3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(6):1560-1569. doi:10.7150/jca.27457 This issue Cite
Research Paper
Epigenetic Field Cancerization in Gastric Cancer: microRNAs as Promising Biomarkers
1. Laboratory of Human and Medical Genetics, Institute of Biological Sciences, Federal University of Pará, Augusto Corrêa Avenue, 66075-110, Belém, Pará, Brazil.
2. Research Center on Oncology, Institute of Health Sciences, Federal University of Pará, Mundurucus Street, 66073-000, Belém, Pará, Brazil.
3. São Camilo and São Luís Hospital, Dr. Marcello Cândia Street, 68901-901, Macapá, Amapá, Brazil.
Received 2018-5-24; Accepted 2018-11-21; Published 2019-2-26
Abstract
Background: The biological role of microRNAs (miRNAs) in field cancerization is unknown. To investigate the involvement of miRNAs in gastric field cancerization, we evaluated the expression profile of ten miRNAs and their diagnostic value.
Methods: We used three groups of FFPE gastric samples: non-cancer (NC), cancer adjacent (ADJ) and gastric cancer (GC). The expression profiles of hsa-miR-10a, -miR-21, -miR-29c, -miR-135b, -miR-148a, -miR-150, -miR-204, -miR-215, -miR-483 and -miR-664a were investigated using qRT-PCR. The results obtained by qRT-PCR were validated in Small RNA-Seq data from the TCGA database. The search for target genes of the studied miRNAs was performed in the miRTarBase public database and miRTargetLink tool, using experimentally validated interactions. In addition, we also performed the functional analysis of these genes using enrichment in KEGG pathways. The potential as biomarker was evaluated using a receiver operating characteristic (ROC) curve and the derived area under the curve (AUC>0.85) analysis.
Results: The miRNAs hsa-miR-10a, -miR-21, -miR-135b, hsa-miR-148a, -miR-150, -miR-215, -miR-204, -miR-483 and -miR-664a were up-regulated in ADJ and GC compared to NC (P<0.03); and hsa-miR-21 and -miR-135b were up-regulated in GC compared to ADJ (P<0.01). Hsa-miR-148a, -miR-150, -miR-215, -miR-483 and -miR-664a were not differentially expressed between GC and ADJ, suggesting that both share similar changes (P>0.1). The TS-miR hsa-miR-29c was up-regulated in ADJ compared to NC and GC (P<0.01); we did not observe a significant difference in the expression of this miRNA between NC and GC. This feature may be an antitumor mechanism used by cancer-adjacent tissue because this miRNA regulates the BCL-2, CDC42 and DMNT3A oncogenes. The expression level of hsa-miR-204 was associated with Helicobacter pylori infection status (P<0.05). Functional analysis using the genes regulated by the studied miRNAs showed that they are involved in biological pathways and cellular processes that are critical for the establishment of H. pylori infection and for the onset, development and progression of GC. hsa-miR-10a, -miR-21, -miR-135b, -miR-148a, -miR-150, -miR-215, -miR-483 and -miR-664a were able to discriminate NC from other tissues with great accuracy (AUC>0.85).
Conclusion: The studied miRNAs are closely related to field cancerization, regulate genes important for gastric carcinogenesis and can be potentially useful as biomarkers in GC.
Keywords: miRNA, field cancerization, epigenetic, gastric cancer, biomarker
Introduction
Gastric cancer (GC) is one of the most common and leading causes of cancer death worldwide [1]. It is a complex and aggressive disease in which the five- year survival rates in the early stages of the disease are ~ 85-100% [2-3] and only 5-30% in advanced stages [3-4]. GC is difficult to manage, especially when the disease is diagnosed in advanced stages, which is reflected in high rates of local recurrence of the disease after surgical resection [5-8], which in part may be associated with field cancerization of the affected region or organ [9-10].
The process of field cancerization is characterized by regions surrounding the tumor that, despite presenting the histological appearance of healthy tissue [11], already have genetic and/or epigenetic alterations that make them prone to tumors or local recurrence [12-13].
Field cancerization has been reported in many types of cancers and has been the subject of many reviews in the literature that seek evidence to prove that this is an important component of carcinogenesis [10, 13-19]. Studies of field cancerization in GC have identified many genetic, epigenetic and chromosomal alterations [19-25]. Changes in gene methylation are the most well-studied and documented epigenetic changes associated with field cancerization in GC [19]. There is also evidence that miRNA expression (e.g., hsa-miR-150, hsa-miR-664a and hsa-miR483) is deregulated during this process [21]. The human stomach has miRNAs that are part of the normal expression of this organ [26-27] and that may be deregulated in GC [21, 28-31].
miRNAs play an important role in cellular and tissue homeostasis [32-36] because they are involved in the post-transcriptional regulation of genes that play key roles in development and proper cellular function [37-38]. This class of molecules is closely related to gastric cancer, and their dysregulation can trigger or contribute to initiation, progression, invasion and metastasis in GC [29, 39-41]. The current knowledge about the involvement of miRNAs in field cancerization is incipient; therefore, new studies are necessary to elucidate the biological role of these molecules during this process.
This study aimed to evaluate the expression profile of ten miRNAs in samples of gastric tissues and their relationship with field cancerization. The miRNAs used were selected from previous studies that suggest their involvement in gastric carcinogenesis [21, 26-28, 30-31]. Our results demonstrate that these miRNAs are significantly deregulated between the investigated groups, suggesting the existence of field cancerization in gastric cancer. In addition, we evaluated the discriminatory ability of these miRNAs and identified eight promising biomarkers of gastric carcinogenesis.
Material and Methods
Ethics statement
This study was reviewed and approved by the Ethics Committee of the Nucleus of Tropical Medicine of the Federal University of Pará (Protocol Number: 1.081.340). All study participants or their legal guardian provided informed written consent in accordance with the Helsinki Declaration of 1964.
Clinical samples
Thirty samples of gastric tissue from patients with non-cancerous gastritis (NC; mean age=51.4 years) were used as controls. The samples of non-cancerous gastritis were as follows: i) seventeen samples of H. pylori-negative chronic gastritis and ii) thirteen samples of H. pylori-positive chronic gastritis (Table S1). NC biopsies were obtained from patients who underwent upper gastrointestinal endoscopy.
We used paired samples of tissues of gastric cancer (GC; ± H pylori; mean age=56.3 years; n=20) and adjacent to the tumor (ADJ; ± H. pylori; n=11) from patients with gastric adenocarcinoma (Table S1).
