3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(15):2895-2907. doi:10.7150/jca.86568 This issue Cite
Research Paper
Expression And Prognostic Role of PRDX1 In Gastrointestinal Cancers
1. Department of Clinical Laboratory, Wuxi Huishan District People's Hospital, Wuxi, Jiangsu Province, 214000, China.
2. College of Basic Medicine, China Medical University, Shenyang, Liaoning province 110000, China.
3. Department of Ultrasound, Wuxi No.2 People's Hospital; Jiangnan University Medical Center, Affiliated Wuxi Clinical College of Nantong University, Wuxi, Jiangsu province 214002, China.
4. Department of Clinical Laboratory, The Affiliated Suqian First People's Hospital of Nanjing Medical University, Suqian, Jiangsu, 223800, China.
* First authors: Zhou Zhang and Pengli Zhou.
Received 2023-5-27; Accepted 2023-7-23; Published 2023-9-11
Abstract
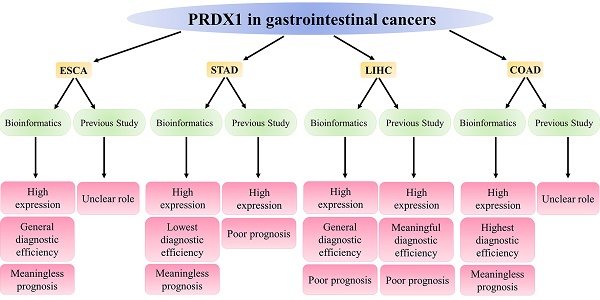
Esophageal, gastric, liver, and colorectal cancers represent four prevalent gastrointestinal cancers that pose substantial threats to global health due to their high morbidity and mortality rates. Peroxiredoxin 1 (PRDX1), a significant component of the PRDXs family, primarily functions to counteract the peroxides produced by metabolic activities in the body, thereby maintaining the dynamic equilibrium of peroxides in vivo. Intriguingly, PRDX1 expression correlates strongly with cancer's onset, progression, and prognosis. This study mainly applied bioinformatics methods to analyze PRDX1's expression, diagnosis, and prognosis in gastrointestinal cancers and to summarize current research advancements. Evidence from the bioinformatics database suggested that the high expression of PRDX1 was a prominent characteristic of these four gastrointestinal cancers, with this observation reaching statistical significance. The high expression of PRDX1 in gastrointestinal cancer cells also confirms this result. Notably, the primary alteration in PRDX1 within these cancers is the presence of genetic mutations. PRDX1 demonstrated the highest diagnostic efficacy for colorectal cancer. Nevertheless, elevated PRDX1 levels only significantly diminished the survival time of liver cancer patients, exerting no statistically significant impact on the survival duration of patients afflicted by the other three types of gastrointestinal cancers. Recent research has indicated variability in PRDX1 expression across different cancer types, with high expression being predominantly observed in these four gastrointestinal cancers and, in most instances, unfavorable prognosis. These findings broadly align with the results derived from bioinformatics. This research underscores the high expression of PRDX1 in gastrointestinal cancers, its relevance to the diagnosis and prognosis monitoring of these cancers, and its potential to guide clinical treatment for these cancers.
Keywords: PRDX1, gastrointestinal cancers, expression, prognosis.
Introduction
Gastrointestinal cancers make up approximately 26% of all cancer incidences and contribute to about 35% of all cancer mortalities, a characteristic duality described as "double high"[1]. This group includes prevalent malignancies such as liver, gastric, esophageal, and colorectal cancer. Notwithstanding significant advances in colorectal cancer screening, the diagnosis of other gastrointestinal cancers, including liver, gastric, and esophageal cancer, often occurs at advanced stages, with patients experiencing a subsequent poor prognosis[2, 3]. Factors such as population growth, aging, and lifestyle changes have led to a continual rise in gastrointestinal cancer incidences, with an accompanying shift toward younger patient demographics[4].
Reactive oxygen species (ROS) are primary oxidative stress products in the body and are widely recognized as critical contributors to cancer development and progression[5] [6]. The accumulation of ROS has a bifaceted impact on cells. A moderate ROS increase can stimulate cell proliferation and differentiation, while an excessive ROS concentration can lead to oxidative damage to DNA, proteins, and lipids[7]. The Peroxiredoxins (PRDXs) family, a group of common peroxidase enzymes, can modulate excessive ROS accumulation, thereby influencing the onset and progression of various diseases, including cancer. Therefore, targeting antioxidant mechanisms in cancer in combination with current cancer therapies may be a potential strategy to treat tumors[8, 9]. PRDX1, a member of the PRDXs family, resides at the 1q34.1 locus, weighs approximately 23 kDa, and is classified as a typical 2-Cys cysteine type. PRDX1 is a member of the 2-Cys PRDXs subfamily and is present mainly in the cytosol[10]. PRDX1 containing a cysteine at the N-terminal Cys52 detoxifies peroxides at the expense of Cys52 oxidation through intermolecular disulphide formation with the other conserved C-terminal Cys173 residue[11].
Within the nucleus, oligomeric PRDX1 can either promote or inhibit cell death by directly binding transcription factors such as p53, c-Myc, NF-κB, and AR, thereby altering their biological activity. In the cytoplasm, PRDX1 in the form of dimers regulates ROS-dependent signaling pathways, thereby influencing cell growth, differentiation, proliferation, and apoptosis[11]. In the presence of reactive oxygen species (ROS) inducers, PRDX1 can promote PCa cell proliferation by increasing AR protein stability and activating AR signaling, even under androgen deprivation conditions[9]. Furthermore, PRDX1, via Toll-like receptors (TLR4), controls inflammation, immunity, and tissue repair processes, thereby augmenting the cytotoxic activity of NK cells against cancer cells[12]. Multiple studies have indicated that PRDX1 is overexpressed in several types of cancers including breast, prostate, lung, and liver cancers[9, 11, 13-16]. The level of its expression has a significant association with tumor cell proliferation, differentiation, metastasis, recurrence, and sensitivity to radiotherapy and chemotherapy[11, 17-19]. Despite these findings, a comprehensive review of PRDX1's role in common gastrointestinal cancers remains elusive.
