3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(18):4333-4340. doi:10.7150/jca.31454 This issue Cite
Research Paper
Elevated MRE11 expression associated with progression and poor outcome in prostate cancer
1. Department of Urology, Fudan University Shanghai Cancer Center, Shanghai 200032
2. Department of Oncology, Shanghai Medical College, Fudan University, Shanghai 20032, P.R. China
* Contribute equally
Received 2018-11-12; Accepted 2019-5-6; Published 2019-7-10
Abstract
Objective: Growing evidence has proved that MRE11, a protein underpinned to be involved in DNA double-strand break (DSB) repair process, is correlated with cancer outcomes. However, its role in prostate cancer (PCa) remains unclear. This study aimed to investigate the expression of MRE11 in tumor tissue and defining its value in predicting prognosis of PCa patients.
Methods: A total of 578 patients from two cohorts were enrolled in this study. Distribution of categorical clinical-pathological data together with levels of MRE11 expression was compared with χ2-test in a contingency table. Immunohistochemical (IHC) staining and evaluation was detected from 78 paired PCa and adjacent normal tissues. Partial likelihood test from univariate and multivariate Cox regression analysis was developed to address the influence of independent factors on disease-free survival (DFS) and overall survival (OS) in two cohorts. The Kaplan-Meier method and log-rank test were performed to assess the survival benefits between discrete levels. Set Enrichment Analysis (GSEA) was performed to select related genes and pathways from The Cancer Genome Atlas (TCGA) database.
Results: In the current study, we demonstrated that MRE11 was highly expressed in PCa compared with normal tissues (P=0.011). In addition, in the TCGA cohort, the median DFS in patients with IHC positive and negative MRE11 expression levels was 24.5 and 30.6 months, and median OS was 28.7 and 33.0 months, respectively. In FUSCC cohort, median DFS in patients with IHC positive and negative MRE11 expression was 28.0 and 35.6 months. Furthermore, survival curves suggested that PCa patients with elevated MRE11 expression levels showed poorer OS (P=0.019) in TCGA cohort and poor DFS (P=0.047) in FUSCC cohort.
Conclusion: In conclusion, our study reveals that elevated MRE11 expression is significantly correlated with cancer progression and poor survival in PCa patients. These data suggest that MRE11 may act as an oncoprotein and a promising prognostic marker for PCa patients.
Keywords: MRE11, DNA repair, homologous recombination, prognosis, prostate cancer
Introduction
Prostate cancer (PCa) has become the most frequently diagnosed cancer and the fifth leading cause of cancer death in men, accounting for approximately 13.5% incidence and 6.7% mortality of all cancer deaths [1]. It is estimated to grow to 1.3 million new cases of PCa in 2018 worldwide, especially in countries with higher socioeconomic development [1-3]. Hence, understanding of the key genes and underlying molecular mechanisms that regulate proliferation and invasion of PCa is significant to promote early diagnosis, develop individual therapies and predict prognosis [4, 5]. However, only limited information of the available researches can be applied to the explanation of aggressive progression in PCa patients [5]. Therefore, new prognostic biomarkers and molecular alterations involved in the initiation and progression of PCa must be urgently identified to realize individualized precise therapeutic regimen.
Meiotic recombination 11 (MRE11), a human ortholog of MRE11A that encodes a nuclear protein, is supposed to be involved in the repair of DNA double-strand break (DSB) [6]. In this organism, MRE11 protein interacts with the Rad50 recombinase and consists of MRE11-RAD50-NBS1 (MRN) complex, which results in active recruitment of DNA damage repair pathways exposed to ionizing radiation [7, 8]. Synergistically with the function of ATM [9], this complex phosphorylates protein H2AX, p53, CHK2 to accelerate DSB repair and active cycle checkpoint [10, 11]. Meanwhile, during DSB repair, MRE11 participates in the amplification of apoptosis signaling if irreversible damage appears. To avoid incomplete repair of DNA damage, body cells developed homologous recombination to ensure the integrity of the genome [12]. Similarly, programmed DSB is required to initiate homologous recombination during meiosis process, in which MRE11 served an essential role for DSB repair [13].
