3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(15):3420-3426. doi:10.7150/jca.31087 This issue Cite
Research Paper
Combination of PET and CXCR4-Targeted Peptide Molecule Agents for Noninvasive Tumor Monitoring
1. Department of Radiology, The Third Affiliated Hospital of Wenzhou Medical University, Wenzhou, Zhejiang, P.R. China
2. Department of Thyroid and Breast Surgery, The Third Affiliated Hospital of Wenzhou Medical University, Wenzhou, Zhejiang, P.R. China
3. Department of Internal Medicine, Clinic of August First Film Studio,301 Hospital, NO.1 Liuli Bridge, Beijing, China
Received 2018-10-30; Accepted 2019-4-25; Published 2019-6-9
Abstract
Precision medicine is emphasizing not only at individual but also at disease molecule level in modern medicine. Therefore, target-specific molecular agents are crucial for precise diagnosis and treatment. We developed a peptide agent that binds a critical chemokine receptor-CXCR4 and could be used to detect tumor status. Confocal images showed binding of the peptide agent to human osteosarcoma cells. Clinical gold-standard molecular imaging agent PET showed tumors had high glucose metabolism, CT showed that these xenograft tumors were calcified and displayed hypervascularity. Peptide imaging demonstrated that these tumors were CXCR4 positive. However, Western blot protein analysis revealed a discordance between the tumor and the CXCR4 targeted agent, suggesting that small changes in peptide sequences have profound effect on binding to their targets. We also demonstrated the molecular screening by modifying the peptide sequence and thereby altering the binding properties of the agent. In conclusion, this study demonstrates that small molecule peptide agents can be used as an additional diagnostic tool for precision medicine.
Keywords: optical imaging, peptide imaging, CXCR4, osteosarcoma, molecular imaging
Introduction
Personalized medicine promises to deliver individualized treatments that are tailored to the genetics, RNA, proteins, and metabolites of each patient's disease [1]. These advances in personalized medicine have the potential to increase the quality of patient care for numerous diseases. For cancer chemotherapy, precision medicine entails administering target-specific agents, including microRNA and antibodies, to a target-positive disease at properly timed and effective biological doses while minimizing systemic toxicity. Successful therapy requires an understanding of ongoing changes as tumors progress and respond to treatments [2]. Therefore, it is crucial to develop new tools, such as noninvasive molecular imaging, for acquiring accurate information about the tumor status [3].
Labeling target-specific agents with imaging reporters offers a tool for repeated and noninvasive evaluation of tumor status. Currently, the molecular imaging agent most commonly used in clinical practice is 18F-fluoro-deoxy-glucose (18F-FDG). Therefore, it is crucial to develop molecular imaging agents that match the targets of precision medicine.
Among the molecular targets, cytokines and chemokines are important intercellular signals in inflammation, immunity, and tumor and endothelial cell processes. These factors have therapeutic potential when exogenously delivered either alone or in combination with biological or chemotherapeutic agents [4-6]. CXCR4 is a chemokine receptor that belongs to the large family of G protein-coupled receptors. CXCR4 plays a crucial role in a number of biological processes, including the trafficking and homeostasis of immune cells such as T lymphocytes. In addition, CXCR4 has been reported to be a prognostic marker in various types of cancer, including leukemia, lung, breast, stomach, pancreas, and prostate cancer [7-11]. CXCR4 is highly expressed on cancer stem cells and plays a crucial role in metastasis to the brain, lymph nodes, lungs, liver, and bone [11, 12]. The metastatic functions are reported through CXCL12 and its receptor CXCR4 [13-18]. Therefore, CXCR4 could be a therapeutic target for tumor progression, angiogenesis, and metastasis, and CXCR4 targeted treatments have the potential to improve overall patient survival [19].
In this study, we developed a peptide-based, CXCR4-targeted agent labeled with a near-infrared (NIR) dye. In cell based studies, we demonstrated that these peptide agents bind to CXCR4 positive human osteosarcoma cells. Whole-body optical imaging demonstrated high NIR signal intensity within CXCR4-positive tumors and CXCR4-positive tissues, suggesting that synthetic peptide agents could be used to evaluate CXCR4 chemokine receptor status in preclinical studies. Furthermore, we demonstrate that target-specific molecular agents can be used with current imaging technologies (18F-FDG PET, CT, or MRI) to noninvasively monitor disease status. The techniques developed here may be useful in developing additional targeted therapeutic agents for personalized medicine.