Bioinformatics, a rapidly evolving discipline, applies methodologies and tools from human genome life science, information science, mathematics, and biology to acquire, process, store, analyze, and interpret vast quantities of biological data, thereby unraveling their biological significance[20-22]. Utilizing various biological databases, bioinformatics analysis can identify key tumor-related biomarkers, thereby establishing a foundation for cancer diagnosis and prognosis [23].
This study applies bioinformatics analysis to illuminate PRDX1 expression, diagnosis, and prognosis in gastrointestinal tumors. Subsequent analysis further scrutinizes the current research progression of PRDX1 in these malignancies, providing insights that may guide future exploration of PRDX1's role in gastrointestinal cancers.
Materials and methods
Expression and Mutation Analysis of PRDX1 in Pan-Cancer
The University of Alabama, Birmingham Cancer Data Analysis Portal website (UALCAN; http://ualcan.path.uab.edu/) was utilized to derive PRDX1 expression levels in pan-cancer and corresponding adjacent tissues. The cBioPortal website (https://www.cbioportal.org/) was subsequently used to acquire data on mutation frequency, mutation type, copy number alterations (CNA), and mutation location of PRDX1 in pan-cancer.
Identification of Differentially Expressed Genes (DEGs) of PRDX1 in Gastrointestinal Cancers
Volcanic maps were created using The Cancer Genome Atlas (TCGA) databases to illustrate the distribution of differentially expressed genes in four types of gastrointestinal cancers. The expression of PRDX1 in gastrointestinal cancers was depicted using the TCGA databases, and a statistical difference analysis was conducted.
Cell culture and reagents
TE-1 (Human esophageal cancer), HepG2 (human hepatocellular carcinoma) and LS513 (Human colorectal cancer) cell lines were purchased from National Collection of Authenticated Cell Cultures. HEEC (Human esophageal epithelium), NCM460 (Human colon epithelium), SGC-7901 (Human gastric cancer) and GES-1(Human gastric epithelium) cell lines were purchased from IMMOCELL. QSG7701 (Human hepatocyte) was purchased from Beyotime. Each cell line is cultured according to the conditions specified in their respective culture instructions.
Reverse transcription-quantitative (RT-q) PCR
Total RNA was extracted from eight cell lines using TRIzol reagent (Beyotime, R0016). Using BeyoRT ™ The II cDNA first strand synthesis kit (Beyotime, D7168M) synthesizes complementary DNA (cDNA). BeyoFast ™ SYBR Green qPCR Mix (Beyotime, D7265) and CFX 96 thermal circulator (Bio Rad) were used for RT-qPCR. Perform RNA extraction, cDNA synthesis and qPCR according to the instructions. RT-qPCR was carried out using the following conditions: preheating for 2 min at 95°C; and then repeating 40 cycles in 95°C for 15 sec and 56°C for 30 sec. The primer sequences of PRDX1 were as follows: Forward, 5'-AGCTGTTATGCCAGATGGTC-3' and Reverse, 5'-TGAGAATCCACAGAAGCACC-3'. Using GAPDH as an internal reference gene and the primer sequence were as follows: Forward, 5'-ATTCCACCCATGGCAAATTC-3' and Reverse, 5'-TGGGATTTCCATTGATGACAAG-3'. The relative expression level of PRDX1 in this study was calculated using the 2-ΔΔCq formula. Repeat the experiment three times.
Gene Mutations and Related Genes Analysis of PRDX1 in Gastrointestinal Cancers
The cBioPortal website was initially employed to investigate the mutation sources and types of PRDX1 in gastrointestinal cancers. Subsequently, the UALCAN website showed the correlation and correlation coefficients of the primary related genes for PRDX1 in common gastrointestinal cancers.
Clinical parameters of patients
We downloaded clinical data of 184 ESCA patients from TCGA, and then deleted 43 samples with incomplete clinical data, resulting in a valid sample of 141; We downloaded clinical data of 415 STAD patients from TCGA, and then deleted 70 samples with incomplete clinical data, resulting in a valid sample of 345; We downloaded clinical data of 371 LIHC patients from TCGA, and then deleted 141 samples with incomplete clinical data, resulting in a valid sample of 230; We downloaded clinical data of 622 COAD patients from TCGA, and then deleted 111 samples with incomplete clinical data, resulting in a valid sample of 511. The specific parameter characteristics are detailed in Table 1.
Clinical parameters of patients.
| ESCA | STAD | LIHC | COAD | |
|---|---|---|---|---|
| N | 141 | 345 | 230 | 511 |
| Age (%) | ||||
| ≤60 | 70(49.6) | 112(32.5) | 121(52.6) | 144(28.2) |
| >60 | 71(50.4) | 233(67.5) | 109(47.4) | 367(71.8) |
| Gender (%) | ||||
| Female | 21(14.9) | 125(36.2) | 73(31.7) | 236(46.2) |
| Male | 120(85.1) | 220(63.8) | 157(68.3) | 275(53.8) |
| T (%) | ||||
| T1 | 24(17.0) | 15(4.3) | 113(49.1) | 17(3.3) |
| T2 | 39(27.7) | 71(20.6) | 51(22.2) | 90(17.6) |
| T3 | 74(52.5) | 162(47.0) | 56(24.3) | 353(69.1) |
| T4 | 4(2.8) | 97(28.1) | 10(4.3) | 51(10.0) |
| N (%) | ||||
| N0 | 67(47.5) | 109(31.6) | 226(98.3) | 301(58.9) |
| N1 | 60(42.6) | 92(26.7) | 4(1.7) | 120(23.5) |
| N2 | 9(6.4) | 71(20.6) | 0(0) | 90(17.6) |
| N3 | 5(3.5) | 73(21.2) | 0(0) | 0(0) |
| M (%) | ||||
| M0 | 132(93.6) | 322(93.3) | 227(98.7) | 429(84.0) |
| M1 | 9(6.4) | 23(6.7) | 3(1.3) | 82(16.0) |
| Stage (%) | ||||
| Stage I | 13(9.2) | 45(13.0) | 111(48.3) | 94(18.4) |
| Stage II | 72(51.1) | 112(32.5) | 50(21.7) | 197(38.6) |
| Stage III | 47(33.3) | 153(44.3) | 65(28.3) | 138(27.0) |
| Stage IV | 9(6.4) | 35(10.1) | 4(1.7) | 82(16.0) |
Expression and Mutation of PRDX1 in Pan-Cancer. (A) Expression of PRDX1 in Pan-Cancer. (B) Expression of PRDX1 in Pan-Cancer and Normal Samples. (C) The alteration frequency for each mutation type and (D) mutation site. *P<0.05; “ns” represents no statistical significance.

