3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(5):1145-1153. doi:10.7150/jca.29049 This issue Cite
Research Paper
Cloning and characterization of the putative AFAP1-AS1 promoter region
1. NHC Key Laboratory of Carcinogenesis, Xiangya Hospital, Central South University, Changsha, Hunan, China
2. The Key Laboratory of Carcinogenesis and Cancer Invasion of the Chinese Ministry of Education, Cancer Research Institute, Central South University, Changsha, Hunan, China
3. Hunan Key Laboratory of Nonresolving Inflammation and Cancer, Disease Genome Research Center, the Third Xiangya Hospital, Central South University, Changsha, Hunan, China
4. Hunan Key Laboratory of Translational Radiation Oncology, Hunan Cancer Hospital and the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, Hunan, China
5. Department of Cancer Biology, Lerner Research Institute, Cleveland Clinic, Cleveland, Ohio, USA
Received 2018-8-7; Accepted 2019-1-4; Published 2019-1-29
Abstract
Actin filament-associated protein 1-antisense RNA1 (AFAP1-AS1), a cancer-related long non-coding RNA, has been found to be upregulated in multiple types of cancers. AFAP1-AS1 is important for the initiation, progression and poor prognosis of many cancers, including nasopharyngeal carcinoma (NPC). However, the mechanism underlying the regulation of AFAP1-AS1 expression is not well-understood. In our study, the potential promoter region of AFAP1-AS1 was predicted by comprehensive bioinformatics analysis. Moreover, promoter deletion analysis identified the sequence between positions -359 and -28 bp as the minimal promoter region of AFAP1-AS1. The ChIP assay results indicate that the AFAP1-AS1 promoter is responsive to the transcription factor c-Myc, which can promote high AFAP1-AS1 expression. This study is the first to clone and characterize the AFAP1-AS1 promoter region. Our findings will help to better understand the underlying mechanism of high AFAP1-AS1 expression in tumorigenesis and to develop new strategies for therapeutic high expression of AFAP1-AS1 in NPC.
Keywords: lncRNA, AFAP1-AS1, promoter, transcription factor, c-Myc
Introduction
Nasopharyngeal carcinoma (NPC) is one of the most common head and neck cancers arising from the nasopharynx epithelium [1-3]. NPC has a distinctive ethnic and geographic distribution and represents a serious health problem in Southeast Asia and southern China, especially in Guangdong Province [4, 5]. The etiology of NPC is multifactorial, including Epstein-Barr virus infection, heredity and environmental factors [6-9]. NPC tumorigenesis often involves inactivation of several tumor suppressor genes and abnormal activation of several oncogenes [10-13], among which an increasing number of long non-coding RNAs (lncRNAs) have been identified in recent years [14-17]. LncRNAs are more than 200 nucleotides in length and have no open reading frame. Dysregulation of lncRNAs has been shown to be involved in the development and progression of NPC [18-20].
Among many cancer-related lncRNAs, the recently identified actin filament-associated protein 1-antisense RNA1 (AFAP1-AS1) has been the focus of great interest [21, 22]. AFAP1-AS1 is derived from the antisense DNA strand of the AFAP1 coding gene locus, and dysregulated expression of AFAP1-AS1 has been found in many types of cancers, including NPC [23, 24]. In our previous study, cDNA microarray analysis of lncRNA expression showed that AFAP1-AS1 is significantly upregulated in NPC. Consistent with this observation, AFAP1-AS1 expression is also upregulated in NPC tissues [25]. In addition, AFAP1-AS1 knockdown significantly suppresses the metastasis and invasion of NPC [26]. Moreover, high AFAP1-AS1 expression is associated with a poor prognosis of NPC patients [27]. However, to our knowledge, a potential mechanism of high AFAP1-AS1 expression in NPC has not been reported.
To understand the mechanism underlying AFAP1-AS1 regulation, we investigated whether AFAP1-AS1 expression was regulated at the level of transcription initiation. Through a comprehensive bioinformatics prediction, the potential promoter region of AFAP1-AS1 was identified and cloned. Furthermore, the transcription factor c-Myc was predicted to be associated with the AFAP1-AS1 promoter and to promote AFAP1-AS1 expression. In addition, deletion analysis identified a 332-bp fragment comprising the minimal promoter region of AFAP1-AS1. This study provides the basis for unraveling the potential mechanism of abnormal AFAP1-AS1 expression in NPC tumorigenesis.
Materials and methods
Cell lines, cell culture and transfection
The NPC cell line HNE2 was maintained in our laboratory and grown in RPMI-1640 medium (Life Technologies, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS; Life Technologies) and 1% penicillin-streptomycin solution (Life Technologies). Cells were incubated at 37ºC in a humidified atmosphere with 5% CO2. Transfection was performed using Lipofectamine 3000 reagent (Life Technologies) according to the manufacturer's protocol.
Cloning of the human AFAP1-AS1 promoter reporter constructs
The potential promoter region -1521/+220 of AFAP1-AS1 and a series of 5'- and 3'-deletion fragments (-1050/+220, -1050/-28, -1050/-80, -1050/-359, -881/-28, -496/-28 and -359/-28) were amplified by PCR using the primers listed in Table 1. All of the primers included 15-bp sequences homologous to the pGL3-enhancer luciferase reporter vector (Promega, Madison, WI, USA). Construct naming is based on the positions of the promoter fragments. The promoter fragments were then inserted into the pGL3-enhancer vector between the NheI and HindIII sites with ClonExpress®II One Step Cloning Kit (Vazyme, Nanjing, China). The recombinants were then transformed into Escherichia coli JM109 and confirmed by DNA sequencing.