DSB repair is an indispensable process to support genomic integrity, and significant genes are activated when DNA damage occurs [14-16]. In most cases, deficient expression of DNA repair genes and inevitable replication error increase the quantity of un-repaired DSB, leading to mutant and further tumorigenesis [17]. Interestingly, Sharma et al. found MRE11A participated in microhomology-mediated alternative end joining (MMEJ), one of the major but error prone DSB repair pathways in higher eukaryotes [18]. Therefore, these observations indicated MRE11 over-mediated DSB repair may lead to imprecise homologous recombination and elevated risk of carcinoma. For example, Yuan et al. suggested that MRE11A was a novel oncoprotein and was significantly associated with malignant behavior trend in breast cancer [19]. However, the prognostic value of MRE11 in PCa was merely documented.
To investigate the MRE11 expression in tumor tissue and defining its value in PCa patients, we enrolled 78 patients who have received previous radical prostatectomy in our institution and validate its role in 500 patients in the Cancer Genome Atlas (TCGA) database. We hypothesized that the oncogenic activity of MRE11 correlated with poor prognosis and might be a potential therapeutic target of PCa.
Materials and Methods
Patients and Variables
The inclusion criteria were as follows: patients were pathologically diagnosed with prostatic adenocarcinoma; patients had not received chemical treatment or physical therapy before surgery. Patients with an age < 18 or a life expectancy < 1 year were excluded in this study. Finally, a total of 78 PCa patients, who have underwent previous radical prostatectomy in the Department of Urology of Fudan University Shanghai Cancer Center (FUSCC) (Shanghai, China) from October 2010 to December 2012, and 500 PCa patients from TCGA database were consecutively recruited in analyses, with electronic medical records or pathology reports available. Clinical and pathological parameters, specifically age at surgery, TNM stage, AJCC stage and Gleason score, in two cohorts were summarized in Table 1. Tissue samples, including PCa and normal tissues, were collected during surgery and available from FUSCC tissue bank. All of the study designs and test procedures were performed in accordance with the Helsinki Declaration II. The Ethics approval and consent to participate of the current study were approved and consented by the ethics committee of FUSCC.
Immunohistochemical (IHC) staining and evaluation
Immunostaining of MRE11 was performed using a mouse monoclonal anti-MRE11 antibody (1:3000, Cat. ab214, Abcam, USA). Positive or negative staining of a certain protein in one FFPE slide was independently assessed by two experienced pathologists, and determined as follows. The staining intensity level was graded from 0 to 3. Samples with no staining, weak, median and strong staining denote to the level of 0, 1, 2 and 3. Based on the coverage percentage of immunoreactive tumor cells (0%, 1-25%, 26-50%, 51-75%, 76-100%), the staining extent was ranging from 0 to 4. The overall IHC score grading from 0 to 12 was evaluated according to the multiply of the staining intensity and extent score. Negative staining represented grade 0 to 3 and positive staining from 4 to 12 for each sample. All examples were classified in three groups (Gleason<7, Gleason=7 and Gleason>7) to confirm differential expression of MRE11.
Statistical analysis
To figure out the associations of different MRE11 mRNA expression sets with clinicopathological characteristics, χ2-test was performed to compare the distribution of categorical data between groups. Scattered plot was utilized to present the differential expression of MRE11 in normal or prostate tissues. The primary end point was overall survival (OS) representing a certain period of time, which was evaluated from the date of radical prostatectomy to the date of death or last follow-up in PCa patients. Disease-free survival (DFS), as the secondary end point, was the length of time from the initiation of curative treatment when no disease can be detected until the date of progression or the start date of a second-line treatment or the date of death, whichever occurred first. The follow-up duration was estimated using the Kaplan-Meier method with 95% confidence interval (CI) and log-rank test in separate curves. Univariate and multivariate analysis were performed with Cox logistic regression models to find independent predictors. Cox regressions on 500 participants enrolled in TCGA cohort were independently analyzed to evaluate confounding covariates including age, TNM stage, Gleason score and MRE11 expression on survival. In FUSCC cohort, AJCC stage was analyzed in evaluation as supplements. Statistics analyses were performed with SPSS software (version 23.0, Inc, Chicago, IL, USA). All hypothetical tests were two-sided and P-values less than 0.05 were considered significant in all tests.
Datasets from the Cancer Genome Atlas (TCGA) database were implemented with GSEA method using the Category version 2.10.1 package. For each separate analysis, Student's-t-test statistical score was performed in consistent pathways and the mean of the differential expression genes was calculated. A permutation test with 1000 times was used to identify the significantly changed pathways. The adjusted P values (adj. P) using Benjamini and Hochberg (BH) false discovery rate (FDR) method by default were applied to correct for the occurrence of false positive results. Significant related genes were defined with an adj. P less than 0.01 and FDR less than 0.25 [20].