Diagnostic Efficacy of PRDX1 in Gastrointestinal Cancers
The TCGA database was used to assess the diagnostic potential of PRDX1 in gastrointestinal cancers. This assessment was followed by plotting the Receiver Operating Characteristic (ROC) curve. The area under the curve (AUC) and the optimal cutoff value were specifically compared to determine the best diagnostic efficacy.
Survival Analysis of PRDX1 in Gastrointestinal Cancers
The TCGA database was used to analyze the survival rates of patients with PRDX1 in gastrointestinal cancers.
Literature Review
To further consolidate understanding of PRDX1's role in common gastrointestinal cancers, relevant literature was examined, focusing on esophageal cancer, gastric cancer, liver cancer, and colorectal cancer. This analysis aimed to summarize the research progress of PRDX1 in these common gastrointestinal cancers.
Statistical Analysis
The R software (4.0.2) and Graphpad 9.0 were employed for data analysis to evaluate statistical differences. Data are expressed as mean ± standard deviation. The unpaired Student's t-test was used for data analysis. ROC curves were utilized to evaluate the diagnostic efficacy of PRDX1 in gastrointestinal cancers. The Kaplan-Meier plots were applied to illustrate the survival curve of PRDX1 in these cancers. All hypothesis tests were two-sided, with P<0.05 being considered indicative of a statistically significant difference. Gene differential analysis [|LogFC|>1, adjusted P-value (FDR) <0.05] was conducted by comparing tumor tissue with controls using the limma R package. The entire gene expression data underwent log2 transformation, with correlations calculated using Spearman's coefficient analysis.
Result
Expression and Mutation of PRDX1 in Pan-Cancer
The expression of PRDX1 in pan- cancer was detected, and the results showed that the expression of PRDX1 in gastrointestinal cancers was higher than the median level of pan-cancer (Figure 1A). When incorporating adjacent tissues for comparison, it was revealed that the expression of PRDX1 in pan-cancer typically exceeds that in corresponding adjacent tissues. Notably in gastrointestinal cancers, PRDX1 expression surpasses that in corresponding adjacent tissues (Figure 1B). The mutation frequency, mutation type, and copy number alterations (CNA) of PRDX1 in pan-cancer were also investigated. Findings highlighted that PRDX1 is primarily characterized by gene mutations in gastrointestinal cancers (46.67% gene mutations, 26.67% gene amplification, 20% structural variant, and 6.67% multiple alterations) (Figure 1C). Further analysis of mutation location based on PRDX1's structure revealed that the E65* alteration in the AhpC-TSA domain is the primary type of genetic alteration of PRDX1 (Figure 1D).
Identification of Differentially Expressed Genes (DEGs) of PRDX1 in Gastrointestinal Cancers
The R software package was used to identify DEGs of gastrointestinal cancers. DEGs were visualized using microarray volcano plots of gastrointestinal cancers (Figure 2A-D). Box-plots were then employed to analyze the expression of PRDX1 in gastrointestinal cancers. When compared to adjacent tissues, PRDX1 is found to be highly expressed in gastrointestinal cancers, with statistical differences noted in all four types of gastrointestinal cancers. This suggests that PRDX1 influences the onset and progression of gastrointestinal cancers (Figure 2E-H).
Relative PRDX1 mRNA Expression in Gastrointestinal Cancers
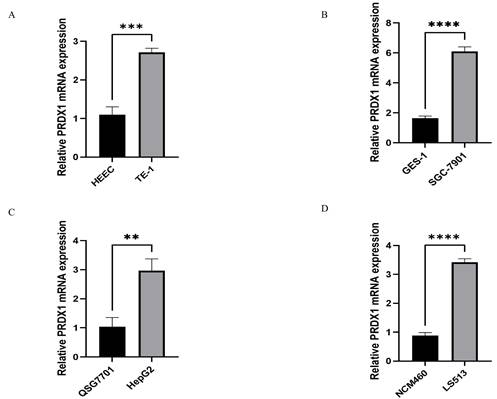
RT-qPCR technology was used to detect the relative expression of PRDX1 in four types of gastrointestinal cancer cells and corresponding normal cells to validate the results of bioinformatics. The following results were found. The expression of PRDX1 in TE-1 cell is higher than that in HEEC cell, and the difference is statistically significant(p<0.001) (Figure 3A); The expression of PRDX1 in SGC-7901 cell is higher than that in GES-1 cell, and the difference is statistically significant(p<0.0001) (Figure 3B); The expression of PRDX1 in HepG2 cell is higher than that in QSG7701 cell, and the difference is statistically significant(p<0.01) (Figure 3C); The expression of PRDX1 in LS513 cell is higher than that in NCM460 cell, and the difference is statistically significant (p<0.0001) (Figure 3D).