The biopsies were fixed in formalin and paraffin-embedded (FFPE). The pathologist evaluated and confirmed the histopathological characteristics of the studied samples, and for GC cases, Lauren's criteria [42] were used. ADJ and GC biopsies were from patients submitted to gastrectomy. Histological sections of the biopsies were stained with hematoxylin-eosin and cresyl violet for rapid detection of H. pylori.
The patients selected for the study were from the Universitary Hospital João de Barros Barreto, Federal University of Pará (Belém City, Pará State, Brazil) and the São Camilo and São Luís Hospital (Macapá City, Amapá State, Brazil).
Total RNA isolation and quantification
Six sections of FFPE tissues with 3 μM thickness were separated from each histopathologically evaluated block and then deparaffinized to extract total RNA. Total RNA (including miRNA) was extracted with the High Pure miRNA Isolation kit (Roche Applied Science, Mannheim, Germany) using the manufacturer's protocol, and then stored at -80°C until analysis. Total RNA concentration was obtained using the Qubit® 2.0 Fluorometer (Thermo Fisher Scientific) using the Qubit® RNA HS Assay kit (Thermo Fisher Scientific).
Quantitative real-time polymerase chain reaction
Approximately 10 ng of total RNA was subjected to reverse transcription polymerase chain reaction using a TaqMan® MicroRNA Reverse Transcription kit (Thermo Fisher Scientific) according to the manufacturer's protocol. Quantitation of miRNA expression was performed using Taqman® MicroRNA Assays (Thermo Fisher Scientific) according to the manufacturer's protocol. The thermocycling occurred under the following conditions: 30 min at 16°C, followed by 30 min at 42°C, 5 min at 85°C and 5 min at 4°C.
Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using the TaqMan® Universal PCR Master Mix kit (Thermo Fisher Scientific) following the manufacturer's protocol and the 7500 Real-Time PCR System (Applied Biosystems). The reactions were performed in triplicate and incubated in 96-well optical plates. The thermocycling conditions were as follows: 95°C for 10 min and 40 cycles of 15 s at 95°C and 1 min at 60°C.
After qRT-PCR experiments, the threshold cycle (CT) data were obtained, and the average values of triplicate reactions were determined (Table S2). The raw data were normalized by the arithmetic mean of the threshold cycles (CTS) of the endogenous controls Z30 and RNU6B (Table S2). The relative expression of the studied miRNAs was obtained by the comparative CT method, and fold change was obtained by the equation 2-ΔΔCT [43].
Statistical analysis
qRT-PCR
Statistical analysis and graphing were developed in the statistical package R v3.4 (http://www.r-project.org/) and RStudio (version 0.99). To evaluate the distribution of the data, we used the Shapiro-Wilk test. One-way ANOVA test was used to evaluate the statistical significance among the groups analyzed. Bonferroni's correction was applied for post hoc analysis, where an adjusted P value<0.05 was considered statistically significant.
RNA-Seq
To validate our results, we analyzed data from 82 samples of small RNA-Seq, downloaded from The Cancer Genome Atlas (TCGA) database. We used pared gastric samples, including 41 adjacent to the tumor and 41 gastric cancer samples. Differential expression analysis of all processed data was performed using DESeq2 library version 1.16.1 [44] in R software, with an expression threshold of 10 counts per miRNA (present at least 10 read counts in at least one of the libraries). To the comparison between adjacent to the gastric cancer (ADJ) and gastric cancer (GC) samples, it was considered statistically significant miRNAs expression with adjusted P values<0.05 and |log2(fold change)|>2.
Receiver operating characteristic (ROC)
Receiver operating characteristic (ROC) and area under the curve (AUC>0.85) analyses were used to evaluate the discriminatory capacity and biomarker potential of the studied miRNAs. A biomedical statistician revised the statistical analysis used in the study.
Search for target driver genes and functional analysis
The search for target driver genes of the studied miRNAs was performed in the miRTarBase [45] public database (http://mirtarbase.mbc.nctu.edu.tw /) using only interactions experimentally validated by strong evidence. One hundred twenty-seven driver genes were prioritized in the searches, all extracted from Table S2A in Vogelstein et al. [46]. MiRTargetLink tool [47] was used to identify genes that are regulated by at least two miRNAs in common among the ten studied.
The functional analysis of the identified target genes was realized using STRING v.10.5 [48], enriched in biological KEGG pathways.
Results
Expression profile of ten miRNAs in the three types of gastric tissues
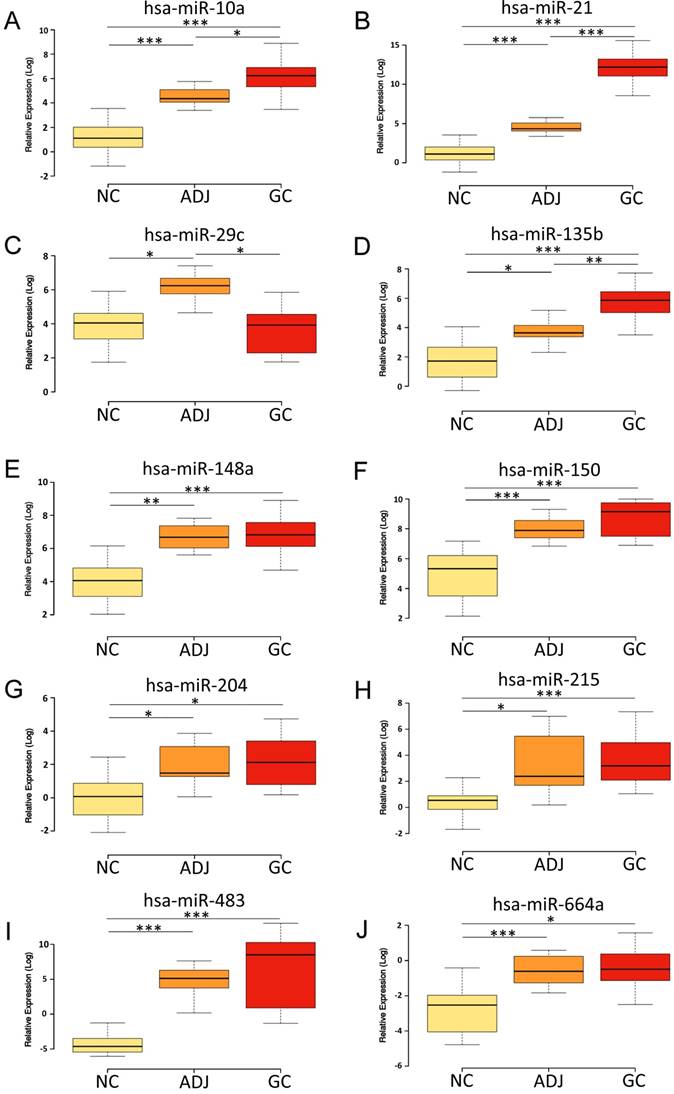
The miRNAs hsa-miR-10a, -miR-21, -miR-135b, hsa-miR-148a, -miR-150, -miR-215, -miR-204, -miR-483 and -miR-664a were up-regulated in ADJ and GC compared to NC (P<0.03; Figure 1; Table S3). Hsa-miR-21 and -miR-135b were also up-regulated in GC compared to ADJ (P<0.01; Figure 1; Table S3). On the other hand, hsa-miR-148a, -miR-150, -miR-215, -miR-483 and -miR-664a were not differentially expressed between ADJ and GC (P>0.1; Figure 1; Table S3), suggesting that both of these tissues share similar changes.