Results
In this study, research was conducted in two series. In the first series, differential expression of MRE11 in normal and prostate tissues was analyzed; in the second series, progression and prognostic value of MRE11 expression in PCa patients were assessed.
Clinicopathological characteristics of the cohorts
As shown in Table 1, patients with increased MRE11 expression significantly correlated with elder patients (P=0.021) in FUSCC cohort and higher Gleason score (P=0.008) in TCGA cohort. Chi-square test showed that baseline data were balanced on the distribution of categorical data, including TNM stage and AJCC stage (P>0.05).
Clinicopathological characteristics in relation to MRE11 expression status in two cohorts
| Characteristics | FUSCC cohort (N=78) | MRE11 expression | χ2 | P | TCGA cohort (N=500) | MRE11 expression | χ2 | P | ||
|---|---|---|---|---|---|---|---|---|---|---|
| IHC positive (N=58) | IHC negative (N=20) | IHC positive (N=319) | IHC negative (N=181) | |||||||
| N (%) | ||||||||||
| Age | 5.287 | 0.021 | 0.772 | 0.380 | ||||||
| <65 years | 20 (25.6) | 11 (19.0) | 9 (45.0) | 333 (66.6) | 208 (65.2) | 125 (69.1) | ||||
| ≥65 years | 58 (74.4) | 47 (81.0) | 11 (55.0) | 167 (33.4) | 111 (34.8) | 56 (30.9) | ||||
| Year of diagnosis | 1.595 | 0.450 | 1.313 | 0.252 | ||||||
| 2010 | 12 (15.4) | 9 (15.5) | 3 (15.0) | - | - | - | ||||
| 2011 | 38 (48.7) | 26 (44.8) | 12 (60.0) | - | - | - | ||||
| 2012 | 28 (35.9) | 23 (39.7) | 5 (25.0) | - | - | - | ||||
| 2000-2006 | - | - | - | 71(14.2) | 41(12.9) | 30(16.6) | ||||
| 2007-2013 | - | - | - | 429(85.8) | 278(87.1) | 151(83.4) | ||||
| Laterality | 1.062 | 0.303 | 2.291 | 0.130 | ||||||
| Left/Right | 17(21.8) | 11(19.0) | 6(21.8) | 65 (13.0) | 36 (11.3) | 29 (16.0) | ||||
| Bilateral | 61(78.2) | 47(81.0) | 14(78.2) | 435 (87.0) | 283 (88.7) | 152 (84.0) | ||||
| pT stage | 0.258 | 0.611 | 2.575 | 0.109 | ||||||
| T1 - T2 | 35 (44.9) | 27 (46.6) | 8 (40.0) | 195(39.0) | 116(36.4) | 79(43.6) | ||||
| T3 - T4 | 43 (55.1) | 31 (53.4) | 12 (60.0) | 305(61.0) | 203(63.6) | 102(56.4) | ||||
| pN stage | 0.729 | 0.393 | 0.043 | 0.836 | ||||||
| N0 | 61 (78.2) | 44 (75.9) | 17 (85.0) | 348 (69.9) | 221 (69.3) | 127 (70.2) | ||||
| N1 | 17 (21.8) | 14 (24.1) | 3 (15.0) | 152 (30.4) | 98 (30.7) | 54 (29.8) | ||||
| pM stage | - | - | 1.401 | 0.237 | ||||||
| M0 | 78 (100.0) | 58 (100.0) | 20 (100.0) | 457 (91.4) | 288 (90.3) | 169 (93.4) | ||||
| M1 | 0 | 0 | 0 | 43 (8.6) | 31 (9.7) | 12 (6.6) | ||||
| AJCC stage † | 0.215 | 0.643 | - | - | ||||||
| I - II | 65 (83.3) | 49 (84.5) | 16 (80.0) | - | - | - | ||||
| III - IV | 13 (16.7) | 9 (15.5) | 4 (20.0) | - | - | - | ||||
| Gleason score | 4.312 | 0.116 | 9.586 | 0.008 | ||||||
| < 7 | 21 (26.9) | 13 (22.4) | 8 (40.0) | 45 (9.0) | 23 (7.2) | 22 (12.2) | ||||
| = 7 | 37 (26.9) | 27 (46.6) | 10 (50.0) | 250 (50.0) | 150 (47.0) | 100 (55.2) | ||||
| ≥ 8 | 20 (25.6) | 18 (31.0) | 2 (10.0) | 205 (14.0) | 146 (45.8) | 59 (32.6) | ||||
† The AJCC staging system is a classification system developed by the American Joint Committee on Cancer for describing the extent of disease progression in cancer patients. It utilizes in part the TNM scoring system: Tumor size, Lymph Nodes affected, Metastases.