Gene Mutations and Related Genes Analysis of PRDX1 in Gastrointestinal Cancers
The gene mutations of PRDX1 in gastrointestinal cancers were scrutinized, revealing that these mutations predominantly involve amino acid changes, with Missense_ Mutation and Single Nucleotide Polymorphism (SNP) being the main mechanisms (Table 2). Following this, related genes analysis of PRDX1 in gastrointestinal cancers was conducted. The analysis indicates that three other types of gastrointestinal cancers exhibit some related genes, but gastric cancer does not (Table 3).
Gene mutations of PRDX1 in gastrointestinal cancers.
| Cancer Type | Sample ID | Protein Change | Mutation Type | Variant Type |
|---|---|---|---|---|
| LIHC | TCGA-CC-A7IE-01 | TESK2-PRDX1 Fusion | Fusion | NA |
| LIHC | TCGA-DD-A73G-01 | TESK2-PRDX1 Fusion | Fusion | NA |
| STAD | TCGA-VQ-A8E0-01 | TESK2-PRDX1 Fusion | Fusion | NA |
| COAD | TCGA-EI-6885-01 | GOLIM4-PRDX1 Fusion | Fusion | NA |
| LIHC | TCGA-DD-A3A3-01 | F195L | Missense_Mutation | SNP |
| ESCA | TCGA-JY-A6FE-01 | S191R | Missense_Mutation | SNP |
| ESCA | TCGA-IG-A3QL-01 | G129D | Missense_Mutation | SNP |
| STAD | TCGA-BR-8680-01 | E193* | Nonsense_Mutation | SNP |
| COAD | TCGA-AG-A002-01 | E65* | Nonsense_Mutation | SNP |
| COAD | TCGA-A6-5661-01 | A63T | Missense_Mutation | SNP |
| COAD | TCGA-DM-A1HB-01 | Y194H | Missense_Mutation | SNP |
Diagnostic Efficacy of PRDX1 in Gastrointestinal Cancers
The Receiver Operating Characteristic (ROC) curve was used to illustrate the diagnostic efficacy of PRDX1 in gastrointestinal cancers. The results showed that the Areas Under the Curve (AUC) for esophageal cancer, gastric cancer, liver cancer, and colorectal cancer were 0.8295, 0.6944, 0.8245, and 0.8849, respectively. The cut-off values for these cancers were 0.7866, 0.7982, 0.7899, and 0.8368, respectively. The specificity for these cancers were 0.692, 0.611, 0.900, and 0.922, while the sensitivities were 0.886, 0.757, 0.743, and 0.767, respectively. When the Areas Under the Curve (AUC) is more than 0.7, we believe that this indicator has general diagnostic value for disease detection. When the Areas Under the Curve (AUC) is more than 0.9, we believe that this indicator has great diagnostic value for disease detection. Obviously, PRDX1 has general diagnostic value for esophageal cancer, liver cancer, and colorectal cancer, while its diagnostic value for gastric cancer is relatively low. Therefore, PRDX1 demonstrates the highest diagnostic efficiency for colorectal cancer and the lowest for gastric cancer (Figure 4).
Survival Analysis of PRDX1 in Gastrointestinal Cancers
The relationship between PRDX1 and survival prognosis of four types of gastrointestinal cancers was investigated (Figure 5). Intriguingly, only in liver cancer does the high expression of PRDX1 significantly reduce patient survival time, with the difference deemed statistically significant (p<0.05). No statistical difference was noted among the three other types of gastrointestinal cancers. This suggests that PRDX1 might hold greater significance in liver cancer (Figure 5C).
Identification of DEGs of PRDX1 in gastrointestinal cancers. (A-D) Volcano plots of gastrointestinal cancers. (E-H) Box-plots of PRDX1 in gastrointestinal cancers. ***P<0.001; ****P<0.0001. DEGs, Differentially Expressed Genes.

Relative PRDX1 mRNA expression in gastrointestinal cancers. (A-D) Relative PRDX1 mRNA expression in gastrointestinal cancers cells. **P<0.01; ***P<0.001; ****P<0.0001.
Related genes analysis of PRDX1 in gastrointestinal cancers
| Cancer Type | Related Gene | Correlation | Coefficient |
|---|---|---|---|
| ESCA | EPHX1 | Postive | 0.81 |
| ESCA | GSTM3 | Postive | 0.77 |
| ESCA | AKR1C1 | Postive | 0.76 |
| ESCA | MICAL1 | Neagive | 0.36 |
| ESCA | SH3KBP1 | Neagive | 0.34 |
| ESCA | CLMN | Neagive | 0.33 |
| COAD | PSMB2 | Postive | 0.65 |
| COAD | EBNA1BP2 | Postive | 0.61 |
| COAD | HSPB11 | Postive | 0.6 |
| COAD | SHC2 | Neagive | 0.35 |
| COAD | GLTSCR2 | Neagive | 0.34 |
| COAD | EEF2 | Neagive | 0.33 |
| LIHC | EIF2B3 | Postive | 0.7 |
| LIHC | PSMA5 | Postive | 0.68 |
| LIHC | EBNA1BP2 | Postive | 0.64 |
| LIHC | COL18A1 | Neagive | 0.33 |
| LIHC | SERPINA5 | Neagive | 0.33 |
| LIHC | TMPRSS6 | Neagive | 0.32 |
| STAD | N/A | N/A | N/A |
Origin and Biological Function of PRDX1
In 1988, a protective protein scavenging sulfur free radicals was first discovered in yeast, even though it did not exhibit any known catalytic antioxidant activity akin to catalase, glutathione peroxidase, or superoxide dismutase[24]. Currently, this protective protein is part of the peroxiredoxin (PRDX) protein family. PRDXs, divided into three types based on the number of cysteine residues involved in redox reactions: typical 2-Cys (PRDX1-4), atypical 2-Cys (PRDX5), and 1-Cys (PRDX6)[9, 25, 26], have six subtypes, PRDX1-6. PRDX1, an important member of the PRDX family, was originally identified as a human erythrokine, which can boost the activity of natural killer cells[27]. Cys52 in PRDX1 reduces H2O2 to H2O and is oxidized to sulfite. Cys173 reacts with sulfite to form intramolecular disulfide bonds, which are reduced by thioredoxin I (Trx I). Following this, Trx I is reduced by NADPH dependent thioredoxin reductase, creating an oxidation-reduction cycle mechanism (Figure 6)[28]. Previous studies have suggested that PRDX or Trx peroxidases are essential endogenous antioxidants that protect cells from oxidative damage by reducing H2O2 and peroxynitrite, as well as by eliminating sulfhydryl radicals[29-31]. Nonetheless, the activity of PRDX1 is closely related to the post-translational modification of its protein. For instance, it has been observed that following acetylation at the K197 site of PRDX1, its ability to eliminate Reactive Oxygen Species (ROS) diminishes [32]. Furthermore, Mammalian sterile tweenty 1 (Mst1) deactivates PRDX1 by phosphorylating the Thr90 and Thr183 sites of PRDX1, leading to the accumulation of hydrogen peroxide in the cell [33]. Chang et al. discovered that cyclin D kinase Cdc2 can also phosphorylate the Thr90 site of PRDX1 and inactivate PRDX1's peroxidase activity[34]. Pin1 binds to PRDX1 through its phospho-Thr90-Pro91 motif. When the Thr90 mutation of PRDX1 occurs, this binding no longer exists[35]. Assays revealed that PRDX1 can be glutathioneized at Cys 52, 83, and 173[36]. This suggests that the post-translational modification of PRDX1 can influence its biological function. Post-translational modification and associated sites of PRDX1 are detailed in Table 4.