Luciferase reporter assay
Promoter activities were detected using the dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions. Briefly, HNE2 cells were transfected with 0.1 μg of Renilla luciferase expression plasmid pRL-TK (internal control for normalizing transfection efficiency; Promega) and 0.4 μg of various AFAP1-AS1 promoter constructs, pGL3-control plasmid (positive control; Promega), or pGL3-enhancer plasmid (negative control). The firefly luciferase readings were normalized by the Renilla luciferase readings to calculate the relative fold-change. Every transfection was independently repeated three times, and the mean ± standard deviation (SD) was used to express the relative fold-change.
Primer pairs used for generating AFAP1-AS1 promoter deletion constructs.
| pGL3-1521/+220 | 5'- CGAGCTCTTACGCGTGCTAGCTGTTTCCCATCCCAATAC -3' |
| 5'- CAGTACCGGAATGCCAAGCTTGCTTTTACCAAGAATCAGC -3' | |
| pGL3-1050/+220 | 5'-CGAGCTCTTACGCGTGCTAGCAAAGTCTTACGGGTGTCG -3' |
| 5'- CAGTACCGGAATGCCAAGCTTGCTTTTACCAAGAATCAGC -3' | |
| pGL3-1050/-80 | 5'-CGAGCTCTTACGCGTGCTAGCAAAGTCTTACGGGTGTCG -3' |
| 5'-CAGTACCGGAATGCCAAGCTTAATAACGGGGAAGACCAG -3' | |
| pGL3-1050/-28 | 5'-CGAGCTCTTACGCGTGCTAGCAAAGTCTTACGGGTGTCG -3' |
| 5'-CAGTACCGGAATGCCAAGCTTGGAACCCTTGATAAACCCT -3' | |
| pGL3-1050/-359 | 5'- CGAGCTCTTACGCGTGCTAGCAAAGTCTTACGGGTGTCG -3' |
| 5'-CAGTACCGGAATGCCAAGCTTTGCAGAAGAAGCAGACCT -3' | |
| pGL3-881/-28 | 5'-CGAGCTCTTACGCGTGCTAGCCCAACATGGAGAAACCTG -3' |
| 5'-CAGTACCGGAATGCCAAGCTTGGAACCCTTGATAAACCCT -3' | |
| pGL3-496/-28 | 5'-CGAGCTCTTACGCGTGCTAGCCCCAAAGAGTTCCCAGTC -3' |
| 5'-CAGTACCGGAATGCCAAGCTTGGAACCCTTGATAAACCCT -3' | |
| pGL3-359/-28 | 5'-CGAGCTCTTACGCGTGCTAGCTGCAGAAGAAGCAGACCT -3' |
| 5'-CAGTACCGGAATGCCAAGCTTGGAACCCTTGATAAACCCT -3' |
RNA extraction and quantitative real-time PCR (qPCR)
Total RNAs were extracted using the TRIzol Extraction Kit (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions. The cDNA was prepared from total RNA using 5X All-In-One RT Master Mix (Applied Biologic Materials (abm), Richmond, Canada), after which real-time qPCR reactions were performed using the Bio-Rad CFX Connect Real-Time system (Bio-Rad, Hercules, CA) with SYBR Green (abm). The expression of each target gene was quantified by the comparative CT method using GAPDH as an endogenous control. The following primers were synthesized by Life Technologies and used to amplify AFAP1-AS1, c-Myc and GAPDH: AFAP1-AS1 forward primer (5'-AAT GGT GGT AGG AGG GAG GA-3'), reverse primer (5'-CAC ACA GGG GAA TGA AGA GG-3'); c-Myc forward primer (5'-CCT ACC CTC TCA ACG ACA GC-3'), reverse primer (5'-TTC CTC CTC AGA GTC GCT GC-3'); and GAPDH forward primer (5'-CAA CGG ATT TGG TCG TAT TGG-3'), reverse primer (5'-TGA CGG TGC CAT GGA ATT T-3'). All reactions were run in triplicate and repeated in three independent experiments.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed in HNE2 cells using a kit from Millipore (Billerica, MA, USA) according to manufacturer's protocol. Cells were fixed in 1% formaldehyde for 10 min at room temperature to crosslink proteins to DNA, after which fixed cells were washed, lysed in cell lysis buffer supplemented with a protease-inhibitor cocktail, and sonicated to shear crosslinked DNA. Then, ~10% of sonicate was saved as an input sample. The crosslinked protein/DNA complexes were immunoprecipitated using the c-Myc antibody, the immunocomplexes were eluted, and the protein/DNA crosslinking was then reversed to release the DNA. The enrichment of purified DNA fragments was determined by real-time PCR using the following two primer sets for AFAP1-AS1: forward primer set 1, TGC ATG ATG ACA CAG AGG GT (start: -1305), reverse primer set 1, GAG GAT ATA GAG GAC TTG GGC T (start: -1166); forward primer set 2, CTC CCG CCA TGA TTC TGA G (start site: +30), and reverse primer set 2, CTT GGC CCA ATT CCT CCT G (start site: +145). Nonspecific antibody (IgG) served as a negative control.
Bioinformatics analysis
The gene sequence of human AFAP1-AS1 was obtained from NCBI. The potential promoter region of the AFAP1-AS1 was predicted using the online promoter prediction software BDGP (http://www.fruitfly.org/seq_tools/promoter.html), Neural Network Promoter Prediction (http://promotor.biosino.org/), and Promoter 2.0 (http://www.cbs.dtu.dk/services/Promoter/). Additionally, CpG Island Searcher (http://www.hugedomains.com/domain_profile.cfm?d=cpgislands&e=com), CpG islands (http://www.ualberta.ca/~stothard/javascript/cpg_islands.html), and CpGProD (http://doua.prabi.fr/software/cpgprod_query) were utilized to find the CpG islands. The potential binding sites of transcription factors in the AFAP1-AS1 gene were identified with the UCSC database.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 5 (GraphPad, La Jolla, CA). Student's t-test was used to evaluate significant differences between two groups of data. All data are represented as the means ± standard deviation. P < 0.05 was considered statistically significant.
