non-coding RNA GAS5 and mircoRNA-221 in colorectal cancer and their effects on colorectal cancer cell proliferation, migration and invasion. Cancer Biomark. 2018;22:283-99
36. Chen Z, Zhang L, Han G. et al. A meta-analysis of the diagnostic accuracy of circular RNAs in digestive system malignancy. Cell Physiol Biochem. 2018;45:962-72
37. Liang WC, Ren JL, Wong CW. et al. LncRNA-NEF antagonized epithelial to mesenchymal transition and cancer metastasis via cis-regulating FOXA2 and inactivating Wnt/beta-catenin signaling. Oncogene. 2018;37:1445-56
38. Wang ZY, Hu M, Dai MH. et al. Upregulation of the long non-coding RNA AFAP1-AS1 affects the proliferation, invasion and survival of tongue squamous cell carcinoma via the Wnt/beta-catenin signaling pathway. Mol Cancer. 2018;17:3
39. Huang Y, Zhang J, Hou L. et al. LncRNA AK023391 promotes tumorigenesis and invasion of gastric cancer through activation of the PI3K/Akt signaling pathway. J Exp Clin Cancer Res. 2017;36:194
40. Yang R, Li P, Zhang G. et al. Long non-coding RNA XLOC_008466 functions as an oncogene in human non-small cell lung cancer by targeting miR-874. Cell Physiol Biochem. 2017;42:126-36
41. Yang Y, Junjie P, Sanjun C. et al. Long non-coding RNAs in colorectal cancer: progression and future directions. J Cancer. 2017;8:3212-25
42. Ma Z, Gu S, Song M. et al. Long non-coding RNA SNHG17 is an unfavourable prognostic factor and promotes cell proliferation by epigenetically silencing P57 in colorectal cancer. Mol Biosyst. 2017;13:2350-61
43. Yu Y, Nangia-Makker P, Farhana L. et al. A novel mechanism of lncRNA and miRNA interaction: CCAT2 regulates miR-145 expression by suppressing its maturation process in colon cancer cells. Mol Cancer. 2017;16:155
44. Zhang J, Jiang Y, Zhu J. et al. Overexpression of long non-coding RNA colon cancer-associated transcript 2 is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. Oncol Lett. 2017;14:6907-14
45. Zhou M, Hu L, Zhang Z. et al. Recurrence-associated long non-coding RNA signature for determining the risk of recurrence in patients with colon cancer. Mol Ther Nucleic Acids. 2018;12:518-29
46. Glas AS, Lijmer JG, Prins MH. et al. The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol. 2003;56:1129-35
47. Jones CM, Athanasiou T. Summary receiver operating characteristic curve analysis techniques in the evaluation of diagnostic tests. Ann Thorac Surg. 2005;79:16-20
48. Liu Z, Zhang Y, Niu Y. et al. A systematic review and meta-analysis of diagnostic and prognostic serum biomarkers of colorectal cancer. PLoS One. 2014;9:e103910
49. Ning S, Wei W, Li J. et al. Clinical significance and diagnostic capacity of serum TK1, CEA, CA 19-9 and CA 72-4 levels in gastric and colorectal cancer patients. J Cancer. 2018;9:494-501
Author contact
![]() Corresponding author: Wenying Zhao, Department of Oncology, The First Affiliated Hospital, Yijishan Hospital of Wannan Medical College, 2 Zheshan West Road, Wuhu 241000, Anhui Province, China. Tel: +86 553 5739070; Email: ahzhaowycom
Corresponding author: Wenying Zhao, Department of Oncology, The First Affiliated Hospital, Yijishan Hospital of Wannan Medical College, 2 Zheshan West Road, Wuhu 241000, Anhui Province, China. Tel: +86 553 5739070; Email: ahzhaowycom

 Global reach, higher impact
Global reach, higher impact