Hsa-miR-29c was significantly up-regulated in ADJ compared to NC and GC (P<0.01; Figure 1; Table S3), while we did not observe a significant difference in the expression of this miRNA between NC and GC (Figure 1; Table S3).
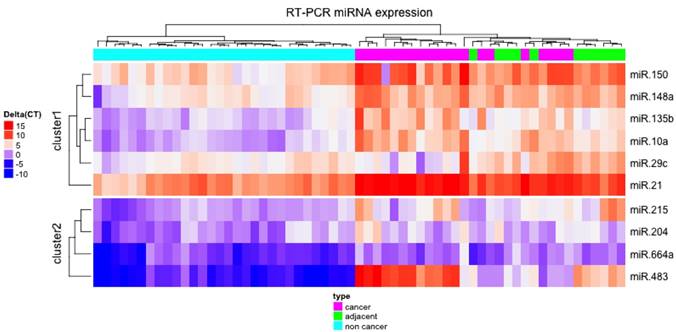
Unsupervised hierarchical clustering of normalized expression of the ten miRNAs studied provided a heatmap that perfectly separated NC from ADJ and GC samples (Figure 2).
Expression of ten miRNAs in small RNA-seq data
To validate the results found by qRT-PCR we analyzed the ten studied miRNAs in the sequencing data from TCGA. Hsa-miR-21, hsa-miR-29c, hsa-miR-135b, hsa-miR-150, hsa-miR-215, hsa-miR-483 and hsa-miR-664a expression levels were similar between ADJ and GC and corroborate the data found by qRT-PCR (Table S3).
Expression levels of the studied miRNAs among the non-cancer, cancer-adjacent and gastric cancer groups. A) hsa-miR-10a; B) hsa-miR-21; C) hsa-miR-29c; D) hsa-miR-135b; E) hsa-miR-148a; F) hsa-miR-150; G) hsa-miR-204; H) hsa-miR-215; I) hsa-miR-483; J) hsa-miR-664a. NC = Non cancer; ADJ = Adjacent to gastric cancer; GC = Gastric cancer. *P value < 0.05; **P value < 0.001; ***P value < 0.0001. P value adjusted by Bonferroni's correction.
Heatmap of normalized expression of the ten studied miRNAs. The unsupervised hierarchical clustering showed that the miRNAs studied were able to separate non-cancer from cancer-adjacent and gastric cancer tissues.
Three miRNAs (hsa-miR-10a, hsa-miR-148a and hsa-miR-204) have an inverse expression profile of the results found by qRT-PCR (Table S2), we attributed this difference to three possible causes: i) sample sizing used during qRT-PCR analysis; ii) genetic characteristics of the Brazilian population, which has a strong genetic substructure and admixture [49]; and/or iii) due to instability and less representativeness of some miRNAs in FFPE samples [50-51].
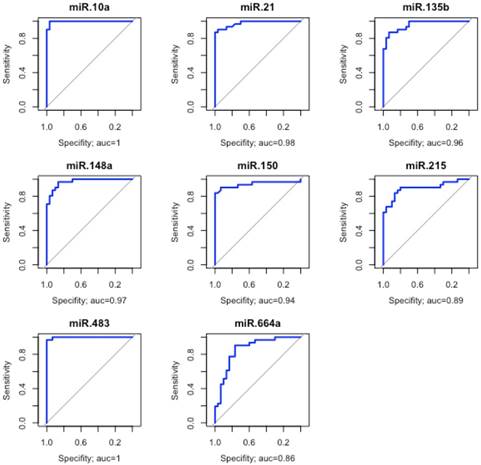
Evaluation of the discriminatory performance of eight miRNAs. These eight miRNAs were able to discriminate the non-cancer from the other tissues with great accuracy. The area under the curve (AUC>0.85).
Evaluation of biomarker potential of the miRNAs
Because hierarchical clustering analysis demonstrated that ADJ and GC samples are very similar, we grouped them into a single group to assess the discriminatory ability of the studied miRNAs. Eight miRNAs (hsa-miR-10a, -miR-21, -miR-135b, -miR-148a, -miR-150, -miR-215, -miR-483 and -miR-664a) were able to discriminate with great accuracy ADJ and GC from NC tissues (AUC>0.85; Figure 3).
Helicobacter pylori infection influence on miRNA expression
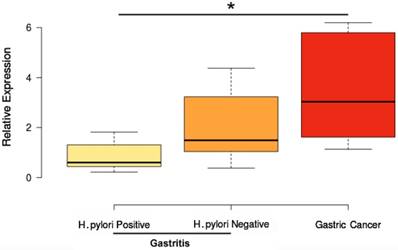
Only hsa-miR-204 was differentially expressed among the analyzed samples regarding H. pylori infection status. Hsa- miR-204 was up-regulated in GC compared to H. pylori-positive gastritis (P value <0.004; Figure 4). This miRNA was not differentially expressed between H. pylori- negative gastritis and GC (Figure 4). These data corroborate studies showing that H. pylori infection can modulate the expression of hsa-miR-204 in gastric tissues [52-55].
MiRNAs, target genes and functional role
One hundred twenty seven driver genes were used during searches within the public miRTarBase database, of which 28 are direct targets of the studied miRNAs (Table S4A). Nine miRNAs regulate ten oncogenes and eighteen tumor suppressors (Table S4A). The PTEN tumor suppressor is a common target of hsa-miR-10a, -miR-21 and -miR-29c, and the BCL-2 oncogene is a common target of hsa-miR-21, -miR-29c, -miR-148a, -miR-150 and -miR-204 (Table S4A). Gene enrichment analysis showed that these genes are involved in 62 biological pathways (Table S5), of which we selected eighteen important pathways for GC (Figure 6A).