Results
Bioinformatics analysis of the AFAP1-AS1 promoter region
To better understand the mechanism involved in the high AFAP1-AS1 expression in NPC, we used various bioinformatics tools to analyze the potential promoter region of AFAP1-AS1. The AFAP1-AS1 sequence was obtained from the National Center for Biotechnology Information (NCBI). The 2000-bp region between positions -1600 bp and +400 bp (the transcription start site was designated as +1) was identified and analyzed. The promoter sequence was analyzed by BDGP, promoter 2.0, and Neural Network Promoter Prediction. A CpG island between positions -1145 and -477 was detected using CpG Islands, CpGProD, and CpG Island Searcher programs. Based on this comprehensive bioinformatics analysis, we chose the region between positions -1521 and +220 for further study (Figure 1).
Construction of the reporter vector for the AFAP1-AS1 promoter
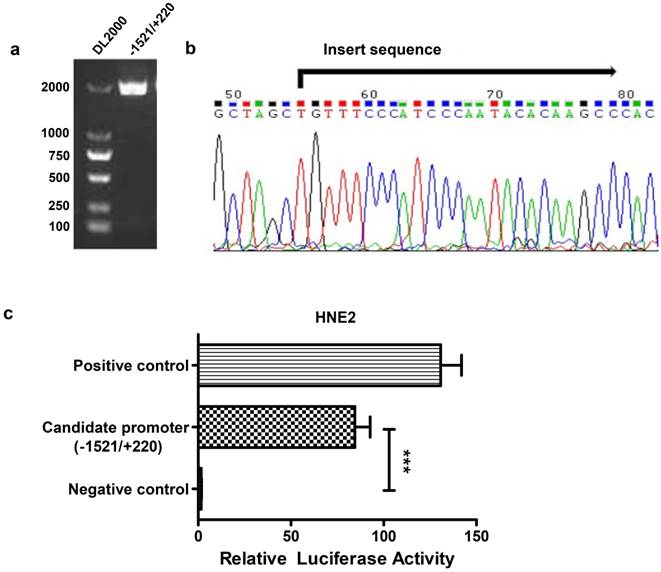
To analyze the transcriptional regulation of AFAP1-AS1, we used human peripheral blood-originated genomic DNA as the template to isolate the AFAP1-AS1 promoter region. A band with an expected size of approximately 1741 bp was successfully amplified and verified by electrophoresis in 1% agarose gel (Figure 2A). The fragment was then cloned into the pGL3-enhancer vector and sequenced (Figure 2B). HNE2 cells were transiently transfected with the AFAP1-AS1 promoter construct (pGL3-1521/+220), and the luciferase activity was then measured. As shown in Figure 2C, the pGL3-1521/+220 construct had a higher luciferase activity than the negative control (relative luciferase activity: candidate promoter, 84.490 ± 8.232 vs. negative control, 1.608 ± 0.175, P<0.001), indicating that the -1521/+220 region of AFAP1-AS1 contains the potential promoter.
Bioinformatics analysis of the AFAP1-AS1 promoter region. Schematic representation of the potential AFAP1-AS1 promoter region and CpG islands. The putative promoter region and CpG islands are shown as rectangular boxes. The transcription start site was designated as +1, the regions before and after the transcription start site are numbered as - or + relative to it. BDGP: Berkeley Drosophila Genome Project; NNPP: Neural Network Promoter Prediction.

Construction of the reporter vector for the -1521/+220 region of the AFAP1-AS1 promoter. A, Gel analysis of the amplification of the -1521/+220 region, lane 1: DL2000 DNA marker, lane 2: the amplification of the -1521/+220 region detected by 1% agarose gel. B, Sequencing results of the pGL3-1521/+220 reporter construct. C, The pGL3-1521/+220 reporter plasmid and pRL-TK were transiently co-transfected into HNE2 cells. Luciferase activities were measured 48 h after transfection. The results are presented as the relative luciferase activity. Positive control: pGL3-control vector, negative control: pGL3-enhancer vector. The deletion samples were compared to the negative control. Data are shown as the means ± SD of at least three independent experiments, ***P< 0.001.
Identification of potential transcription factor binding sites in the AFAP1-AS1 promoter region in NPC cells
We used the UCSC website to predict potential transcription factor binding sites in the region between positions -1521 and +220 of AFAP1-AS1. Several putative binding sites for transcription factors were identified in this region, including c-Myc, ZBTB33, and CHD2. Considering that c-Myc is a very common transcriptional regulator [28, 29], we selected this transcription factor for subsequent validation (Figure 3A).
Identification of the potential transcription factor binding sites in the AFAP1-AS1 promoter region. A, Schematic representation of the predicted transcription factor binding in the -1521/+220 region using the UCSC website. B, Expression of c-Myc was measured in HNE2 cells transfected with the c-Myc overexpression vector by qPCR. C, c-Myc upregulates AFAP1-AS1 expression in HNE2 cells transfected with the c-Myc overexpression vector, compared with the control group (NC). D, ChIP assay in HNE2 cells using a c-Myc antibody, followed by quantitative real-time PCR with two sets of primers designed for two c-Myc binding sites of the AFAP1-AS1 promoter region. All ChIP data are shown relative to 10% input (defined as 1). Nonspecific antibody (IgG) served as a negative control. *P< 0.05, ** P< 0.01, ***P< 0.001.