Expression of MRE11 in FUSCC
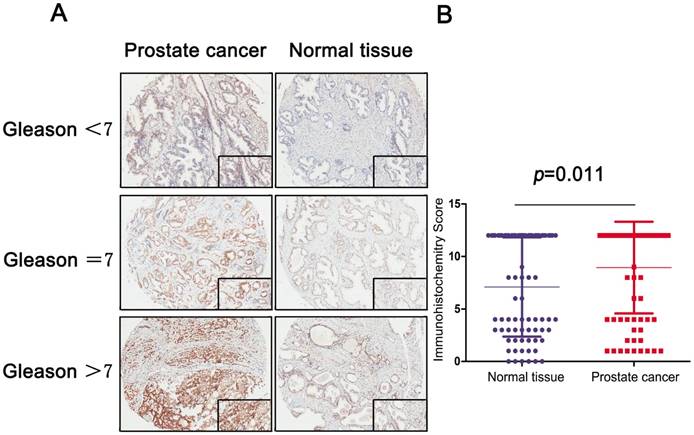
To analyze the MRE11 expression profile of PCa tissue, IHC revealed staining distribution concerning different Gleason score between normal and tumor tissues. The scatter plot of IHC score revealed that elevated MRE11 was significantly expressed in prostate tumor tissues (P=0.011).
Cox regression analyses and survival outcomes of two cohorts
In the multivariate models of FUSCC and TCGA cohort in Table 2, traditional prognostic factors, specifically pathological T stage, was still relevant to DFS in PCa patients, indicating a fine representativeness of the population in the cohort of current research. In FUSCC cohort, high Gleason score was significant correlated with poor DFS (P<0.001) in multivariate model of Cox regression analyses. Importantly, subgroups of MRE11 expression (IHC negative vs. IHC positive) showed that MRE11 amplification significantly associated with poor DFS (P=0.048) for PCa patients in FUSCC cohort (Table 2). As shown in Table 3, univariate and multivariate analysis of OS in TCGA cohort indicated that Gleason score was significantly associated with OS (P=0.019) merely in univariate Cox regression. Increased MRE11 expression profiles was suggested to have prognostic value in both univariate (P=0.047) and multivariate (P=0.046) analysis. The other factors, including age and TNM stage, were not assessed as prognostic indicators of OS in our study (P>0.05) (Table 2 and Table 3).
Multivariate Cox logistic regression analysis of DFS in FUSCC and TCGA cohorts (DFS: disease-free survival; FUSCC: Fudan University Shanghai Cancer Center; TCGA: the Cancer Genome Atlas)
| Covariates | FUSCC | TCGA | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | ||
| Age (ref. <65 years) | 1.756 | 0.433-7.125 | 0.431 | 1.005 | 0.972-1.038 | 0.780 | |
| pT stage (ref. T1 - T2) | 18.506 | 2.706-126.551 | 0.003 | 2.026 | 1.101-3.728 | 0.023 | |
| pN stage (ref. N0) | 0.256 | 0.034-1.933 | 0.186 | 0.929 | 0.592-1.458 | 0.929 | |
| pM stage (ref. M0) | - | - | - | 0.475 | 0.149-1.514 | 0.208 | |
| AJCC stage (ref. I - II) | 0.724 | 0.137-4.124 | 0.724 | - | - | - | |
| Gleason score (ref. <7) | 2.518 | 0.797-7.959 | 0.116 | 3.202 | 3.202-2.013 | <0.001 | |
| MRE11 expression (ref. negative) | 8.588 | 1.015-72.667 | 0.048 | 1.071 | 0.687-1.669 | 0.763 | |
Univariate and multivariate Cox logistic regression analysis of OS in TCGA cohort (OS: overall survival; TCGA: the Cancer Genome Atlas)
| Covariates | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | ||
| Age (ref. <65 years) | 1.053 | 0.955-1.160 | 0.300 | 1.064 | 0.952-1.188 | 0.276 | |
| pT stage (ref. T1 - T2) | 3.228 | 0.598-17.426 | 0.173 | 0.739 | 0.077-7.078 | 0.793 | |
| pN stage (ref. N0) | 2.408 | 0.629-9.226 | 0.200 | 1.775 | 0.365-8.636 | 0.477 | |
| pM stage (ref. M0) | 1.958 | 0.241-15.934 | 0.530 | - | - | 0.989 | |
| Gleason score (ref. <7) | 6.139 | 1.341-28.100 | 0.019 | 5.333 | 0.574-49.553 | 0.141 | |
| MRE11 expression (ref. negative) | 8.318 | 1.031-67.131 | 0.047 | 9.933 | 1.042-94.665 | 0.046 | |
A: IHC stain of prostate and normal tissue in different Gleason score subgroups (Gleason<7, Gleason=7 and Gleason>7); B: Scatter plot of IHC score between normal and tumor tissues (P=0.011).