The search for target genes using the miRTargetLink tool provided fourteen genes (Table S4B) that are regulated by at least two miRNAs among the ten studied ones. The enrichment analysis using these fourteen genes demonstrated that they participate in 44 biological pathways (Table S6), from which we selected nineteen important pathways for the establishment and development of GC (Figure 5B). Interestingly, the two enrichment analyzes basically point to the same biological pathways (Figure 5A-B), reinforcing the biological role of these target genes.
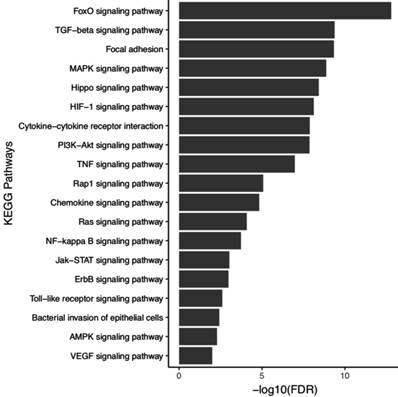
Studies shows that five miRNAs analyzed in this study (hsa-miR-21, -miR-135b, -miR-148a, -miR-150 and -miR-204) have their expression levels changed by H. pylori [56-59]. To assess the biological role of five miRNAs that may undergo influence on their expression levels during H. pylori infection, we performed enrichment in biological KEGG pathways using a 199 experimentally validated target genes (Table S7). These genes are involved in 88 biological pathways (Table S8), from which we selected 19 pathways important for the establishment and progression of H. pylori infection (Figure 6).
Discussion
In this study, we evaluated the expression profile of ten miRNAs in three different groups of FFPE gastric samples, aiming to test the involvement of these molecules in field cancerization in GC and its biomarker performance.
Our data showed overexpression of hsa-miR-10a, -miR-21, -miR-135b, -miR-148a, -miR-150, -miR-204, -miR-215, -miR-483 and -miR-664a in ADJ and GC compared to NC tissues (Figure 1; Table S3). These results support previous studies [21, 28-31], which demonstrate the up-regulation of many of these miRNAs in GC. These miRNAs regulate 18 driver genes (tumor suppressor genes; Table S4A), including APC [60], RUNX1 [61], EP300 [62-63], KLF4 [64-65], PTEN [66-67], TP53 [68], RB1 [69], SMAD2 [70-71] and SMAD4 [72-73], which have already been reported to be down-regulated in GC. Studies has shown that hsa-miR-10a, -miR-21, -miR-135b, -miR-148a, -miR-150, -miR-215 and -miR-483 regulate important processes for the establishment and progression of gastric tumors, such as cell cycle progression, migration, proliferation, invasion, apoptosis and metastasis [56, 61, 74-81].
Influence of H. pylori infection on hsa-miR-204 expression. *P value<0.05. P value adjusted by Bonferroni's correction.
Functional enrichment in KEGG pathways using the target genes of the studied miRNAs. In highlight pathways relevant for establishment and progression of GC. A) Enrichment using driver genes. Information based on table SA2 in Vogelstein et al. [46]. B) Enrichment using genes provided by the miRTargetLink tool.
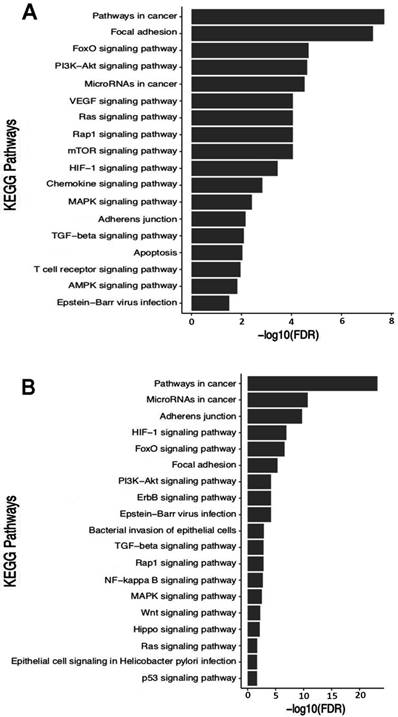
Functional enrichment in KEGG pathways using the target genes of the five miRNAs associated to H. pylori infection. In highlight some KEGG pathways relevant for establishment and progression of infection.
Gene enrichment analysis in KEGG pathways revealed that these miRNAs regulate genes involved in the FoxO, PI3K-Akt, HIF-1, Ras, Rap1, ErbB, TGF-β, AMPK, MAPK, Wnt, mTOR signaling pathways, and Epstein-Barr Virus and H. pylori infections (Figure 5A-B). These pathways control the tissue and/or organ homeostasis and, when deregulated, can contribute to the development and progression of GC.
When ADJ was compared to GC, we observed that the expression levels of hsa-miR-10a, -miR-148a, -miR-150, -miR-215, -miR-483 and -miR-664a were very similar, and both tissues differed significantly from NC tissue (Figure 1; Table S3). These results suggest that the adjacent tissue has alterations similar to those found in tumors, which makes it prone to onset tumors and/or local recurrence. These findings also corroborate the existence of field cancerization in gastric cancer and lead us to agree with Assumpção et al. [82], who argue that the use of adjacent tissue for comparison purposes can lead to biases regarding the true gene expression profile associated with GC. Thus, the use of ADJ tissue as a control should be performed with caution in studies that evaluate gene expression in GC because ADJ tissue already has epigenetic changes that are present in the tumor. This characteristic clearly distinguishes ADJ from a truly normal tissue.
When evaluating the differences in expression of hsa-miR-21 and hsa-miR-135b among the investigated groups, we observed that they were smaller between NC and ADJ tissues (3.87-fold; 3.31-fold) than between ADJ and GC (10.39-fold; 9.04-fold) or between NC and GC (40.19-fold; 29.96-fold) tissues (Table S3). Thus, unlike that observed for the other miRNAs, the expression of hsa-miR-21 and hsa-miR-135b in ADJ is more similar to NC than to GC tissues. The similarity between these two tissues can be attributed to the fact that the NC samples are composed of gastritis and, like the ADJ tissues, are under the influence of an intense inflammatory process.