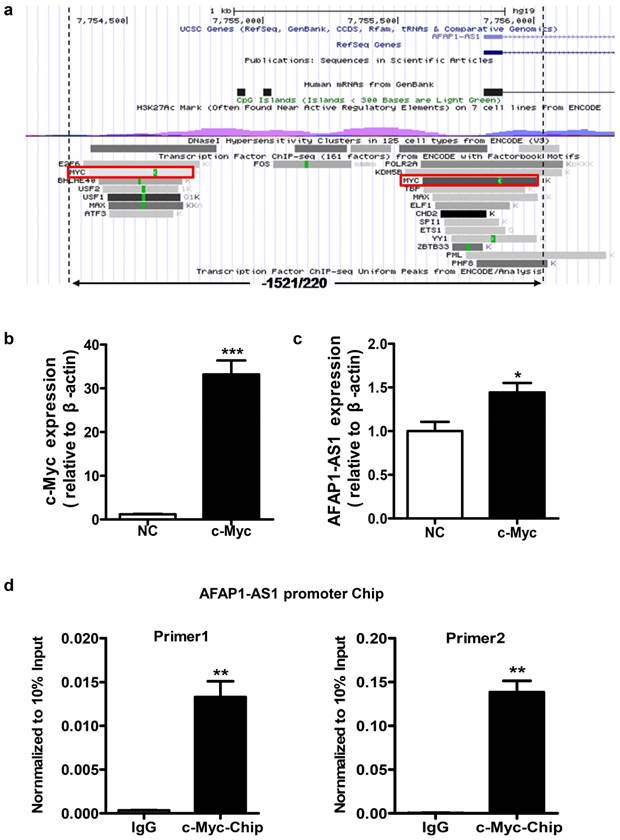
To verify whether c-Myc has a regulatory function in AFAP1-AS1 expression, a c-Myc overexpression vector was transiently transfected to express c-Myc in HNE2 cells (Figure 3B). As shown in Figure 3C, c-Myc overexpression can increase AFAP1-AS1 expression (P=0.027). To determine whether c-Myc binds specifically to the AFAP1-AS1 promoter, we performed chromatin immunoprecipitation (ChIP) assays in HNE2 cells. Two sets of primers for the AFAP1-AS1 promoter in close proximity to two predicted binding sites in the -1521/+220 region were used for real-time PCR. ChIP with an antibody against c-Myc resulted in a significant enrichment of the AFAP1-AS1 promoter region using both primer sets, compared to the control IgG (P=0.010 and 0.009, Figure 3D). Thus, these data indicate that c-Myc is the main transcription factor that promotes high AFAP1-AS1 expression in NPC cells.
Luciferase reporter assay for the AFAP1-AS1 promoter. A, Schematic illustration of the 5'-deletion and 3'-deletion constructs of the AFAP1-AS1 promoter. B, The pGL3-enhancer and the deletion constructs were transiently transfected into HNE2 cells with pRL-TK. Luciferase activities were measured 48 h after transfection. The results are presented as the relative luciferase activity. Positive control: pGL3-control vector, negative control: pGL3-enhancer vector. The deletion samples were compared to the negative control. Data are shown as the means ± SD of at least three independent experiments. ***P< 0.001, ns: no significance.
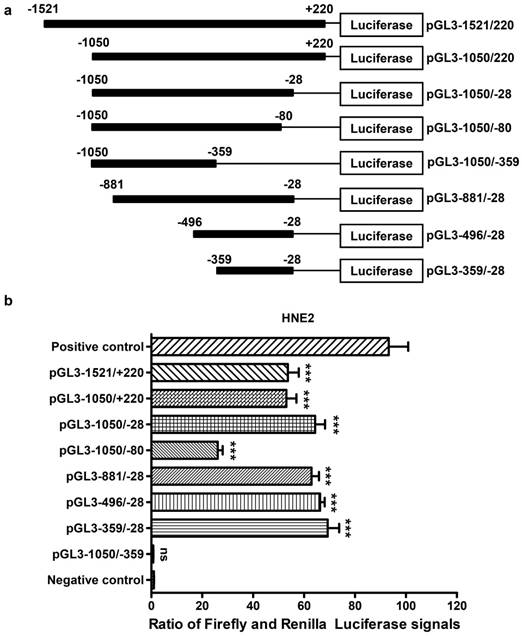
Deletion analysis of the AFAP1-AS1 promoter in NPC cells
To identify the core promoter region of AFAP1-AS1, a series of progressive deletion fragments were generated and subcloned into the pGL3-enhancer vector. These constructs (plasmids pGL3-1521/+220, pGL3-1050/+220, pGL3-1050/-28, pGL3-1050/-80, pGL3-1050/-359, pGL3 -881/-28, pGL3-496/-28 and pGL3-359/-28, depicted in Figure 4A) were transiently transfected into HNE2 cells, and the luciferase activities driven by the AFAP1-AS1 promoter constructs were measured.
As shown in Figure 4B, we standardized the negative control (pGL3-enhancer) to 1, and the luciferase activity from constructs pGL3-1050/-28 (60.560 ± 3.898), pGL3-881/-28 (63.520 ± 2.889) and pGL3-496/-28 (64.220 ± 1.774) exhibited similar levels in HNE2 cells. These levels were higher than those from constructs pGL3-1521/+220 (54.410 ± 4.313) and pGL3-1050/+220 (51.670 ± 3.945). The reporters driven by the shorter 332-bp fragment (PGL3-359/-28) showed the highest luciferase activity (65.740 ± 4.514, P<0.001, in comparison with a negative control). However, when the region between -80 and -28 was deleted, there was a remarkable reduction in luciferase activity (26.190 ± 1.936). Moreover, the pGL3-1050/-359 construct showed nearly no luciferase expression (0.741 ± 0.074, P=0.683, in comparison with a negative control), and it displayed a significantly lower luciferase activity compared to that of pGL3-1050/-28. In conclusion, the above data show that the region between positions -359 and -28 contains the core promoter that activates AFAP1-AS1 transcription and is necessary and required for basal transcriptional activity of AFAP1-AS1.