Results
NIR-labeled CXCR4 agent binding
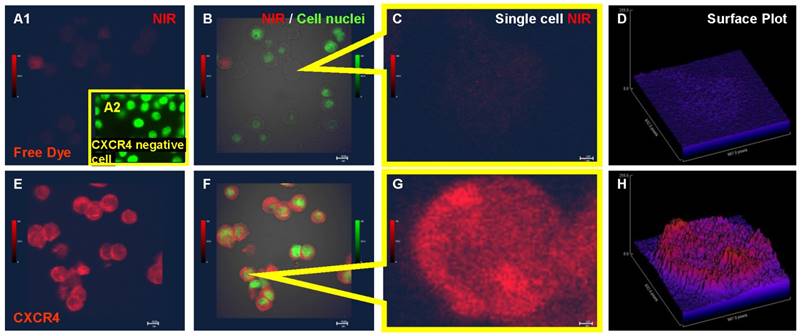
In an in vitro binding assay, no detectable NIR signal was emitted from the Saos-2 cells incubated with free NIR dye (Fig. 1A1, B and C) and the NIR signal intensity was near background levels in a quantitative 3D plot (Fig. 1D). There was no detectable signal when CXCR4 agent co-incubated with CXCR4 negative nasal cancer cell SUNE-1 (Fig. 1A2). In contrast, the NIR-labeled CXCR4 agent bound to the all of the osteosarcoma cells when processed in parallel (Fig. 1E-F). Merged images of the NIR signal, the cell nuclei, and the differential interference contrast (DIC) confirmed that the NIR signal did not colocalize with the cell nucleus (Fig. 1F). The uneven intensity of the NIR signal in single cell images and in corresponding quantitative signal intensity plots suggests that the peptide agent may bind to CXCR4 in specific compartments within the cell (Fig. 1G-H).
Confocal images demonstrating uptake of the CXCR4 peptide agent by human osteosarcoma cells. A. Saos-2 cells incubated with free near-infrared (NIR) dye. B. Merged image of the NIR signal, cell nuclei, and bright field shows cell morphology and lack of NIR signal. C. High-magnification image of free NIR dye uptake by a single cell. D. Quantitative 3D plot of the NIR signal intensity showing free dye signal near background levels. E. NIR-labeled CXCR4 agent binds to Saos-2 osteosarcoma cells. F. Merged image of the NIR signal on the CXCR4 agent, cell nuclei, and bright field. G. High-magnification image of a single cell binding to the NIR-labeled CXCR4 agent. H. Quantitative 3D plot of the NIR signal showing the CXCR4 agent bound to a single cell.
In vivo molecular imaging
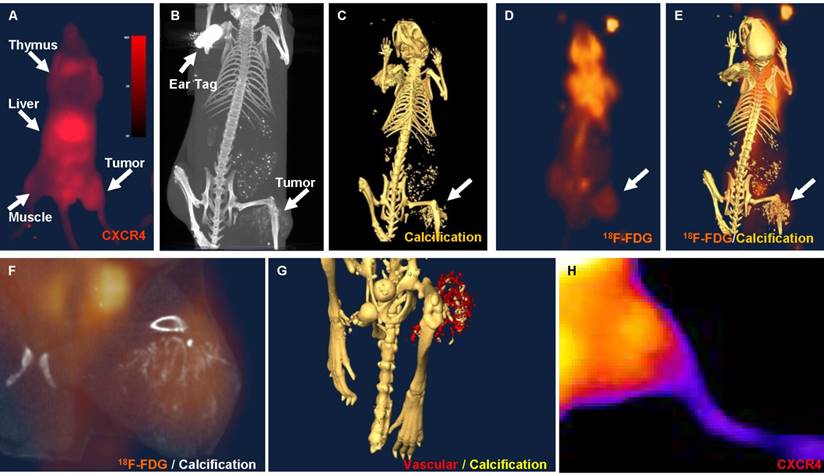
Using the NIR-labeled CXCR4 agent, an increase in NIR signal intensity in osteosarcomas xenografts could be detected in subcutaneous model as early as 7 days after the inoculation of Saos-2 cells. NIR imaging illustrates the binding of the CXCR4 agent within the tumor, as well as known CXCR4-positive tissues, including the thymus and liver (Fig. 2A). Tumor-to-background ratios ranged from 1.01 to 1.75 during a 48-hour period (n=8). Whole-body CT imaging confirmed the size and location of the tumor (Fig. 2B). Skeletal CT imaging demonstrated calcification of the tumors and revealed that the bony component of the tumor had invaded beyond the tumor mass (Fig. 2C). 18F-FDG PET imaging demonstrated high glucose metabolism within the center of the tumor (Fig. 2D). Merged 18F-FDG PET and skeletal CT images illustrated the anatomical relationship between the tumor, glucose uptake, and calcification (Fig. 2E-F). Merged vasculature contrast and skeletal CT images show the hypervascularity of the tumor (Fig. 2G). Finally, high-magnification optical NIR images demonstrate the binding strength of the CXCR4 agent within the tumor (Fig. 2H).
