3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(13):2399-2409. doi:10.7150/jca.86974 This issue Cite
Research Paper
Bioinformatics and Expression Analyses of miR-639, miR-641, miR-1915-3p and miR-3613-3p in Colorectal Cancer Pathogenesis
1. Department of Biology, Faculty of Art and Science, Gaziantep University, 27310, Gaziantep, Turkey.
2. Department of General Surgery, Faculty of Medicine, Gaziantep University, 27310, Gaziantep, Turkey.
Abstract
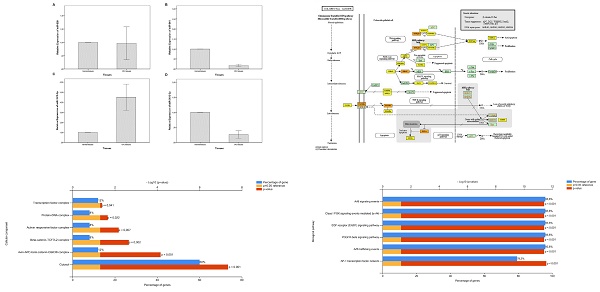
Objectives: MicroRNAs (miRNAs) have important function in cancer development and progression. This study aims to determine the expression levels of miR-639, miR-641, miR-1915-3p, and miR-3613-3p in tissues of colorectal cancer (CRC) patients and the role of these miRNAs in the CRC pathogenesis.
Methods: Tumor and non-tumor tissues were collected from a total of 59 CRC patients. qRT-PCR was used to identify the expressions of miR-639, miR-641, miR-1915-3p and miR-3613-3p. Through bioinformatics analysis, the target genes of miRNAs were identified by using DIANA mirPath v.3. Signaling pathways were generated using KEGG pathway database. Biological pathway, cellular component analysis, and analysis of Protein-Protein Interactions (PPI) Networks were performed using FunRich and STRING database.
Results: Our findings revealed that miR-639, miR-641 and miR-3613-3p were significantly downregulated, and miR-1915-3p was significantly upregulated in tumor tissues compared to non-tumor tissues (p˂0.05). Furthermore, MAPK signaling pathway was the most enriched KEGG pathway regulated by miR-639, miR-641, miR-1915-3p and miR-3613-p. According to the FunRich, it was demonstrated that the targeted genes by miRNAs related to the cellular component and biological pathways such as beta-catenin-TCF7L2, axin-APC-beta-catenin-GSK3B complexes, Arf6 signaling, Class I PI3K signaling, etc. And, by the PPI analysis, it was established that the target genes were clustered on CTNNB1 and KRAS.
Conclusions: These outcomes imply that miR-639, miR-641 and miR-3613-3p have tumor suppressor roles, while miR-1915-3p has an oncogenic role in the pathogenesis of CRC. According to the results of the current study, dysregulated miR-639, miR-641, miR-1915-3p, and miR-3613-3p might contribute to the development of CRC.
Keywords: colorectal carcinoma, expression, microRNA, bioinformatics analysis, biomarker

 Global reach, higher impact
Global reach, higher impact