Discussion
Long non-coding RNAs (lncRNAs) are often defined as transcribed non-coding RNAs that are longer than 200 nucleotides and have no significant protein-coding potential [30-32]. In the past, lncRNAs were mistaken for transcriptional “noise”. However, with the advancement of high-throughput technologies, an increasing number of studies have focused on important roles of lncRNAs in a wide range of physiological and pathological processes, especially in cancers [33-35]. LncRNAs may act as tumor suppressors or oncogenes by regulating target gene expression at the transcriptional, posttranscriptional, and epigenetic levels. LncRNAs can affect the initiation, progression, and prognosis of various cancers, including NPC [36-39]. A growing number of lncRNAs have been reported to be closely associated with proliferation, apoptosis, metastasis, and radiation response of NPC cells, including MALAT-1 [40], LINC01420 [41], LOC553103 [42], LOC401317 [43], and AFAP1-AS1 [27].
Long non-coding RNA AFAP1-AS1 is located in the 4p16.1 region of human chromosome 4, derives from the antisense DNA strand of the AFAP1 gene, and can regulate the expression of AFAP1 [27]. AFAP1-AS1 was initially discovered in Barrett's esophagus and esophageal adenocarcinoma in 2013 [44]. Our in-depth study of AFAP1-AS1 functions in NPC and lung cancer progression showed that high AFAP1-AS1 expression might promote cell metastasis and invasion via regulation of the small GTPase Rho/Rac signaling pathway and actin filament integrity [23, 27]. In recent years, numerous studies have demonstrated that high AFAP1-AS1 levels are found in a variety of cancers, including esophageal squamous cell carcinoma [45], non-small cell lung cancer [46], pancreatic ductal adenocarcinoma [47], hepatocellular carcinoma [48], colorectal [49], cholangiocarcinoma [50], gastric [51], and gallbladder cancer [52]. In addition, the dysregulated expression of AFAP1-AS1 is often associated with cell proliferation, cell cycle progression, angiogenesis, invasion, metastasis, radioresistance and poor prognosis of cancers [53, 54].
The mechanisms behind the abnormal AFAP1-AS1 expression in cancers are not well-understood. A single study on AFAP1-AS1 showed that it was hypomethylated and overexpressed in Barrett's esophagus and esophageal adenocarcinoma [44]. Considering that transcriptional regulation is one of the most important components of gene expression regulation [55], we investigated the mechanisms regulating AFAP1-AS1 expression at the level of transcription. Our comprehensive analysis of the CpG island and promoter prediction showed that the -1521/+220 region of AFAP1-AS1 contains the potential promoter, and a luciferase reporter driven by the promoter of AFAP1-AS1 showed high expression in HNE2 cells.
Analysis of the -1521/+220 promoter region of AFAP1-AS1 by bioinformatics methods showed putative binding sites for transcription factors, such as c-Myc, ZBTB33, and CDH2. It is noteworthy that c-Myc has two binding sites in this region of AFAP1-AS1. The c-Myc proto-oncogene is a vital member of the Myc/Mad/Max transcription factor network, which regulates nearly 15% of genes in the human genome [56-58], and thereby participates in the regulation of various pathological processes in cancers [59, 60]. Thus, c-Myc was selected as a possible candidate transcription factor for regulation of AFAP1-AS1 expression. Our research demonstrated that the AFAP1-AS1 promoter is responsive to c-Myc and that c-Myc overexpression can increase the expression of AFAP1-AS1, indicating that c-Myc may be the main transcription factor that promotes high AFAP1-AS1 expression in NPC.
To identify the necessary and required region for the basal transcriptional activity of AFAP1-AS1, a series of deletion constructs based on pGL3-1521/+220 were generated and transfected in HNE2 cells. We found that the reporters driven by the shorter 332-bp fragment (PGL3-359/-28) showed the highest luciferase activity, and the deletion between positions -80 and -28 caused a significant decrease in promoter activity. Moreover, the luciferase expression was almost undetectable with the pGL3-1050/-359 construct. Thus, a region from -359 to -28 was identified as the minimal sequence that contains the core promoter and controls promoter activity. In addition to c-Myc, we also predicted several other putative transcription factor binding sites in the promoter region of AFAP1-AS1. Among these transcription factors, CDH2 [61], ZBTB33 [62], POLR2A [63], KDM5B [64], MAX [28], and ETS1 [65] have been proven to be upregulated in different types of cancers and are closely related to tumorigenesis and tumor progression [66-69]. Remarkably, the ETS1 and Myc promoter signals are well-correlated and share many properties; for example, Myc and ETS1 can collaborate to increase expression of some oncogenic genes [65]. In addition, Myc and KDM5B can form a nuclear complex that promotes expression of several genes associated with tumorigenesis [70]. Furthermore, the Myc/MAX network comprises a group of transcription factors whose distinct interactions result in gene-specific transcriptional activation [71]. Therefore, Myc and other predicted transcription factors may exert a synergistic effect on the regulation of high AFAP1-AS1 expression.