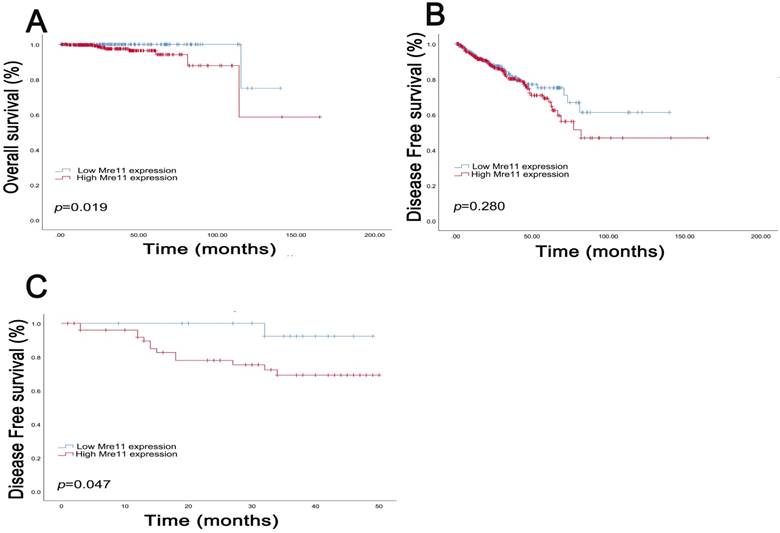
Survival curves suggested that PCa patients with elevated MRE11 expression levels showed poorer OS (P=0.019) in TCGA cohort and poor DFS (P=0.047) in FUSCC cohort. In addition, in TCGA cohort, median DFS was 26.9 months and OS was 30.5 months respectively. The median DFS in patients with IHC positive and negative MRE11 expression levels were 24.5 and 30.6 months, and median OS was 28.7 and 33.0 months, respectively. In FUSCC cohort, data on OS was not available due to the favorable prognosis of PCa patients, while median DFS was 30.7 months. Median DFS in patients with IHC positive and negative MRE11 expression was 28.0 and 35.6 months.
A total of 100 significant genes were obtained from GSEA, and the genes with positive correlation were plotted. Besides, MRE11 was found involved in the most significant pathways including mitotic spindle, UV response and transforming growth factor beta (TGF-β) signaling pathways. The details were illustrated in Figure 3.
Discussion
In our present study, we performed IHC and detected survival benefits to investigate whether MER11 has potential prognostic value in PCa. We observed MRE11 expression markedly enhanced in prostate cells was associated with malignant behavior. Collectively, our data demonstrated that high level of MRE11 expression was associated with high risk of recurrence rate and decrease patient survival. It opens up a novel way for MRE11 expression to affect the pathogenesis of PCa by underlying DNA damage variation.
In human malignancy, the MRN (MRE11-RAD50-NBS1) protein complex plays a vital role in DSB repair foci and cell cycle control, which makes it responsible for the genome stability [8]. Relatively, heterozygous mutation only contributes a limited fraction of tumorigenesis, and aberrant molecular variation of MRN is more frequent and of great clinical significance [21, 22]. Previous studies had reported the role of MRE11 in cell survival and proliferation [23], and malignant cancer behavior was significantly correlated with elevated MRE11 expression phenotypes in breast [19, 24, 25], gastric [26] and rectal cancer [27, 28]. Similarly, it was noteworthy that increased recurrence rate and chemoresistance of certain cancers correlated with high expression level of MRN complex [28-30]. Moreover, deleterious mutation-induced over-expressed RAD50 and NBS1 were associated with undesirable survival benefits in PCa patients, while independent prognostic value of MRE11 was rarely documented [31, 32].