Peptide sequence alteration analysis
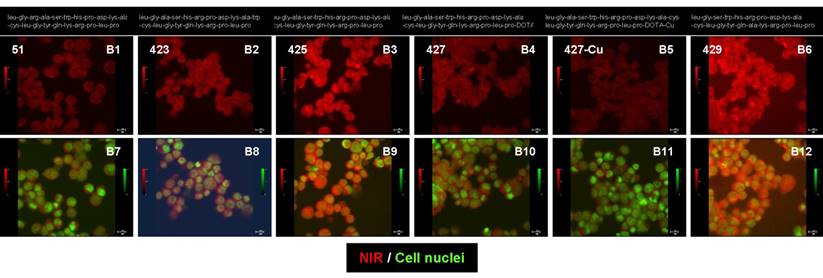
The sequence of the NIR-labeled CXCR4 binding peptide (agent 425) was altered and these new agents were compared in a cell-binding assay (Figure 3). The optical signal intensities of the different peptide sequences and structural alterations are presented at the same scale (B1 to B6) and merged with cell nuclei (B7 to B12). Agents 51 (B1 and B7), 423 (B2 and B8), and 429 (B6 and B12) are peptides with different sequences from agent 425 (B3 and B9). Agent 425 was modified with a radio-chelator to generate agent 427 (B4 and B10) and then further coupled with an isotope to produce agent 427-cu (B5 and B11). The altered cell binding of these agents confirms that changing the peptide sequence, adding a radio-chelator, or coupling to an isotope affects the agent binding and signal intensity of the NIR label.
In vivo images of osteosarcoma xenografts in nude mice. A. NIR image showing the distribution of CXCR4 agent in the thymus, liver, and tumor. B. Whole-body computed tomography (CT) image showing the location of the tumor. C. CT image of the skeleton demonstrating calcification in the tumor region (arrow). D. 18F-fluoro-deoxy-glucose positron emission tomography (18F-FDG PET) image showing high glucose metabolism in the tumor region (arrow). E. Merged CT and 18F-FDG PET images showing the anatomical distribution of the 18F-FDG-PET signal. F. High-magnification of merged CT and 18F-FDG PET images showing calcification and high glucose metabolism in the tumor region. G. Merged skeletal CT images and images taken after the addition of vasculature contrast showing hypervascularity and calcification of the tumor region. H. High-magnification optical NIR imaging showing high signal intensity in the tumor region.