In conclusion, to our knowledge, this study is the first to clone and characterize the promoter region of AFAP1-AS1. We found that the transcriptional factor c-Myc plays an important role in enhancing AFAP1-AS1 expression. Our findings provide new insights into unraveling the underlying mechanism of high AFAP1-AS1 expression in tumorigenesis.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81572787, 81672683, 81672993, 81772901, 81702907, 81772928, 81872278 and 81803025), the Natural Science Foundation of Hunan Province (2016JC2035, 2017SK2015, 2018JJ3704, 2018JJ3815, 2018SK21210, and 2019JJ50778).
Abbreviations
AFAP1-AS1, actin filament-associated protein 1-antisense RNA1; NPC, nasopharyngeal carcinoma; lncRNA, long non-coding RNAs; qPCR, quantitative real-time PCR; SD, standard deviation; NCBI, National Center for Biotechnology Information; ChIP, chromatin immunoprecipitation.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Liao Q, Zeng Z, Guo X, Li X, Wei F, Zhang W. et al. LPLUNC1 suppresses IL-6-induced nasopharyngeal carcinoma cell proliferation via inhibiting the Stat3 activation. Oncogene. 2013;33:2098-109
2. Zeng Z, Huang H, Huang L, Sun M, Yan Q, Song Y. et al. Regulation network and expression profiles of Epstein-Barr virus-encoded microRNAs and their potential target host genes in nasopharyngeal carcinomas. Sci China Life Sci. 2014;57:315-26
3. Fan C, Tang Y, Wang J, Xiong F, Guo C, Wang Y. et al. The emerging role of Epstein-Barr virus encoded microRNAs in nasopharyngeal carcinoma. J Cancer. 2018;9:2852-64
4. Wei F, Wu Y, Tang L, Xiong F, Guo C, Li X. et al. Trend analysis of cancer incidence and mortality in China. Sci China Life Sci. 2017;60:1-4
5. Yan QJ, Zeng ZY, Gong ZJ, Zhang WL, Li XY, He BY. et al. EBV-miR-BART10-3p facilitates epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma by targeting BTRC. Oncotarget. 2015;6:41766-82
6. Zeng Z, Huang H, Zhang W, Xiang B, Zhou M, Zhou Y. et al. Nasopharyngeal carcinoma: advances in genomics and molecular genetics. Sci China Life Sci. 2011;54:966-75
7. Tu C, Zeng Z, Qi P, Li X, Yu Z, Guo C. et al. Genome-Wide Analysis of 18 Epstein-Barr Viruses Isolated from Primary Nasopharyngeal Carcinoma Biopsy Specimens. J Virol. 2017;91:e00301-e17
8. Zeng Z, Fan S, Zhang X, Li S, Zhou M, Xiong W. et al. Epstein-Barr virus-encoded small RNA 1 (EBER-1) could predict good prognosis in nasopharyngeal carcinoma. Clin Transl Oncol. 2015;18:206-11
9. Tu C, Zeng Z, Qi P, Li X, Guo C, Xiong F. et al. Identification of genomic alterations in nasopharyngeal carcinoma and nasopharyngeal carcinoma-derived Epstein-Barr virus by whole-genome sequencing. Carcinogenesis. 2018;39:1517-28
10. Song Y, Li X, Zeng Z, Li Q, Gong Z, Liao Q. et al. Epstein-Barr virus encoded miR-BART11 promotes inflammation-induced carcinogenesis by targeting FOXP1. Oncotarget. 2016;7:36783-99
11. Wei F, Wu YF, Tang L, He Y, Shi L, Xiong F. et al. BPIFB1 (LPLUNC1) inhibits migration and invasion of nasopharyngeal carcinoma by interacting with VTN and VIM. Brit J Cancer. 2018;118:233-47
12. Yang Y, Liao Q, Wei F, Li X, Zhang W, Fan S. et al. LPLUNC1 Inhibits Nasopharyngeal Carcinoma Cell Growth via Down-Regulation of the MAP Kinase and Cyclin D1/E2F Pathways. PLoS ONE. 2013;8:e62869
13. Wang Y, Mo Y, Gong Z, Yang X, Yang M, Zhang S. et al. Circular RNAs in human cancer. Mol Cancer. 2017;16:25
14. Yu J, Liu Y, Gong Z, Zhang S, Guo C, Li X. et al. Overexpression long non-coding RNA LINC00673 is associated with poor prognosis and promotes invasion and metastasis in tongue squamous cell carcinoma. Oncotarget. 2017;8:16621-32
15. Yu J, Liu Y, Guo C, Zhang S, Gong Z, Tang Y. et al. Upregulated long non-coding RNA LINC00152 expression is associated with progression and poor prognosis of tongue squamous cell carcinoma. J Cancer. 2017;8:523-30
16. Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F. et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79
17. Zhou R, Wu Y, Wang W, Su W, Liu Y, Wang Y. et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134-42
18. Gong Z, Yang Q, Zeng Z, Zhang W, Li X, Zu X. et al. An integrative transcriptomic analysis reveals p53 regulated miRNA, mRNA, and lncRNA networks in nasopharyngeal carcinoma. Tumour Biol. 2016;37:3683-95
19. Tang Y, He Y, Zhang P, Wang J, Fan C, Yang L. et al. LncRNAs regulate the cytoskeleton and related Rho/ROCK signaling in cancer metastasis. Mol Cancer. 2018;17:77
20. Fan C, Wang J, Tang Y, Wang Y, Xiong F, Zhang S. et al. Long non-coding RNA LOC284454 promotes migration and invasion of nasopharyngeal carcinoma via modulating the Rho/Rac signaling pathway. Carcinogenesis. 2018 Epub ahead of print