Post-translational modification of PRDX1 in point mutation.
| Records, n | ||||
|---|---|---|---|---|
| Mutation | Flanking sequence | Modification | LTP | HTP |
| K7 | MSSGNAkIGHPAPN | Acetylation | 0 | 4 |
| K7 | MSSGNAkIGHPAPN | Ubiquitylation | 0 | 6 |
| K7 | MSSGNAkIGHPAPN | Sumoylation | 0 | 1 |
| K7 | MSSGNAkIGHPAPN | Succinylation | 0 | 1 |
| K16 | GHPAPNFkAtAVMPD | Acetylation | 0 | 13 |
| K16 | GHPAPNFkAtAVMPD | Ubiquitylation | 0 | 8 |
| K16 | GHPAPNFkAtAVMPD | Sumoylation | 0 | 1 |
| K16 | GHPAPNFkAtAVMPD | Methylation | 0 | 1 |
| K16 | GHPAPNFkAtAVMPD | Succinylation | 0 | 1 |
| T18 | PAPNFkAtAVMPDGQ | Phosphorylation | 0 | 1 |
| K27 | VMPDGQFkDIsLsDy | Acetylation | 0 | 15 |
| K27 | VMPDGQFkDIsLsDy | Ubiquitylation | 0 | 16 |
| K27 | VMPDGQFkDIsLsDy | Methylation | 0 | 1 |
| K27 | VMPDGQFkDIsLsDy | Succinylation | 0 | 1 |
| S30 | DGQFkDIsLsDykGk | Phosphorylation | 0 | 6 |
| S32 | QFkDIsLsDykGkYV | Phosphorylation | 1 | 16 |
| Y34 | kDIsLsDykGkYVVF | Phosphorylation | 0 | 8 |
| K35 | DIsLsDykGkYVVFF | Acetylation | 0 | 79 |
| K35 | DIsLsDykGkYVVFF | Ubiquitylation | 0 | 38 |
| K35 | DIsLsDykGkYVVFF | Succinylation | 0 | 1 |
| K37 | sLsDykGkYVVFFFY | Acetylation | 0 | 1 |
| K37 | sLsDykGkYVVFFFY | Ubiquitylation | 0 | 7 |
| T90 | CHLAWVNtPkkQGGL | Phosphorylation | 3 | 2 |
| K92 | LAWVNtPkkQGGLGP | Acetylation | 0 | 2 |
| K92 | LAWVNtPkkQGGLGP | Succinylation | 0 | 1 |
| K93 | AWVNtPkkQGGLGPM | Acetylation | 0 | 1 |
| K93 | AWVNtPkkQGGLGPM | Ubiquitylation | 0 | 7 |
| K93 | AWVNtPkkQGGLGPM | Sumoylation | 0 | 1 |
| K109 | IPLVsDPkRtIAQDy | Acetylation | 0 | 67 |
| K109 | IPLVsDPkRtIAQDy | Ubiquitylation | 0 | 14 |
| K109 | IPLVsDPkRtIAQDy | Succinylation | 0 | 1 |
| K120 | AQDyGVLkADEGIsF | Ubiquitylation | 0 | 15 |
| K120 | AQDyGVLkADEGIsF | Succinylation | 0 | 1 |
| K136 | GLFIIDDkGILRQIT | Acetylation | 0 | 5 |
| K136 | GLFIIDDkGILRQIT | Ubiquitylation | 0 | 18 |
| K168 | QAFQFtDkHGEVCPA | Acetylation | 0 | 7 |
| K168 | QAFQFtDkHGEVCPA | Ubiquitylation | 0 | 7 |
| K178 | EVCPAGWkPGsDtIk | Acetylation | 0 | 2 |
| K178 | EVCPAGWkPGsDtIk | Ubiquitylation | 0 | 18 |
| K185 | kPGsDtIkPDVQkSk | Acetylation | 0 | 2 |
| K185 | kPGsDtIkPDVQkSk | Ubiquitylation | 0 | 8 |
| K185 | kPGsDtIkPDVQkSk | Sumoylation | 0 | 1 |
| K192 | kPDVQkSkEyFskQK | Acetylation | 0 | 3 |
| K192 | kPDVQkSkEyFskQK | Ubiquitylation | 0 | 9 |
| K192 | kPDVQkSkEyFskQK | Methylation | 0 | 4 |
LTP (Low Throughput Papers), the number of records in which this modification site was determined using methods other than discovery mass spectrometry; HTP (High Throughput Papers), the number of records in which this modification site was assigned using only proteomic discovery mass spectrometry.