CXCR4 expression in tissues
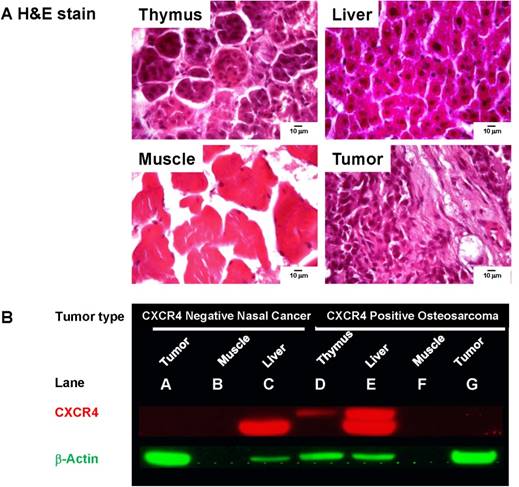
To validate our whole-body imaging results, we dissected the organs exhibiting high NIR signal intensity (thymus, liver, and tumor) and muscle tissue as a negative control. Hematoxylin and eosin (H&E) staining confirmed the tissue origination (Fig. 4A). As an additional negative control, we harvested tissues from mice bearing CXCR4-negative nasal tumors. By Western blot analysis, high CXCR4 expression was detected in the liver of both CXCR4-negative and positive tumor xenografts (Fig. 4B, lanes C and E). Surprisingly, the CXCR4 antibody did not detect CXCR4 expression in either tumor lysate (lanes A and G) or the muscle negative control (lanes B and F). However, a higher molecular weight CXCR4 protein was detected only in the thymus and liver of mice bearing the CXCR4-positive tumor xenograft (lanes D and E), suggesting that the peptide-imaging agent and CXCR4 antibody have species- and tissue-specific binding properties.
Discussion
One of the current goals of our research is to develop molecular imaging agents as tools to validate disease targets. Such direct visualization could be used to evaluate disease markers and provide the information necessary for developing personalized treatment plans using biological target-specific therapeutic agents. CXCR4 is an important therapeutic target in many diseases and several CXCR4 antagonists are being evaluated clinically [13, 15, 20-33]. Therefore, development of a noninvasive method for evaluating and monitoring biological changes in CXCR4 status is vital for both preclinical studies and ongoing clinical trials. Using a human osteosarcoma model, we validated a readily produced peptide agent targeting CXCR4. Using confocal microscopy, we demonstrated that the target-specific component of the CXCR4 peptide agent is important for tumor cell binding, as the NIR dye alone did not bind cells. In vivo optical imaging confirmed high NIR signal intensity within the CXCR4-positive tumor and within CXCR4 receptor-positive organs. Furthermore, we demonstrated that small molecule, target-specific peptide agents in combination with advanced structural, metabolic, and vasculature imaging technologies can be used to define disease status in vivo.
A. Pathology stains verify tissues originating from thymus, liver, muscle, and xenograft tumor. B. CXCR4 expression was probed in tissue lysates from mice inoculated with CXCR4 negative nasal cancer cells and CXCR4-positive Saos-2 cells. No CXCR4 was detected in the CXCR4-negative tumor (A), CXCR4-positive tumor (G), or muscle tissue from tumor-bearing mice (B, F). Low molecular weight CXCR4 was detected in the liver of tumor-bearing mice (C and E). High molecular weight CXCR4 was only detected in the thymus (D) and liver (E) of a mouse bearing the CXCR4 positive tumor. β-actin was used as protein loading control.
Peptide sequence and structural changes altered the binding of NIR-labeled CXCR4 agents to the Saos-2 cells in vitro. The NIR signal intensities are presented at the same scale (B1 to B6) and merged with cell nuclei (B7 to B12) for each of the peptide agents.
The largest challenge for this study was in validating CXCR4 expression in tumor lysates by Western blotting. We selected the antibody used in this study based on its well-established use, published results [34], and availability. Western blot analysis clearly demonstrated CXCR4 expression in the mouse liver, but did not detect any CXCR4 in the tumors. However, high molecular weight CXCR4 was only detected in the thymus and liver from the mouse with the CXCR4-positive Saos-2 xenograft. This high molecular weight form of CXCR4 was not detected in tissues from the mouse bearing the CXCR4-negative nasal tumor. Results from additional CXCR4 antibodies suggested that the CXCR4 positive osteosarcoma stimulates the host thymus and may produce another form of CXCR4 protein not detected by the currently available CXCR4 antibodies. Despite the failure of the antibody validation step, our peptide-imaging agent has been used to detect CXCR4 in several other disease models, such as leukemia and liver cancer (data not shown). The findings of our Western blot analyses highlight the challenges associated with antibody-based agents and suggest that each target-specific agent exhibits selective binding in different animal models [35-37].