Studies have shown that IL-6 cytokines, through STAT3 activation (signal transducers and activators of transcription 3) [83-84], induce the expression of hsa-miR-21 [84-87] and hsa-miR-135b [88] oncomiRs, miRNAs that inhibit the expression of tumor suppressor genes. IL-6 is a pro-inflammatory cytokine that is expressed in gastric tissues submitted to intense or chronic inflammatory processes, such as gastritis [89-90] and GC [89-91]. These two miRNAs were found to be down-regulated in healthy gastric tissues [31, 92], where IL-6 activity is lower or non-existent [89]. Thus, we hypothesized that high expression level of hsa-miR-21 and hsa-miR-135b in gastritis samples (NC) are a consequence of high levels of IL-6 expression. This complex interaction network induced by the inflammatory process can alter the microenvironment of the healthy gastric tissue, providing an environment prone to the appearance of epigenetic alterations (e. g., miRNA deregulation) that contribute to the development of this type of tumor.
Studies demonstrated that H. pylori infection can inhibit the expression of hsa-miR-204 [52-55]. Although hsa-miR-204 demonstrates dual role (TS-miR or oncomiR) in different types of cancer [93], its ability to suppress the migration/invasion and proliferation of gastric cancer cells [54] suggests that it acts as a tumor suppressor miRNA in GC. HOXA10 [94], SOX4 [54] and IL-11 [95] are targets of this miRNA. The over-expression of these genes in GC promotes migration, invasion, proliferation and mesenchymal epithelial transition in GC cells [54, 96-99]. Curiously, Zhou et al. [54] identified over-expression of SOX4 as a consequence of down-regulation of hsa-mir-204 during H. pylori infection. Thus, the HOXA10, SOX4 and IL-11 genes may have their levels of expression increased in gastric tissues affected by H. pylori infection (e.g., chronic gastritis), as a consequence of hsa-miR-204 suppression.
Other four miRNAs analyzed in this study (up-regulated: hsa-miR-21 and -miR-135b; down-regulated: hsa-miR-148a and -miR-150) can be modulated and aid in the establishment and progression of H. pylori pathogenesis. For example, overexpression of hsa-miR-21 [56-57] and hsa-miR-135b [57-58] is induced during H. pylori infection. Hsa-miR-21 is able to alter the homeostasis of the PI3K/Akt signaling pathway, leading to imbalance between apoptosis and cell proliferation in GC [100]. The expression of hsa-miR-135b is induced by the activation of the NF-kβ signaling pathway (proinflammatory signaling), which is induced by H. pylori infection or by the activation of tumor necrosis factor alpha (TNF- α). This miRNA reduces KLF4 activity causing inhibition of apoptosis and increases resistance to cisplatin [58]. H. pylori infection is also capable of inducing the down-regulation of hsa-miR-148a [59] and hsa-miR-150 [57, 101]. Hsa-miR-148a inhibits the activity of MMP-7, a matrix metalloproteinase responsible for inducing migration and invasion of GC cells by E-cadherin cleavage [59]. Studies suggest that hsa-miR-150 is involved in the development of MALT gastric lymphoma associated with H. pylori infection by acting on DNA mismatch repair pathway [57, 101].
Our functional analysis using genes regulated by the five miRNAs (hsa-miR-21, -miR-135b, -miR-148a, -miR-150 and -miR-204) associated with H. pylori infection demonstrated that these genes are involved in important pathways for the establishment and promotion of infection and GC (e.g., MAPK, HIF-1, Cytokine-cytokine receptor interaction, PI3K-Akt, TNF, Rap1, Chemokine, NF-kappa B, Jak-STAT, ErbB, Toll-like receptor, Bacterial invasion of epithelial cells, AMPK and VEGF signaling pathways). These pathways regulate cell cycle, inflammation, immune response, migration, proliferation, growth, survival, apoptosis, differentiation, transcription and cell adhesion. In this context, H. pylori infection can modulate these pathways by creating a complex cascade of deregulation that provides an epigenetically and genetically altered microenvironment (field cancerization) useful to its establishment, which over time favors and contributes to gastric carcinogenesis.
We observed that the expression of hsa-miR-29c was higher in ADJ than in NC and GC (Figure 1; Table S3). This expression profile is very similar to that reported for healthy gastric tissues [26-27, 30-31]. Since hsa-miR-29c induces apoptosis, inhibits cell growth and invasion and suppresses metastasis in GC [102-105], we speculate that this tissue utilizes the overexpression of this TS-miR as an antitumor mechanism.
The use of FFPE samples in clinical routine has become an easy low-cost practice to preserve the histological integrity of the tissue for long periods of time. However, this method decreases the quality and integrity of nucleic acids (DNA and RNA) and some miRNAs may lose integrity and representativeness in this type of sample [50-51]. Treece et al. [106] were able to identify 13 miRNAs differentially expressed in samples preserved in FFPE of gastric adenocarcinoma and that have potential in classifying gastric cancer subtypes. We were able to identify 10 miRNAs with good representativeness in this type of samples. The ROC curve and AUC analyses identified hsa-miR-10a, -miR-21, -miR-135b, -miR-150, -miR-148a, -miR-215, -miR-483 and -miR-664a as GC biomarkers because they showed high accuracy (AUC>0.90; Figure 3) in discriminating NC tissues from other tissues (ADJ and GC). Thus, these miRNAs can be considered potentially useful as biomarkers for GC. Future studies using a larger number of patients are necessary to validate the utility of these markers, especially for the Brazilian population where GC cases are elevated.