Expression and Role of PRDX1 in Esophageal Cancer
Esophageal cancer, a malignancy localized in the esophageal epithelium, constitutes 2% of all malignant tumors. This carcinoma chiefly manifests as either esophageal squamous cell carcinoma (ESCC) or esophageal adenocarcinoma, with ESCC being the predominant type[37]. Several studies report an overexpression of PRDX1 in ESCC cells compared to adjacent non-cancerous tissues[38-41]. Enhanced PRDX1 expression is implicated in the onset and advancement of ESCC, while diminished PRDX1 expression appears to curb tumorigenesis [42]. These observations suggest a possible oncogenic role of PRDX1 in ESCC. Wu et al. [43] reported that miR-375, by directly targeting PRDX1 mRNA, reduces PRDX1 expression levels, thereby inhibiting the proliferation and migration of ESCC cells. A study by Chen et al. [42] identified PRDX1 as a regulatory factor influencing the formation of primary cilia in ESCC cells. PRDX1, by modulating the HEF1/Aurora A/HDAC6 signaling pathway, fosters the loss of primary cilia, contributing to ESCC tumorigenesis and tumor growth. Inhibiting PRDX1, in this context, exerts anti-tumor effects. Song et al. [44] demonstrated high PRDX1 expression in ESCC, revealing that PRDX1 silencing curtails ESCC proliferation and enhances cell apoptosis. The underlying mechanism potentially involves the obstruction of the PI3K/AKT signaling pathway. However, histone deacetylase inhibitor FK228 is known to activate Prdx1, partly via the regulation of promoter histone acetylation, thereby inducing apoptosis and repressing tumor growth [45]. Interestingly, another study indicated a low PRDX1 expression in ESCC, exhibiting a significant negative correlation with lymph node metastasis and TNM staging. In contrast to patients with elevated Prdx1 expression, those exhibiting low Prdx1 expression presented shorter overall survival durations. This suggests a potential tumor-suppressive role for PRDX1, with its downregulation contributing to the progression of esophageal squamous cell carcinoma and serving as a significant prognostic marker [46]. The exact role of PRDX1 in esophageal cancer, therefore, remains ambiguous, warranting further investigation.
Diagnostic efficacy of PRDX1 in gastrointestinal cancers.

Survival analysis of PRDX1 in gastrointestinal cancers. (A-D) Effect of PRDX1 expression level on gastrointestinal cancers patient survival.

Expression and Role of PRDX1 in Gastric Cancer
Gastric cancer (GC), ranking as one of the most prevalent malignancies globally, stands as the fourth primary cause of cancer-related fatalities [47]. Numerous studies have indicated that PRDX1 mRNA and protein expression levels in GC tissue exceed those in surrounding non-cancerous tissues. High PRDX1 expression positively correlates with lymph node invasion and unfavorable prognosis, making PRDX1 a potential therapeutic and prognostic target for gastric cancer patients [48-50]. Wei et al. identified that Prdx1 regulates the invasion and metastasis of GC cell lines by inhibiting E-Ca expression [48]. Zhang et al. [51] discovered that in GC, while PRDX1 is overexpressed, miR-596 levels are reduced. Importantly, miR-596 can directly target PRDX1 mRNA to downregulate PRDX1 expression levels, exerting an inhibitory effect on GC cells. Dehydroharmine hydrochloride (HH), an alkaloid compound exhibiting anti-tumor activity, was found by Tan et al. [52] to significantly decrease the invasion and migration ability of GC cells following HH treatment. This decrease correlated with the reduction of MMP-2 and HIF-1 levels, as well as with the expression of PRDX1. These findings suggest that elevated PRDX1 expression in gastric cancer could serve as a reference indicator for poor prognosis.
Expression and Role of PRDX1 in Liver Cancer
Liver cancer continues to pose a substantial global health threat, with projections estimating over 1 million cases by 2025 [53]. The most prevalent radical interventions encompass surgical resection and liver transplantation. Yet, most hepatocellular carcinoma (HCC) patients are often diagnosed at an advanced stage, rendering them unsuitable for surgical interventions. Prognosis for patients undergoing chemotherapy and radiation therapy typically remains poor. Thus, exploring the molecular mechanism of HCC pathogenesis, identifying specific biomarkers, and targeting liver cancer cells provide feasible diagnostic and therapeutic strategies [54-56]. Various studies reveal high PRDX1 expression in liver cancer, indicating an inverse correlation with the prognosis of HCC patients. Consequently, PRDX1 may serve as a potential biomarker for HCC detection and prognosis, and its inhibitors could be candidate drugs for liver cancer treatment [57-61]. Sun et al. [13] observed that PRDX1 knockout activated cleaved caspase-3, caspase-9, Bax protein, and PARP-1, leading to a reduction in Bcl-2 expression and subsequent induction of apoptosis. This also triggered the expression of dynein related protein1 (Drp1), fission1 (Fis1), and dynein2 (Dyn2), promoting mitochondrial fission and inhibiting the proliferation of hepatoma cells. Lou et al. [62] found that PRDX1 silencing elevated the accumulation of ferrous ions and lipid peroxidation in HepG2 cells, thus fostering iron-induced apoptosis in liver cancer. Patricia et al. [63] determined that PRDX1 silencing led to the down-regulation of AFP, osteopontin, and β-catenin transcripts, while γ-E-cadherin and proapoptotic proteins (Bax, caspase-3) transcripts were upregulated. Further, the activity of glutamyl transpeptidase decreased, and alkaline phosphatase and Casp3 activity increased, collectively inhibiting cell proliferation and growth. Kang et al. [64] discovered that an extract of Auricularia auricula, by inhibiting PRDX1, diminished the levels of total glutathione (GSSG/GSH) and superoxide dismutase (SOD), thereby inactivating antioxidant enzymes, impeding cells from processing reactive oxygen species, and promoting apoptosis of liver cancer cells. Consequently, high PRDX1 expression in liver cancer could serve as a diagnostic marker for liver cancer and is linked to poor prognosis, corroborating the findings of bioinformatics.