Kaplan-Meier survival analyses on different MRE11 expression groups with OS (A) and DFS (B) in the included 500 PCa patients from TCGA database. Kaplan-Meier survival analyses on different MRE11 expression groups with DFS (C) in the included 78 PCa patients from FUSCC cohort.
Datasets from TCGA database were implemented with GSEA method. For each separate analysis, Student's-t-test statistical score was performed in consistent pathways and the mean of the differential expression genes was calculated. A permutation test with 1000 times was used to identify the significantly changed pathways. The adjusted P values (adj. P) using Benjamini and Hochberg (BH) false discovery rate (FDR) method by default were applied to correct for the occurrence of false positive results. The significant related genes were defined with an adj. P less than 0.01 and a FDR less than 0.25.
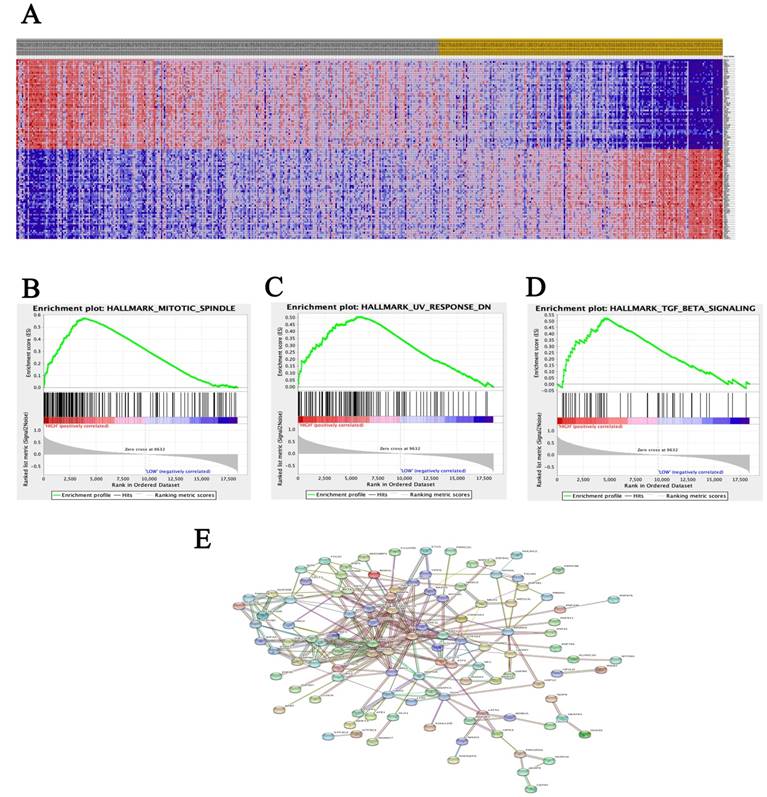
In this study, data analysis form two cohorts support our hypothesis and clearly detect high expression of MRE11 in tumor tissues for PCa patients, leading to increased disease recurrence rates and decreased overall survival. Meanwhile, GSEA analysis illustrated that MRE11 involved in the most significant pathways including mitotic spindle, UV response and transforming growth factor beta (TGF-β) signaling pathways were enriched in PCa samples. Besides participating in DSB repair pathways, MRE11 also interacts with MMAP, expressed by mitosis-specific MRN complex, to maintain optimal genome stability during mitosis [33]. In addition, the ionizing-radiation induced DSB activates a complex co-network of proteins. MRN complex results in active recruitment of DNA damage repair pathways [7, 8]. Recruitment of repair factors is a prerequisite for initiation of DNA damage repair by the homologous recombination pathway. It was reported that Smad7 and SPTBN1 have independent effect on driving DNA repair process, and regulates TGF-β signaling as well [34, 35].
Strength of our study lies in our first attempt to investigate the role of MRE11 as a prognostic factor of PCa. Relationship between MRE11 and PCa was rarely documented, while it is noteworthy that MRE11 is a confirmed repair factor in DSB response process and estimated highly expressed in many cancer patients [11]. With this in mind, in the FUSCC cohort of our study, we found dramatic MRE11 IHC score contrast between 78 paired tumor and adjacent normal tissues, and first demonstrated that PCa patients with elevated MRE11 expression had shorter DFS, which was similar to the validation in the TCGA database with 500 PCa patients. On the other hand, to further explain the underlying ability of invasion and metastasis of MRE11, data from public database were implemented with GSEA analysis to identify significant genes and pathways, which might clarify the correlation triggering carcinogenesis.