We developed several dozen agents with different sequences and structures, but only presented four modified CXCR4 agents here. The CXCR4 peptide agent used for the in vivo studies exhibited the most conserved binding affinity for diverse human disease models. The binding data from the complete set of peptide agents will be summarized in a journal that focuses on chemistry. The binding affinity and signal intensity for these peptide agents varied for each of the cell lines and tumor models studied, confirming that minor structural modifications significantly affect the binding affinity of peptide agents [38]. By including a NIR label on the CXCR4 peptide agent, we were able to combine NIR/single photon emission CT (NIR/ SPECT) and NIR/PET imaging for evaluating the progression of the osteosarcoma xenograft. The results presented here confirm that NIR-labeled peptide agents can be easily modified for disease target specificity and have the potential to be utilized in precision medicine for noninvasive imaging of molecular markers.
Materials and Methods
CXCR4 agent synthesis
(NIR dye- leu-gly-ala-ser-trp-his-arg-pro-asp-lys-ala-cys-leu-gly-tyr-gln-lys-arg-pro-leu-pro-NH2): d-amino-peptide LGASWHRPDKACLGYQKRPLP (DVI-C11A [39]) was prepared following standard procedures for Fmoc solid-phase peptide synthesis on a Symphony synthesizer (Protein Technologies, Inc., Tucson, AZ). The conjugation of NIR-dye-NHS [40] to the peptide was carried out on the solid support in presence of N, N-diisopropylethylamine (10%) in DMF. The compound was cleaved from the support using trifluoroacetic acid/water/triethylsilane (94/2/4) and purified by reverse phase HPLC. The product was validated by mass and analytic HPLC. Matrix- assisted laser desorption/ionization (MALDI) for C153H221N36O33S4+ calculated as 3218.56 [M]+, and found to be 3218.56; HPLC (water and acetonitrile containing 0.1% TFA from10 to 80% in 30 min) retention time was 18.41 min and purity was over 95%. The alternate peptide sequences used in the cell-binding assay were synthesized in the same manner and the sequences were presented in the results.
Tumor cell lines
Saos-2, a metastatic human osteosarcoma cancer cell line that expresses high levels of CXCR4, was purchased from the American Type Culture Collection (ATCC, Manassas, VA). CXCR4 negative nasal cancer cell line SUNE-1 was a kind gift from Sun Yat-sen University. Both cells were cultured in Dulbecco's Modified Eagle's medium supplemented with high glucose and F12 nutrient (Invitrogen, Carlsbad, CA) and 10% fetal bovine serum (HyClone; Thermo Fisher Scientific Inc., Waltham, MA) in a humidified incubator maintained at 37°C with 5% CO2.
Tumor xenografts
To generate single cells suspensions, Saos-2 cells cultured near confluence were incubated with 0.05% trypsin-EDTA buffer, followed by centrifugation at 130 × g for 5 min. The cell pellet was resuspended in sterile PBS, and then approx. One million tumor cells were implanted subcutaneously into the hind region of four- to six-week-old male athymic nude mice (18-22 g) (Harlan Sprague Dawley, Inc., Indianapolis, IN). Before and after surgery, the mice were housed in a pathogen-free mouse colony and given sterilized pellet chow and sterilized water.
Fluorescent confocal imaging
Cultured tumor cells were harvested and incubated with NIR-labeled CXCR4 agent or NIR free dye for 60 min at 37ºC. To fix the cells and stain the nuclei, cells were then washed in PBS and incubated with Sytox green (Molecular Probes; Invitrogen) diluted in 95% ethanol for 15 min at 4ºC. Stained cells were mounted onto microscope slides and signal intensities of labeled cells were recorded from one z-stack slice on an Olympus confocal microscope (FluoView 1000; Olympus America, Center Valley, PA). Sytox green signals (ex/em 488/510 nm) were pseudocolored green; CXCR4 agent or NIR dye signals (ex/em 765/810 nm) were pseudocolored red.
In vivo imaging
Five nmol NIR-labeled CXCR4 was injected via the tail vein into nude mice bearing human osteosarcoma xenografts. Each mouse was imaged for up to 3 days after the injection with a Bruker In-Vivo Multispectral System FX Pro instrument (Bruker Preclinical Imaging, Billerica, MA). For comparative purposes, all images were captured over time using the same standard protocol and were saved as uncompressed 16-bit tagged image file format (TIFF) images for further analysis. Tumor vasculature was assessed by CT imaging (Siemens MicroCAT II; Siemens Medical Solutions, Malvern, PA) after the injection of Ominipaque (GE Healthcare, Princeton, NJ). The glucose metabolic status of tumors was determined by PET imaging (Siemens Inveon) after the injection of 18F-FDG (Cyclotope, Houston, TX). ImageJ (National Institutes of Health, Bethesda, MD) and Amira (Visage Imaging, Inc., Carlsbad, CA) were used for three-dimensional (3D) reconstructions, volume rendering, and tumor-to-background ratio analysis.