21. Tang Y, He Y, Shi L, Yang L, Wang J, Lian Y. et al. Co-expression of AFAP1-AS1 and PD-1 predicts poor prognosis in nasopharyngeal carcinoma. Oncotarget. 2017;8:39001-11
22. He B, Zeng J, Chao W, Chen X, Huang Y, Deng K. et al. Serum long non-coding RNAs MALAT1, AFAP1-AS1 and AL359062 as diagnostic and prognostic biomarkers for nasopharyngeal carcinoma. Oncotarget. 2017;8:41166-77
23. Zeng Z, Bo H, Gong Z, Lian Y, Li X, Li X. et al. AFAP1-AS1, a long noncoding RNA upregulated in lung cancer and promotes invasion and metastasis. Tumor Biol. 2015;37:729-37
24. Bo H, Fan L, Li J, Liu Z, Zhang S, Shi L. et al. High Expression of lncRNA AFAP1-AS1 Promotes the Progression of Colon Cancer and Predicts Poor Prognosis. J Cancer. 2018;9:4677-83
25. Wang Y, Mo Y, Yang X, Zhou R, Wu Z, He Y. et al. Long non-coding RNA AFAP1-AS1 is a novel biomarker in various cancers: a systematic review and meta-analysis based on the literature and GEO datasets. Oncotarget. 2017;8:102346-60
26. Lian Y, Xiong F, Yang L, Bo H, Gong Z, Wang Y. et al. Long noncoding RNA AFAP1-AS1 acts as a competing endogenous RNA of miR-423-5p to facilitate nasopharyngeal carcinoma metastasis through regulating the Rho/Rac pathway. J Exp Clin Cancer Res. 2018;37:253
27. Bo H, Gong Z, Zhang W, Li X, Zeng Y, Liao Q. et al. Upregulated long non-coding RNA AFAP1-AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget. 2015;6:20404-18
28. Jiang X, Wang J, Deng X, Xiong F, Ge J, Xiang B. et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol Cancer. 2019;18:10
29. Dang CV, O'Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253-64
30. Yang LT, Tang YY, Xiong F, He Y, Wei F, Zhang SS. et al. LncRNAs regulate cancer metastasis via binding to functional proteins. Oncotarget. 2018;9:1426-43
31. He Y, Jing Y, Wei F, Tang Y, Yang L, Luo J. et al. Long non-coding RNA PVT1 predicts poor prognosis and induces radioresistance by regulating DNA repair and cell apoptosis in nasopharyngeal carcinoma. Cell Death Dis. 2018;9:235
32. He R, Liu P, Xie X, Zhou Y, Liao Q, Xiong W. et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36:145
33. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253-61
34. Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest. 2016;126:2775-82
35. Tang Y, Wang J, Lian Y, Fan C, Zhang P, Wu Y. et al. Linking long non-coding RNAs and SWI/SNF complexes to chromatin remodeling in cancer. Mol Cancer. 2017;16:42
36. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452-63
37. Yang L, Froberg JE, Lee JT. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem Sci. 2014;39:35-43
38. Fan C, Tang Y, Wang J, Xiong F, Guo C, Wang Y. et al. Role of long non-coding RNAs in glucose metabolism in cancer. Mol Cancer. 2017;16:130
39. Wang J, Tang Y, Fan C, Xiong F, Guo C, Zhou Y. et al. The role of exosomal non-coding RNAs in cancer metastasis. Oncotarget. 2017;9:12487-502
40. Wang Y, Xue D, Li Y, Pan X, Zhang X, Kuang B. et al. The Long Noncoding RNA MALAT-1 is A Novel Biomarker in Various Cancers: A Meta-analysis Based on the GEO Database and Literature. J Cancer. 2016;7:991-1001
41. Yang L, Tang Y, He Y, Wang Y, Lian Y, Xiong F. et al. High Expression of LINC01420 indicates an unfavorable prognosis and modulates cell migration and invasion in nasopharyngeal carcinoma. J Cancer. 2017;8:97-103
42. He B, Li W, Wu Y, Wei F, Gong Z, Bo H. et al. Epstein-Barr virus-encoded miR-BART6-3p inhibits cancer cell metastasis and invasion by targeting long non-coding RNA LOC553103. Cell Death Dis. 2016;7:e2353
43. Gong Z, Zhang S, Zeng Z, Wu H, Yang Q, Xiong F. et al. LOC401317, a p53-regulated long non-coding RNA, inhibits cell proliferation and induces apoptosis in the nasopharyngeal carcinoma cell line HNE2. PLoS One. 2014;9:e110674
44. Wu W, Bhagat TD, Yang X, Song JH, Cheng Y, Agarwal R. et al. Hypomethylation of Noncoding DNA Regions and Overexpression of the Long Noncoding RNA, AFAP1-AS1, in Barrett's Esophagus and Esophageal Adenocarcinoma. Gastroenterology. 2013;144:956-66
45. Luo H, Huang M, Guo J, Fan R, Xia X, He J. et al. AFAP1-AS1 is upregulated and promotes esophageal squamous cell carcinoma cell proliferation and inhibits cell apoptosis. Cancer Med-US. 2016;5:2879-85
46. Deng J, Liang Y, Liu C, He S, Wang S. The up-regulation of long non-coding RNA AFAP1-AS1 is associated with the poor prognosis of NSCLC patients. Biomed Pharmacoth. 2015;75:8-11
47. Ye Y, Chen J, Zhou Y, Fu Z, Zhou Q, Wang Y. et al. High expression of AFAP1-AS1 is associated with poor survival and short-term recurrence in pancreatic ductal adenocarcinoma. J Transl Med. 2015;13:137