Recently Vidal et al. [25] identified the involvement of circular RNAs in gastric field cancerization, however, further studies are needed to address and evaluate the role of these non-coding RNAs in this process. Among the ten miRNAs studied, only three (hsa-miR-150, hsa-miR-483 and hsa-miR-664a) had been validated and associated with this model of carcinogenesis [21]. Our study is significant and adds new data on the epigenetics of gastric field cancerization since we validated and demonstrated the involvement of seven new miRNAs. Thus, we demonstrate that miRNAs are closely related to field cancerization in GC and can be potentially useful as biomarkers, therapeutic targets and for the predictive purposes of occurrence and/or recurrence of this type of tumor.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
We would like to thank Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - CAPES (Bio. Computacional, No. 3381/2013); Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq; Fundação Amazônia Paraense de Amparo a Pesquisa - FAPESPA and PROPESP/UFPA for the financial support and fellowships.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ferlay J, Soerjomataram I, Dikshit R. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-86
2. Alfaro EE, Lauwers GY. Early gastric neoplasia: diagnosis and implications. Adv Anat Pathol. 2011;18:268-80
3. Correa P. Gastric cancer: Overview. Gastroenterol Clin North Am. 2013;42:211-217
4. Yakirevich E, Resnick MB. Pathology of gastric cancer and its precursor lesions. Gastroenterol Clin North Am. 2013;42:261-84
5. Papachristou DN, Fortner JG. Local recurrence of gastric adenocarcinomas after gastrectomy. J Surg Oncol. 1981;18:47-53
6. D'Angelica M, Gonen M, Brennan MF. et al. Patterns of initial recurrence in completely resected gastric adenocarcinoma. Ann Surg. 2004;240:808-16
7. Wang SY, Yeh CN, Lee HL. et al. Clinical impact of positive surgical margin status on gastric cancer patients undergoing gastrectomy. Ann Surg Oncol. 2009;16:2738-43
8. Waddell T, Verheij M, Allum W. et al. Gastric cancer: ESMO-ESSO-ESTRO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Annals of Oncology. 2013;24(Suppl6):vi57-vi63
9. Nakajima T, Maekita T, Oda I. et al. Higher methylation levels in gastric mucosae significantly correlate with higher risk of gastric cancers. Cancer Epidemiol Biomarkers Prev. 2006;15:2317-2321
10. Bernstein C, Bernstein H, Payne CM. et al. Field defects in progression to gastrointestinal tract cancers. Cancer Lett. 2008;260:1-10
11. Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963-8
12. Braakhuis BJ, Tabor MP, Kummer JA. et al. A genetic explanation of Slaughter's concept of field cancerization: evidence and clinical implications. Cancer Res. 2003;63:1727-30
13. Chai H, Brown RE. Field effect in cancer-an update. Ann Clin Lab Sci. 2009;39:331-7
14. Rivenbark AG, Coleman WB. Field cancerization in mammary carcinogenesis - Implications for prevention and treatment of breast cancer. Exp Mol Pathol. 2012;93:391-8
15. Dotto GP. Multifocal epithelial tumors and field cancerization: stroma as a primary determinant. J Clin Invest. 2014;124:1446-53
16. Hawthorn L, Lan L, Mojica W. Evidence for field effect cancerization in colorectal cancer. Genomics. 2014;103:211-21
17. Mohan M, Jagannathan N. Oral field cancerization: an update on current concepts. Oncol Rev. 2014;8:244
18. Patel A, Tripathi G, Gopalakrishnan K. et al. Field cancerisation in colorectal cancer: A new frontier or pastures past? World J Gastroenterol. 2015;21:3763-3772
19. Baba Y, Ishimoto T, Kurashige J. et al. Epigenetic field cancerization in gastrointestinal cancers. Cancer Lett. 2016;375:360-6
20. Ivanauskas A, Hoffmann J, Jonaitis LV. et al. Distinct TPEF/HPP1 gene methylation patterns in gastric cancer indicate a field effect in gastric carcinogenesis. Dig Liver Dis. 2008;40:920-6
21. Assumpção MB, Moreira FC, Hamoy IG. et al. High-Throughput miRNA Sequencing Reveals a Field Effect in Gastric Cancer and Suggests an Epigenetic Network Mechanism. Bioinform Biol Insights. 2015;9:111-7
22. Takeshima H, Niwa T, Takahashi T. et al. Frequent involvement of chromatin remodeler alterations in gastric field cancerization. Cancer Lett. 2015;357:328-38
23. Tahara T, Shibata T, Kawamura T. et al. Telomere length shortening in gastric mucosa is a field effect associated with increased risk of gastric cancer. Virchows Arch. 2016;469:19-24
24. Yoshida S, Yamashita S, Niwa T. et al. Epigenetic inactivation of FAT4 contributes to gastric field cancerization. Gastric Cancer. 2017;20:136-145
25. Vidal AF, Ribeiro-dos-Santos AM, Vinasco-Sandoval T. et al. The comprehensive expression analysis of circular RNAs in gastric cancer and its association with feld cancerization. Scientific Reports. 2017;7:14551
26. Ribeiro-dos-Santos Â, Khayat AS, Silva A. et al. Ultra-deep sequencing reveals the microRNA expression pattern of the human stomach. PLoS One. 2010;5:e13205
27. Moreira FC, Assumpção M, Hamoy IG. et al. MiRNA Expression Profile for the Human Gastric Antrum Region Using Ultra-Deep Sequencing. PLoS One. 2014;9:e92300
28. Gomes LL, Moreira FC, Hamoy IG. et al. Identification of miRNAs Expression Profile in Gastric Cancer Using Self-Organizing Maps (SOM). Bioinformation. 2014;10:246-50
29. Zhang Z, Li Z, Li Y. et al. MicroRNA and signaling pathways in gastric cancer. Cancer Gene Ther. 2014;21:305-16
30. Darnet S, Moreira FC, Hamoy IG. et al. High-Throughput Sequencing of miRNAs Reveals a Tissue Signature in Gastric Cancer and Suggests Novel Potential Biomarkers. Bioinform Biol Insights. 2015;9:1-8
31. Vidal AF, Cruz AMP, Magalhães L. et al. hsa-miR-29c and hsa-miR-135b differential expression as potential biomarker of gastric carcinogenesis. World J Gastroenterol. 2016;22:2060-2070
32. Schneider MR. MicroRNAs as novel players in skin development, homeostasis and disease. Br J Dermatol. 2012;166:22-8
33. Danger R, Braza F, Giral M. et al. MicroRNAs, Major Players in B Cells Homeostasis and Function. Front Immunol. 2014;5:98