Involvement of PRDX1 in redox cycle mechanism.

Expression and Role of PRDX1 in Colorectal Cancer
Colorectal cancer stands as one of the common gastrointestinal cancers and is the second leading cause of cancer mortality [65]. Recent years have witnessed a global increase in the incidence of colorectal cancer, attributable to changes in lifestyle and dietary patterns, with an emerging trend towards younger age at onset [66]. Tumor metastasis accounts for a significant cause of mortality in colorectal cancer patients. Therefore, studying the pathogenesis and metastasis mechanism of colorectal cancer, discovering highly specific and sensitive molecular markers, provides significant new targets for colorectal cancer treatment and prognosis assessment, which is crucial for enhancing the prognosis of colorectal cancer [67]. Several studies have identified a positive correlation between PRDX1 expression and tumor grade, lymph node metastasis, and vascular density [68, 69]. This variability in the ability to invade and migrate tumor cells across different CRC cell lines suggests that PRDX1 might predict prognosis and serve as a potential therapeutic target, regulating tumor metastasis and angiogenesis in colorectal cancer. It could also serve as a biomarker for early detection of cancer progression and identification of potential pathogenesis [70, 71]. Xu et al. [72] proposed that the side effects of Celastrol might be mitigated through structural modification, and PRDX1 inhibition appears promising for treating colorectal cancer. Nonetheless, Tai et al. [73] reported that Gynostemma pentaphyllum saponins (GpS) hinder the development of colorectal cancer by upregulating PRDX1 and PRDX2, inhibiting the Ras, RAF/MEK/ERK/STAT, PI3K/AKT/mTOR signaling pathways, and regulating the JNK/p38 MAPK signaling pathway. These findings indicate potential controversies concerning the expression and role of PRDX1 in colorectal cancer.
Expression and Role of PRDX1 in Other Gastrointestinal Cancers
Some studies indicate a close relationship between PRDX1 and other gastrointestinal cancers such as pancreatic cancer [74], cholangiocarcinoma [75], and oral cancer [76]. In pancreatic cancer, there is a significant elevation in PRDX1 expression levels, making it a viable therapeutic target. Park et al. found that naringenin could diminish PRDX1 expression in pancreatic cancer cells, escalate ROS levels in cancer cells, and subsequently upregulate apoptosis signal-regulated kinase 1 (ASK1) to induce apoptosis [77]. In cholangiocarcinoma, PRDX1 expression also significantly increases, influencing the occurrence and development of cholangiocarcinoma by regulating SNAT1 expression[75]. In oral cancer, PRDX1 is highly expressed and is significantly correlated with tumor staging, lymph node metastasis, and pathological grading, making it a promising biomarker for predicting lymph node metastasis and prognosis in oral cancer patients[76]. However, these gastrointestinal cancers have a relatively low incidence rate. Moreover, existing studies on PRDX1 in these cancers are limited, warranting further research for more conclusive insights.
Discussion
Gastrointestinal cancers, encompassing esophageal cancer, gastric cancer, liver cancer, and colorectal cancer, constitute a primary category of malignancies posing significant threats to human health due to their high incidence and mortality rates. Unraveling the molecular mechanisms that underpin the diagnosis and treatment of gastrointestinal cancers is imperative for facilitating early detection and forecasting prognostic outcomes. As a vital component of the PRDXs family, the peroxidase activity of PRDX1 plays a key role in maintaining the equilibrium of intracellular reactive oxygen species (ROS), thereby influencing the onset, progression, and prognosis of cancer [78-80].
This investigation primarily focuses on reporting the expression and mechanism of PRDX1 in gastrointestinal cancers via bioinformatics methods and review of existing literature, with the goal of offering guidance for the diagnosis and prognosis of gastrointestinal cancers. Initial bioinformatics analysis indicates that PRDX1 expression in gastrointestinal cancers exceeds the median level across pan-cancer. Furthermore, the expression of PRDX1 in the majority of cancers is typically higher than in the corresponding adjacent tissues, with this trend being especially pronounced in gastrointestinal cancers. This forms the groundwork for studying the diagnosis and prognosis of PRDX1 in gastrointestinal cancers, aligning with the research findings of Gao et al [81]. Subsequent utilization of the TCGA database enabled the creation of volcanic maps for gastrointestinal cancers to highlight differentially expressed genes (DEGs). To further elucidate the expression of PRDX1 in gastrointestinal cancers, box-plots were generated to display PRDX1 expression in four types of gastrointestinal cancers and their adjacent tissues. Results indicated high PRDX1 expression in all four types of gastrointestinal cancers, all of which were statistically significant. And we used RT-qPCR technology to validate the cells of gastrointestinal cancers, and the results were consistent with the box-plot. However, literature review disclosed some discordance regarding the expression level of PRDX1 in esophageal cancer [38, 41, 45, 46]and colorectal cancer (68, 71, 73). The heightened PRDX1 expression in gastric cancer [48, 51] and liver cancer [57, 60] is consistent with the box-plot results.
In effort to delve deeper into the molecular mechanism of PRDX1 in gastrointestinal cancers, a statistical analysis of gene mutations and related genes of PRDX1 in gastrointestinal cancers was conducted. These mutations were characterized primarily by amino acid changes, with Missense_Mutation and SNP being the predominant forms. Additionally, the analysis suggested that three other types of gastrointestinal cancers exhibit related genes, with the exception of gastric cancer. This implies that PRDX1 mutations may be significant in the onset and progression of gastrointestinal cancers. The related genes of PRDX1 set the stage for future exploration of specific PRDX1 molecules in gastrointestinal cancers.