48. Zhang J, Weng M, Song F, Xu Y, Liu Q, Wu J. et al. Long noncoding RNA AFAP1-AS1 indicates a poor prognosis of hepatocellular carcinoma and promotes cell proliferation and invasion via upregulation of the RhoA/Rac2 signaling. Int J Oncol. 2016;48:1590-8
49. Wang F, Ni H, Sun F, Li M, Chen L. Overexpression of lncRNA AFAP1-AS1 correlates with poor prognosis and promotes tumorigenesis in colorectal cancer. Biomed Pharmacoth. 2016;81:152-9
50. Shi X, Zhang H, Wang M, Xu X, Zhao Y, He R. et al. LncRNA AFAP1-AS1 promotes growth and metastasis of cholangiocarcinoma cells. Oncotarget. 2017;8:58394-404
51. Guo J, Li S, Guo G. Long Noncoding RNA AFAP1-AS1 Promotes Cell Proliferation and Apoptosis of Gastric Cancer Cells via PTEN/p-AKT Pathway. Digest Dis Sci. 2017;62:2004-10
52. Ma F, Wang S, Cai Q, Zhang M, Yang Y, Ding J. Overexpression of LncRNA AFAP1-AS1 predicts poor prognosis and promotes cells proliferation and invasion in gallbladder cancer. Biomed Pharmacoth. 2016;84:1249-55
53. He R, Hu Z, Wang Q, Luo W, Li J, Duan L. et al. The role of long non-coding RNAs in nasopharyngeal carcinoma: As systemic review. Oncotarget. 2017;8:16075-83
54. Zhang F, Li J, Xiao H, Zou Y, Liu Y, Huang W. AFAP1-AS1: A novel oncogenic long non-coding RNA in human cancers. Cell Proliferat. 2017;51:e12397
55. Bonasio R, Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu Rev Genet. 2014;48:433-55
56. Duan S, Guo W, Xu Z, He Y, Liang C, Mo Y. et al. Natural killer group 2D receptor and its ligands in cancer immune escape. Mol Cancer. 2019;18:29
57. Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2:764-76
58. Kenneth NS, White RJ. Regulation by c-Myc of ncRNA expression. Curr Opin Genet Dev. 2009;19:38-43
59. Dang CV. MYC on the Path to Cancer. Cell. 2012;149:22-35
60. Lin CJ, Malina A, Pelletier J. c-Myc and eIF4F Constitute a Feedforward Loop That Regulates Cell Growth: Implications for Anticancer Therapy. Cancer Res. 2009;69:7491-4
61. Ma T, Zhao Y, Wei K, Yao G, Pan C, Liu B. et al. MicroRNA-124 Functions as a Tumor Suppressor by Regulating CDH2 and Epithelial-Mesenchymal Transition in Non-Small Cell Lung Cancer. Cell Physiol Bioche. 2016;38:1563-74
62. Bo H, Fan L, Gong Z, Liu Z, Shi L, Guo C. et al. Upregulation and hypomethylation of lncRNA AFAP1-AS1 predicts a poor prognosis and promotes the migration and invasion of cervical cancer. Oncol Rep. 2019 doi: 10.3892/or.2019.7027. [Epub ahead of print]
63. Liu Y, Zhang X, Han C, Wan G, Huang X, Ivan C. et al. TP53 loss creates therapeutic vulnerability in colorectal cancer. Nature. 2015;520:697-701
64. Wang X, Oishi N, Shimakami T, Yamashita T, Honda M, Murakami S. et al. Hepatitis B virus X protein induces hepatic stem cell-like features in hepatocellular carcinoma by activating KDM5B. World J Gastroenterol. 2017;23:3252-61
65. Chen J, Fu Y, Day DS, Sun Y, Wang S, Liang X. et al. VEGF amplifies transcription through ETS1 acetylation to enable angiogenesis. Nat Commun. 2017;8:383
66. Zhang Y, Xia M, Jin K, Wang S, Wei H, Fan C. et al. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol Cancer. 2018;17:45
67. Deng X, Xiong F, Li X, Xiang B, Li Z, Wu X. et al. Application of atomic force microscopy in cancer research. J Nanobiotechnology. 2018;16:102
68. Wang YA, Li XL, Mo YZ, Fan CM, Tang L, Xiong F. et al. Effects of tumor metabolic microenvironment on regulatory T cells. Mol Cancer. 2018;17:168
69. Tang L, Wei F, Wu Y, He Y, Shi L, Xiong F. et al. Role of metabolism in cancer cell radioresistance and radiosensitization methods. J Exp Clin Cancer Res. 2018;37:87
70. Wong PP, Miranda F, Chan KV, Berlato C, Hurst HC, Scibetta AG. Histone Demethylase KDM5B Collaborates with TFAP2C and Myc To Repress the Cell Cycle Inhibitor p21cip (CDKN1A). Mol Cell Biol. 2012;32:1633-44
71. Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653-99
Author contact
![]() Corresponding authors: Fang Xiong and Zhaoyang Zeng. Tel.: 86-731-8480-5412; Fax: 86-731-8480-5383; Email: xiongfedu.cn OR zengzhaoyangedu.cn; Address: NHC Key Laboratory of Carcinogenesis, Xiangya Hospital, Central South University, Xiangya road 110, Changsha 410008, Hunan, P. R. China
Corresponding authors: Fang Xiong and Zhaoyang Zeng. Tel.: 86-731-8480-5412; Fax: 86-731-8480-5383; Email: xiongfedu.cn OR zengzhaoyangedu.cn; Address: NHC Key Laboratory of Carcinogenesis, Xiangya Hospital, Central South University, Xiangya road 110, Changsha 410008, Hunan, P. R. China

 Global reach, higher impact
Global reach, higher impact