Western blot analysis
Tissues were homogenized in cold RIPA buffer with protease inhibitor cocktails (Sigma-Aldrich, St. Louis, MO). Tissue debris was removed by centrifugation. Equal amounts of 1× Laemmli sample buffer (Bio-Rad Laboratories, Hercules, CA) were added to the samples, and the mixtures were heated to 95°C. The samples were then subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose membranes. Finally, each membrane was incubated with anti-human CXCR4 antibody according to the manufacture instructions (R&D Systems, Minneapolis, MN) for 1 h and developed by using the ECL chemiluminescence detection kit (GE Healthcare Life Sciences, Piscataway, NJ). Membranes were incubated with β-actin antibody (Sigma-Aldrich, St. Louis, MO) as a loading control.
Abbreviations
CT: Computed tomography; FDG or 18F-FDG: fluoro-deoxy-glucose; MRI: magnetic resonance imaging; NIR: near-infrared; PET: positron emission tomography; SDS-PAGE: sodium dodecyl sulfate polyacrylamide gel electrophoresis; ex: excitation; em: emission; nm: nanometer; DIC: differential interference contrast; GAPDH: glyceraldehyde 3-phosphate dehydrogenases; H&E: hematoxylin and eosin; SPECT: single photo emission computed tomography.
Acknowledgements
We thank BioScience Writers and Dr. Flo Witte for scientific editing.
Authors' contributions statement
All authors read the manuscript and contributed to the final version.
Ethics Statement
All animals were handled in accordance with good animal practice as defined by the relevant national and/or local animal welfare bodies. All animal work was approved by the Wenzhou Medical University Institutional Animal Care and Use Committee. The animal facilities are operated in compliance with the Guide for the Care and Use of Laboratory Animals.
Availability of data and materials
The datasets used and/or analyzed in the current study are available by request from the corresponding author.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Davis JC, Furstenthal L, Desai AA, Norris T, Sutaria S, Fleming E. et al. The microeconomics of personalized medicine: today's challenge and tomorrow's promise. Nat Rev Drug Discov. 2009;8:279-86
2. Langenkamp E, Molema G. Microvascular endothelial cell heterogeneity: general concepts and pharmacological consequences for anti-angiogenic therapy of cancer. Cell Tissue Res. 2009;335:205-22
3. Guan G, Lu Y, Zhu X, Liu L, Chen J, Ma Q. et al. CXCR4-targeted near-infrared imaging allows detection of orthotopic and metastatic human osteosarcoma in a mouse model. Scientific reports. 2015;5:15244
4. Margolin K. Cytokine therapy in cancer. Expert Opin Biol Ther. 2008;8:1495-505
5. Ortiz-Sanchez E, Helguera G, Daniels TR, Penichet ML. Antibody-cytokine fusion proteins: applications in cancer therapy. Expert Opin Biol Ther. 2008;8:609-32
6. Kim-Schulze S, Taback B, Kaufman HL. Cytokine therapy for cancer. Surg Oncol Clin N Am. 2007;16:793-818 viii
7. van der Meulen AA, Biber K, Lukovac S, Balasubramaniyan V, den Dunnen WF, Boddeke HW. et al. The role of CXC chemokine ligand (CXCL)12-CXC chemokine receptor (CXCR)4 signalling in the migration of neural stem cells towards a brain tumour. Neuropathol Appl Neurobiol. 2009;35:579-91
8. Lee JI, Jin BH, Kim MA, Yoon HJ, Hong SP, Hong SD. Prognostic significance of CXCR-4 expression in oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2009;107:678-84