At the same time, there are some limitations proposed in this study as follows. Firstly, retrospective nature of the data set is inevitable, including relatively small sample size, poor population variety, and selected bias from FUSCC cohort. However, realizing this limitation of our demographic group, we collected data from TCGA to provide more plausible evidence for our conclusion. Secondly, our research failed to deeply clarify the underlying mechanism of MRE11 involved in PCa. Thirdly, data from FUSCC cohort miss the overall survival values until the last follow-up.
Conclusion
In conclusion, our study first reveal that elevated MRE11 expression is significantly correlated with cancer progression and poor survival in PCa patients. These data suggest that MRE11 may act as an oncoprotein and a promising prognostic marker for PCa patients. In this regard, more validation cohorts and further elucidation are required to identify all value of MRE11 and its clinical application for PCa patients.
Abbreviations
PCa: Prostate cancer; MRE11: meiotic recombination 11; DSB: DNA double-strand break; IHC: immunohistochemical; TCGA: the Cancer Genome Atlas; FUSCC: Fudan University Shanghai Cancer Center; DFS: disease-free survival; OS: overall survival; HR: hazard ratio; CI: confidence interval; AJCC: American Joint Committee on Cancer.
Acknowledgements
Ethics approval and consent to participate
The Ethics approval and consent to participate of the current study was approved and consented by the ethics committee of Fudan University Shanghai Cancer center.
Availability of data and material
The datasets during and/or analyzed during the current study available from the corresponding author on reasonable request.
Funding
This work is supported by Grants from the National Natural Science Foundation of China (No. 81202004, 81802525, 81472377), Shanghai Natural Science Foundation of China (No. 16ZR1406400).
Authors' contributions
The work presented here was carried out in collaboration between all authors. YDW defined the research theme, discussed analyses, interpretation, and presentation. QXJ and ZHL developed the algorithm and performed the statistical analysis. XWH and WJ drafted the manuscript, carried out to record the clinical data, analyzed the IHC data, and interpreted the results. WY and ZY participated in reviewing all clinical records, and co-worked on associated data collection. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, KD Miller, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7-30
2. Wong MC. et al. Global Incidence and Mortality for Prostate Cancer: Analysis of Temporal Patterns and Trends in 36 Countries. Eur Urol. 2016;70(5):862-874
3. Bray F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394-424
4. Rodrigues DN. et al. Molecular pathology and prostate cancer therapeutics: from biology to bedside. J Pathol. 2014;232(2):178-184
5. DeMarzo AM. et al. Pathological and molecular aspects of prostate cancer. Lancet. 2003;361(9361):955-964
6. Petrini JH. et al. Isolation and characterization of the human MRE11 homologue. Genomics. 1995;29(1):80-86
7. Quaiser A. et al. The Mre11 protein interacts with both Rad50 and the HerA bipolar helicase and is recruited to DNA following gamma irradiation in the archaeon Sulfolobus acidocaldarius. BMC Mol Biol. 2008;9:25
8. van den Bosch M, Bree RT, Lowndes NF. The MRN complex: coordinating and mediating the response to broken chromosomes. EMBO Rep. 2003;4(9):844-849
9. Lavin MF. et al. ATM-Dependent Phosphorylation of All Three Members of the MRN Complex: From Sensor to Adaptor. Biomolecules. 2015;5(4):2877-2902
10. Lavin MF. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26(56):7749-7758
11. Jackson S, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071-1078
12. Myler LR. et al. Single-Molecule Imaging Reveals How Mre11-Rad50-Nbs1 Initiates DNA Break Repair. Mol Cell. 2017;67(5):891-898
13. Lukaszewicz A. et al. MRE11 and COM1/SAE2 are required for double-strand break repair and efficient chromosome pairing during meiosis of the protist Tetrahymena. Chromosoma. 2010;119(5):505-518