34. Runtsch MC, Round JL, O'Connell RM. MicroRNAs and the regulation of intestinal homeostasis. Front Genet. 2014;5:347
35. Vickers KC, Landstreet SR, Levin MG. et al. MicroRNA-223 coordinates cholesterol homeostasis. Proc Natl Acad Sci USA. 2014;111:14518-23
36. Moresi V, Marroncelli N, Coletti D. et al. Regulation of skeletal muscle development and homeostasis by gene imprinting, histone acetylation and microRNA. Biochim Biophys Acta. 2015;1849:309-16
37. Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642-55
38. Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. Int J Genomics. 2014;2014:970607
39. Wu HH, Lin WC, Tsai KW. Advances in molecular biomarkers for gastric cancer: miRNAs as emerging novel cancer markers. Expert Rev Mol Med. 2014;16:e1
40. Song J, Bai Z, Zhang Z. MicroRNAs are implicated in the initiation and progression of gastric cancer. Chin Med J (Engl). 2014;127:554-9
41. Jiang C, Chen X, Alattar M. et al. MicroRNAs in tumorigenesis, metastasis, diagnosis and prognosis of gastric cancer. Cancer Gene Ther. 2015;22:291-301
42. Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. an attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31-49
43. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101-8
44. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550
45. Chou CH, Chang NW, Shrestha S. et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44:D239-47
46. Vogelstein B, Papadopoulos N, Velculescu VE. et al. Cancer genome landscapes. Science. 2013;339:1546-58
47. Hamberg M, Backes C, Fehlmann T. et al. MiRTargetLink-miRNAs, Genes and Interaction Networks. Int J Mol Sci. 2016;17:564
48. Szklarczyk D, Morris JH, Cook H. et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362-D368
49. Santos NP, Ribeiro-Rodrigues EM, Ribeiro-Dos-Santos AK. et al. Assessing individual interethnic admixture and population substructure using a 48-insertion-deletion (INSEL) ancestry-informative marker (AIM) panel. Hum Mutat. 2010;31:184-90
50. Kakimoto Y, Tanaka M, Kamiguchi H, Ochiai E, Osawa M. MicroRNA Stability in FFPE Tissue Samples: Dependence on GC Content. PLoS One. 2016;11:e0163125
51. Zhang P, Lehmann BD, Shyr Y, Guo Y. The Utilization of Formalin Fixed-Paraffin-Embedded Specimens in High Throughput Genomic Studies. Int J Genomics. 2017;2017:1926304
52. Matsushima K, Isomoto H, Inoue N. et al. MicroRNA signatures in Helicobacter pylori-infected gastric mucosa. Int J Cancer. 2011;128:361-70
53. Shiotani A, Uedo N, Iishi H. et al. H. pylori eradication did not improve dysregulation of specific oncogenic miRNAs in intestinal metaplastic glands. J Gastroenterol. 2012;47:988-98
54. Zhou X, Li L, Su J. et al. Decreased miR-204 in H. pylori-associated gastric cancer promotes cancer cell proliferation and invasion by targeting SOX4. PLoS One. 2014;9:e101457
55. Chang H, Kim N, Park JH. et al. Different microRNA expression levels in gastric cancer depending on Helicobacter pylori infection. Gut Liver. 2015;9:188-96
56. Zhang Z, Li Z, Gao C. et al. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest. 2008;88:1358-66
57. Floch P, Capdevielle C, Staedel C. et al. Deregulation of MicroRNAs in Gastric Lymphomagenesis Induced in the d3Tx Mouse Model of Helicobacter pylori Infection. Front Cell Infect Microbiol. 2017;7:185
58. Shao L, Chen Z, Soutto M. et al. Helicobacter pylori-induced miR-135b-5p promotes cisplatin resistance in gastric cancer. FASEB J. 2018;9:fj201701456RR
59. Yang Y, Li X, Du J, Yin Y, Li Y. Involvement of microRNAs-MMPs-E-cadherin in the migration and invasion of gastric cancer cells infected with Helicobacter pylori. Experimental Cell Research. 2018;367:196-204
60. Wang LP, Ma XQ, Cai JC. Clinicopathological significance and function of miR-135b in the occurrence and development of gastric cancer. Zhonghua Yi Xue Za Zhi. 2012;92:3269-73
61. Li N, Zhang QY, Zou JL. et al. miR-215 promotes malignant progression of gastric cancer by targeting RUNX1. Oncotarget. 2016;7:4817-28
62. Koshiishi N, Chong JM, Fukasawa T. et al. p300 gene alterations in intestinal and diffuse types of gastric carcinoma. Gastric Cancer. 2004;7:85-90
63. Kim MS, Lee SH, Yoo NJ. et al. Frameshift mutations of tumor suppressor gene EP300 in gastric and colorectal cancers with high microsatellite instability. Hum Pathol. 2013;44:2064-70
64. Wei D, Gong W, Kanai M. et al. Drastic down-regulation of Krüppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res. 2005;65:2746-54
65. Zhang N, Zhang J, Wang ZW. et al. Altered expression of Krüppel-like factor 4 and β-catenin in human gastric cancer. Oncol Lett. 2012;3:1017-1022
66. Fei G, Ebert MP, Mawrin C. et al. Reduced PTEN expression in gastric cancer and in the gastric mucosa of gastric cancer relatives. Eur J Gastroenterol Hepatol. 2002;14:297-303
67. Xu WT, Yang Z, Lu NH. Roles of PTEN (Phosphatase and Tensin Homolog) in gastric cancer development and progression. Asian Pac J Cancer Prev. 2014;15:17-24
68. Ferraz MA, Zabaglia LM, Pereira WN. et al. Downregulated Expression of E-cadherin and TP53 in Patients with Gastric Diseases: the Involvement of H. pylori Infection and Its Virulence Markers. J Gastrointest Cancer. 2016;47:20-6
69. Deng Y, Huang Z, Xu Y. et al. MiR-215 modulates gastric cancer cell proliferation by targeting RB1. Cancer Lett. 2014;342:27-35
70. Wu Y, Li Q, Zhou X. et al. Decreased levels of active SMAD2 correlate with poor prognosis in gastric cancer. PLoS One. 2012;7:e35684
71. Wang C, Li Y, Zhang H. et al. Oncogenic PAK4 regulates Smad2/3 axis involving gastric tumorigenesis. Oncogene. 2014;33:3473-84
72. Wang LH, Kim SH, Lee JH. et al. Inactivation of SMAD4 tumor suppressor gene during gastric carcinoma progression. Clin Cancer Res. 2007;13:102-10
73. Leng A, Liu T, He Y. et al. Smad4/Smad7 balance: a role of tumorigenesis in gastric cancer. Exp Mol Pathol. 2009;87:48-53
74. Wu Q, Jin H, Yang Z. et al. MiR-150 promotes gastric cancer proliferation by negatively regulating the pro-apoptotic gene EGR2. Biochem Biophys Res Commun. 2010;392:340-5