9. Nimmagadda S, Pullambhatla M, Stone K, Green G, Bhujwalla ZM, Pomper MG. Molecular imaging of CXCR4 receptor expression in human cancer xenografts with [64Cu]AMD3100 positron emission tomography. Cancer research. 2010;70:3935-44
10. Gao M, Dong Q, Yao H, Zhang Y, Yang Y, Dang Y. et al. Induced neural stem cells modulate microglia activation states via CXCL12/CXCR4 signaling. Brain, behavior, and immunity. 2016
11. Zeng Z, Shi YX, Samudio IJ, Wang RY, Ling X, Frolova O. et al. Targeting the leukemia microenvironment by CXCR4 inhibition overcomes resistance to kinase inhibitors and chemotherapy in AML. Blood. 2009;113:6215-24
12. van den Oord J. The CCR9-CCL25 axis mediates melanoma metastasis to the small intestine. Nat Clin Pract Oncol. 2008;5:440-1
13. Hattori Y, Miyata T. Microglia extensively survey the developing cortex via the CXCL12/CXCR4 system to help neural progenitors to acquire differentiated properties. Genes Cells. 2018
14. Martinez-Ordonez A, Seoane S, Cabezas P, Eiro N, Sendon-Lago J, Macia M. et al. Breast cancer metastasis to liver and lung is facilitated by Pit-1-CXCL12-CXCR4 axis. Oncogene. 2018;37:1430-44
15. Peled A, Klein S, Beider K, Burger JA, Abraham M. Role of CXCL12 and CXCR4 in the pathogenesis of hematological malignancies. Cytokine. 2018;109:11-6
16. Uchida D, Kuribayashi N, Kinouchi M, Sawatani Y, Shimura M, Mori T. et al. Effect of a novel orally bioavailable CXCR4 inhibitor, AMD070, on the metastasis of oral cancer cells. Oncology reports. 2018;40:303-8
17. Zhu Y, Tang L, Zhao S, Sun B, Cheng L, Tang Y. et al. CXCR4-mediated osteosarcoma growth and pulmonary metastasis is suppressed by MicroRNA-613. Cancer Sci. 2018;109:2412-22
18. Liu Y, Ren CC, Yang L, Xu YM, Chen YN. Role of CXCL12-CXCR4 axis in ovarian cancer metastasis and CXCL12-CXCR4 blockade with AMD3100 suppresses tumor cell migration and invasion in vitro. Journal of cellular physiology. 2019;234:3897-909
19. Stanisavljevic L, Assmus J, Storli KE, Leh SM, Dahl O, Myklebust MP. CXCR4, CXCL12 and the relative CXCL12-CXCR4 expression as prognostic factors in colon cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37:7441-52
20. Amarante MK, Vitiello GAF, Rosa MH, Mancilla IA, Watanabe MAE. Potential use of CXCL12/CXCR4 and sonic hedgehog pathways as therapeutic targets in medulloblastoma. Acta Oncol. 2018:1-9
21. Benedicto A, Romayor I, Arteta B. CXCR4 receptor blockage reduces the contribution of tumor and stromal cells to the metastatic growth in the liver. Oncology reports. 2018;39:2022-30
22. Bonham LW, Karch CM, Fan CC, Tan C, Geier EG, Wang Y. et al. CXCR4 involvement in neurodegenerative diseases. Transl Psychiatry. 2018;8:73
23. Breun M, Schwerdtfeger A, Martellotta DD, Kessler AF, Perez JM, Monoranu CM. et al. CXCR4: A new player in vestibular schwannoma pathogenesis. Oncotarget. 2018;9:9940-50
24. Derlin T, Hueper K. CXCR4-targeted therapy in breast cancer. Lancet Oncol. 2018;19:e370
25. Gao PT, Ding GY, Yang X, Dong RZ, Hu B, Zhu XD. et al. Invasive potential of hepatocellular carcinoma is enhanced by loss of selenium-binding protein 1 and subsequent upregulation of CXCR4. Am J Cancer Res. 2018;8:1040-9
26. Hirano Y, Ode Y, Ochani M, Wang P, Aziz M. Targeting junctional adhesion molecule-C ameliorates sepsis-induced acute lung injury by decreasing CXCR4(+) aged neutrophils. Journal of leukocyte biology. 2018
27. Jin CH, Li Y, Xia J, Li Y, Chen M, Hu Z. et al. CXCR4 blockade improves leukemia eradication by allogeneic lymphocyte infusion. Am J Hematol. 2018;93:786-93