14. Hakem R. DNA-damage repair; the good, the bad, and the ugly. Embo j. 2008;27(4):589-605
15. Scott S, Pandita TK. The cellular control of DNA double-strand breaks. J Cell Biochem. 2006;99(6):1463-1475
16. Helleday T. et al. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst). 2007;6(7):923-935
17. O'Connor MJ. Targeting the DNA Damage Response in Cancer. Mol Cell. 2015;60(4):547-560
18. Sharma S. et al. Homology and enzymatic requirements of microhomology-dependent alternative end joining. Cell Death Dis. 2015;6(3):e1697
19. Yuan SS. et al. Role of MRE11 in cell proliferation, tumor invasion, and DNA repair in breast cancer. J Natl Cancer Inst. 2012;104(19):1485-1502
20. Subramanian A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545-15550
21. Dzikiewicz-Krawczyk A. The importance of making ends meet: mutations in genes and altered expression of proteins of the MRN complex and cancer. Mutat Res. 2008;659(3):262-273
22. Gao J. et al. RAD50/MRE11/NBS1 proteins in relation to tumour development and prognosis in patients with microsatellite stable colorectal cancer. Histol Histopathol. 2008;23(12):1495-1502
23. Deng Y. et al. Multiple roles for MRE11 at uncapped telomeres. Nature. 2009;460(7257):914-918
24. Kim H. et al. Frequency of pathogenic germline mutation in CHEK2, PALB2, MRE11, and RAD50 in patients at high risk for hereditary breast cancer. Breast Cancer Res Treat. 2017;161(1):95-102
25. Yang CH. et al. Interaction of MRE11 and Clinicopathologic Characteristics in Recurrence of Breast Cancer: Individual and Cumulated Receiver Operating Characteristic Analyses. Biomed Res Int. 2017;2017:2563910
26. Altan B. et al. High Expression of MRE11-RAD50-NBS1 Is Associated with Poor Prognosis and Chemoresistance in Gastric Cancer. Anticancer Res. 2016;36(10):5237-5247
27. Ho V. et al. MRE11 and ATM Expression Levels Predict Rectal Cancer Survival and Their Association with Radiotherapy Response. PLoS One. 2016;11(12):e0167675
28. Ho V. et al. Overexpression of the MRE11-RAD50-NBS1 (MRN) complex in rectal cancer correlates with poor response to neoadjuvant radiotherapy and prognosis. BMC Cancer. 2018;18(1):869
29. Kuo KT. et al. Prognostic significance of NBS1 and Snail expression in esophageal squamous cell carcinoma. Ann Surg Oncol. 2012;19(Suppl 3):S549-S557
30. Soderlund K. et al. Intact Mre11/Rad50/Nbs1 complex predicts good response to radiotherapy in early breast cancer. Int J Radiat Oncol Biol Phys. 2007;68(1):50-58
31. Grupp K. et al. The prognostic impact of high Nijmegen breakage syndrome (NBS1) gene expression in ERG-negative prostate cancers lacking PTEN deletion is driven by KPNA2 expression. Int J Cancer. 2014;135(6):1399-1407
32. Jividen K. et al. Genomic analysis of DNA repair genes and androgen signaling in prostate cancer. BMC Cancer. 2018;18(1):960
33. Xu R. et al. Mitosis-specific MRN complex promotes a mitotic signaling cascade to regulate spindle dynamics and chromosome segregation. Proc Natl Acad Sci U S A. 2018;115(43):E10079-E10088
34. Park S. et al. Smad7 enhances ATM activity by facilitating the interaction between ATM and Mre11-Rad50-Nbs1 complex in DNA double-strand break repair. Cell Mol Life Sci. 2015;72(3):583-596
35. Horikoshi N. et al. beta2-spectrin depletion impairs DNA damage repair. Oncotarget. 2016;7(23):33557-33570
Author contact
![]() Corresponding authors: Xiao-Jian Qin, M.D. (Email: xiaojianqincom) & Hai-Liang Zhang, M.D. (Email: zhanghl918com) & Ding-Wei Ye, M.D. (Email: dwyeliecom). Department of Urology, Fudan University Shanghai Cancer Center, No. 270 Dong'an Road, Shanghai, 200032, People's Republic of China. Tel: 86-21-64175590-2805
Corresponding authors: Xiao-Jian Qin, M.D. (Email: xiaojianqincom) & Hai-Liang Zhang, M.D. (Email: zhanghl918com) & Ding-Wei Ye, M.D. (Email: dwyeliecom). Department of Urology, Fudan University Shanghai Cancer Center, No. 270 Dong'an Road, Shanghai, 200032, People's Republic of China. Tel: 86-21-64175590-2805

 Global reach, higher impact
Global reach, higher impact