75. Guo S-L, Peng Z, Yang X. et al. miR-148a Promoted Cell Proliferation by Targeting p27 in Gastric Cancer Cells. Int J Biol Sci. 2011;7:567-574
76. Jin Z, Selaru FM, Cheng Y. et al. MicroRNA-192 and -215 are upregulated in human gastric cancer in vivo and suppress ALCAM expression in vitro. Oncogene. 2011;30:1577-1585
77. Chen W, Tang Z, Sun Y. et al. miRNA expression profile in primary gastric cancers and paired lymph node metastases indicates that miR-10a plays a role in metastasis from primary gastric cancer to lymph nodes. Exp Ther Med. 2012;3:351-356
78. Zhang BG, Li JF, Yu BQ. et al. microRNA-21 promotes tumor proliferation and invasion in gastric cancer by targeting PTEN. Oncol Rep. 2012;27:1019-26
79. Xu YJ, Fan Y. MiR-215/192 participates in gastric cancer progression. Clin Transl Oncol. 2015;17:34-40
80. Li L, Zhou L, Li Y. et al. MicroRNA-21 stimulates gastric cancer growth and invasion by inhibiting the tumor suppressor effects of programmed cell death protein 4 and phosphatase and tensin homolog. J Buon. 2014;19:228-36
81. Wu K, Ma L, Zhu J. miR-483-5p promotes growth, invasion and self-renewal of gastric cancer stem cells by Wnt/β-catenin signaling. Mol Med Rep. 2016;14:3421-8
82. Assumpção PP, Dos Santos SE, Dos Santos ÂK. et al. The adjacent to tumor sample trap. Gastric Cancer. 2016;19:1024-5
83. Hodge DR, Hurt EM, Farrar WL. The role of IL-6 and STAT3 in inflammation and cancer. Eur J Cancer. 2005;41:2502-12
84. Löffler D, Brocke-Heidrich K, Pfeifer G. et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330-1333
85. Iliopoulos D, Jaeger SA, Hirsch HA. et al. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493-506
86. Rozovski U, Calin GA, Setoyama T. et al. Signal transducer and activator of transcription (STAT)-3 regulates microRNA gene expression in chronic lymphocytic leukemia cells. Mol Cancer. 2013;12:50
87. Singh M, Garg N, Venugopal C. et al. STAT3 pathway regulates lung-derived brain metastasis initiating cell capacity through miR-21 activation. Oncotarget. 2015;6:27461-77
88. Matsuyama H, Suzuki HI, Nishimori H. et al. miR-135b mediates NPM-ALK-driven oncogenicity and renders IL-17-producing immunophenotype to anaplastic large cell lymphoma. Blood. 2011;118:6881-92
89. Kinoshita H, Hirata Y, Nakagawa H. et al. Interleukin-6 mediates epithelial-stromal interactions and promotes gastric tumorigenesis. PLoS One. 2013;8:e60914
90. Rossi AF, Cadamuro AC, Biselli-Périco JM. et al. Interaction between inflammatory mediators and miRNAs in Helicobacter pylori infection. Cell Microbiol. 2016;18:1444-5
91. Wang Z, Si X, Xu A. et al. Activation of STAT3 in Human Gastric Cancer Cells via Interleukin (IL)-6-Type Cytokine Signaling Correlates with Clinical Implications. PLoS One. 2013;8:e75788
92. Link A, Schirrmeister W, Langner C. et al. Differential expression of microRNAs in preneoplastic gastric mucosa. Scientific Reports. 2015;5:8270
93. Li T, Pan H, Li R. The dual regulatory role of miR-204 in cancer. Tumour Biol. 2016;37:11667-11677
94. Garzon R, Garofalo M, Martelli MP. et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci USA. 2008;105:3945-50
95. Pollari S, Leivonen SK, Perälä M. et al. Identification of microRNAs inhibiting TGF-β-induced IL-11 production in bone metastatic breast cancer cells. PLoS One. 2012;7:e37361
96. Nakayama T, Yoshizaki A, Izumida S. et al. Expression of interluekin-11 (IL-11) and IL-11 receptor alpha in human gastric carcinoma and IL-11 upregulates the invasive activity of human gastric carcinoma cells. Int J Oncol. 2007;30:825-33
97. Zhang Z, Zhang J, Miao L. et al. Interleukin-11 promotes the progress of gastric carcinoma via abnormally expressed versican. Int J Biol Sci. 2012;8:383-93
98. Peng X, Liu G, Peng H. SOX4 contributes to TGF-b-induced epithelialemesenchymal transition and stem cell characteristics of gastric cancer cells. Genes & Diseases. 2018;5:49-61
99. Han Y, Lu S, Wen YG. et al. Overexpression of HOXA10 promotes gastric cancer cells proliferation and HOXA10(+)/CD44(+) is potential prognostic biomarker for gastric cancer. Eur J Cell Biol. 2015;94:642-52
100. Noto JM, Peek RM. The role of microRNAs in Helicobacter pylori pathogenesis and gastric carcinogenesis. Front Cell Infect Microbiol. 2012;1:21
101. Santos JC, Brianti MT, Almeida VR. et al. Helicobacter pylori infection modulates the expression of miRNAs associated with DNA mismatch repair pathway. Mol Carcinog. 2017;56:1372-1379
102. Matsuo M, Nakada C, Tsukamoto Y. et al. MiR-29c is downregulated in gastric carcinomas and regulates cell proliferation by targeting RCC2. Mol Cancer. 2013;12:15
103. Saito Y, Suzuki H, Imaeda H. et al. The tumor suppressor microRNA-29c is downregulated and restored by celecoxib in human gastric cancer cells. Int J Cancer. 2013;132:1751-60
104. Gong J, Li J, Wang Y. et al. Characterization of microRNA-29 family expression and investigation of their mechanistic roles in gastric cancer. Carcinogenesis. 2014;35:497-506
105. Wang Y, Liu C, Luo M. et al. Chemotherapy-Induced miRNA-29c/Catenin-δ Signaling Suppresses Metastasis in Gastric Cancer. Cancer Res. 2015;75:1332-44
106. Treece AL, Duncan DL, Tang W. et al. Gastric adenocarcinoma microRNA profiles in fixed tissue and in plasma reveal cancer-associated and Epstein-Barr virus-related expression patterns. Lab Invest. 2016;96:661-71
Author contact
![]() Corresponding author: Ândrea Kely Campos Ribeiro-dos-Santos, PhD., Laboratory of Human and Medical Genetics, Institute of Biological Sciences, Federal University of Pará, Augusto Corrêa Avenue, 66075-110, Belém, Pará, Brazil. Telephone: +55-91-32017843; Fax: +55-91-32017843; E-mail: akelyufpacom
Corresponding author: Ândrea Kely Campos Ribeiro-dos-Santos, PhD., Laboratory of Human and Medical Genetics, Institute of Biological Sciences, Federal University of Pará, Augusto Corrêa Avenue, 66075-110, Belém, Pará, Brazil. Telephone: +55-91-32017843; Fax: +55-91-32017843; E-mail: akelyufpacom

 Global reach, higher impact
Global reach, higher impact