28. Kircher M, Herhaus P, Schottelius M, Buck AK, Werner RA, Wester HJ. et al. CXCR4-directed theranostics in oncology and inflammation. Ann Nucl Med. 2018
29. Komorowski M, Tisonczyk J, Kolakowska A, Drozdz R, Kozbor D. Modulation of the Tumor Microenvironment by CXCR4 Antagonist-Armed Viral Oncotherapy Enhances the Antitumor Efficacy of Dendritic Cell Vaccines against Neuroblastoma in Syngeneic Mice. Viruses. 2018:10
30. Lapa C, Hanscheid H, Kircher M, Schirbel A, Wunderlich G, Werner R. et al. Feasibility of CXCR4-directed radioligand therapy in advanced diffuse large B cell lymphoma. Journal of nuclear medicine: official publication, Society of Nuclear Medicine. 2018
31. Luo Y, Pan Q, Feng J, Cao X, Li F. Chemokine Receptor CXCR4-Targeted PET/CT With 68Ga-Pentixafor Shows Superiority to 18F-FDG in a Patient With Waldenstrom Macroglobulinemia. Clinical nuclear medicine. 2018;43:548-50
32. Neri M, Sansone L, Pietrasanta L, Kisialiou A, Cabano E, Martini M. et al. Gene and protein expression of CXCR4 in adult and elderly patients with chronic rhinitis, pharyngitis or sinusitis undergoing thermal water nasal inhalations. Immun Ageing. 2018;15:10
33. Tahirovic YA, Truax VM, Wilson RJ, Jecs E, Nguyen HH, Miller EJ. et al. Discovery of N-Alkyl Piperazine Side Chain Based CXCR4 Antagonists with Improved Drug-like Properties. ACS Med Chem Lett. 2018;9:446-51
34. Bertolini F, Dell'Agnola C, Mancuso P, Rabascio C, Burlini A, Monestiroli S. et al. CXCR4 Neutralization, a Novel Therapeutic Approach for Non-Hodgkin's Lymphoma. Cancer Res. 2002;62:3106-12
35. Tanaka T, Nomura W, Narumi T, Esaka A, Oishi S, Ohashi N. et al. Structure-activity relationship study on artificial CXCR4 ligands possessing the cyclic pentapeptide scaffold: the exploration of amino acid residues of pentapeptides by substitutions of several aromatic amino acids. Organic & biomolecular chemistry. 2009;7:3805-9
36. Zhang Y, Patel S, Abdelouahab H, Wittner M, Willekens C, Shen S. et al. CXCR4 inhibitors selectively eliminate CXCR4-expressing human acute myeloid leukemia cells in NOG mouse model. Cell death & disease. 2012;3:e396
37. Ueda S, Oishi S, Wang ZX, Araki T, Tamamura H, Cluzeau J. et al. Structure-activity relationships of cyclic peptide-based chemokine receptor CXCR4 antagonists: disclosing the importance of side-chain and backbone functionalities. Journal of medicinal chemistry. 2007;50:192-8
38. Poschenrieder A, Schottelius M, Schwaiger M, Kessler H, Wester HJ. The influence of different metal-chelate conjugates of pentixafor on the CXCR4 affinity. EJNMMI research. 2016;6:36
39. Zhou N, Luo Z, Luo J, Fan X, Cayabyab M, Hiraoka M. et al. Exploring the stereochemistry of CXCR4-peptide recognition and inhibiting HIV-1 entry with D-peptides derived from chemokines. J Biol Chem. 2002;277:17476-85
40. Wang W, Ke S, Kwon S, Yallampalli S, Cameron AG, Adams KE. et al. A new optical and nuclear dual-labeled imaging agent targeting interleukin 11 receptor alpha-chain. Bioconjug Chem. 2007;18:397-402
Author contact
![]() Corresponding author: Kun Luo, M.D., Department of Internal Medicine, Clinic of August First Film Studio, 301 Hospital, NO.1 Liuli Bridge, Beijing, China; Tel: +86 577 6586 6436; Fax: +86 577 6586 6436; E-mail: luokunedu.cn
Corresponding author: Kun Luo, M.D., Department of Internal Medicine, Clinic of August First Film Studio, 301 Hospital, NO.1 Liuli Bridge, Beijing, China; Tel: +86 577 6586 6436; Fax: +86 577 6586 6436; E-mail: luokunedu.cn

 Global reach, higher impact
Global reach, higher impact