ROC curves were used to demonstrate the diagnostic efficacy of PRDX1 in gastrointestinal cancers. PRDX1 demonstrated the highest diagnostic efficiency for colorectal cancer and the lowest for gastric cancer. No current literature provides clear evidence regarding the diagnostic efficacy of PRDX1 in gastrointestinal cancers.
The prognostic role of PRDX1 in gastrointestinal cancers was examined by presenting the survival curve of PRDX1 in gastrointestinal cancers through the TCGA database. Interestingly, high PRDX1 expression only decreased the survival time in liver cancer patients. No statistical difference was observed among the three other types of gastrointestinal cancers. Literature review also indicated that high PRDX1 expression promotes the proliferation, invasion, and metastasis of liver cancer cells, thereby fostering the onset and progression of liver cancer [57, 60, 82]. These findings align with the results of the survival analysis.
However, our study still has some limitations. In this study, we mainly summarized the role of PRDX1 in gastrointestinal cancers by bioinformatics and previous study. In general, our exploration of PRDX1 in gastrointestinal cancers is meaningful. It is a pity that we did not conduct extensive molecular and cellular experiments to confirm the role of PRDX1 in gastrointestinal cancers apart from RT-qPCR technology. Moreover, we did not rely on real patients of gastrointestinal cancers to improve our study. In the future, we will validate and refine our study from these aspects.
In summary, this study provides evidence that PRDX1 is highly expressed in gastrointestinal cancers and is associated with the diagnosis and prognosis of these malignancies, thus offering potential guidance in the clinical management of gastrointestinal cancers.
Abbreviations
AUC: area under the curve; PRDX1: peroxiredoxin 1; TCGA: the cancer genome atlas; GTEx: genotype-tissue expression; ACC: adrenocortical carcinoma; BLCA: bladder urothelial carcinoma; BRCA: breast invasive carcinoma; CESC: cervical squamous cell carcinoma; CHOL: cholangiocarcinoma; COAD: colorectal adenocarcinoma; DLBC: Lymphiod neoplasm diffuse large B-cell lymphoma; ESCA: esophageal carcinoma; GBM: glioblastoma multiforme; HNSC: head and Neck squamous cell carcinoma; KICH: kidney chromophobe; KIRC: kidney renal clear cell carcinoma; KIRP: kidney renal papillary cell carcinoma; LGG: brain lower grade glioma; OV: ovarian serous cystadenocarcinoma; MESO: mesothelioma; LIHC: liver hepatocellular carcinoma; LUAD: lung adenocarcinoma; LUSC: lung squamous cell carcinoma; PAAD: pancreatic adenocarcinoma; PRAD: prostate adenocarcinoma; PCPG: pheochromocytoma and Paraganglioma; READ: Rectum adenocarcinoma; SARC: sarcoma; SKCM: skin cutaneous melanoma; LAML: acute myeloid leukemia; TGCT: testis germ cell tumors; THCA: thyroid carcinoma; THYM: thymoma; STAD: stomach adenocarcinomna; UCEC: uterine corpus endometrial carcinoma; UCS: uterine carcinosarcoma; UVM: uveal melanoma.
Acknowledgements
Funding
This research was supported by the Suqian Key Laboratory of Science and Technology Project (Suqian Clinical Medical Laboratory) (Grant No. M201902), Maternal and Child Scientific Research Project of Jiangsu Province (Grant No. F201868), Jiangsu Province Six Talents Project (YY-230).
Author contributions
ZZ and BP were responsible for the conception of the study; ZZ, PZ and ML were responsible for bioinformatics analysis, RT-qPCR and literature review; ZZ and BP wrote the original draft; ZZ and BP reviewed and edited the final manuscript. All authors have read and approved the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7-33
2. Joshi SS, Badgwell BD. Current treatment and recent progress in gastric cancer. CA Cancer J Clin. 2021;71:264-79
3. Zheng Y, Li Z, Wang Y, Chen W, Lin Y, Guo J. et al. CircRNA: A new class of targets for gastric cancer drug resistance therapy. Pathol Oncol Res. 2023;29:1611033
4. Huang J, Lucero-Prisno DE 3rd, Zhang L, Xu W, Wong SH, Ng SC. et al. Updated epidemiology of gastrointestinal cancers in East Asia. Nat Rev Gastroenterol Hepatol. 2023;20:271-87
5. Harris IS, DeNicola GM. The Complex Interplay between Antioxidants and ROS in Cancer. Trends Cell Biol. 2020;30:440-51
6. Cheung EC, Vousden KH. The role of ROS in tumour development and progression. Nat Rev Cancer. 2022;22:280-97
7. Yang S, Lian G. ROS and diseases: role in metabolism and energy supply. Mol Cell Biochem. 2020;467:1-12
8. Liu Y, Wang P, Hu W, Chen D. New insights into the roles of peroxiredoxins in cancer. Biomed Pharmacother. 2023;164:114896
9. Aki T, Unuma K, Uemura K. The Role of Peroxiredoxins in the Regulation of Sepsis. Antioxidants (Basel). 2022 11
10. Lee YJ. Knockout Mouse Models for Peroxiredoxins. Antioxidants (Basel). 2020 9
11. Ding C, Fan X, Wu G. Peroxiredoxin 1 - an antioxidant enzyme in cancer. J Cell Mol Med. 2017;21:193-202
12. Min Y, Kim MJ, Lee S, Chun E, Lee KY. Inhibition of TRAF6 ubiquitin-ligase activity by PRDX1 leads to inhibition of NFKB activation and autophagy activation. Autophagy. 2018;14:1347-58
13. Sun HH, Li YL, Jiang H, Yin XH, Jin XL. PRDX1 Influences The Occurrence and Progression of Liver Cancer by Inhibiting Mitochondrial Apoptosis Pathway. Cell J. 2022;24:657-64
14. Zhou X, Li L, Guo X, Zhang C, Du Y, Li T. et al. HBXIP induces anoikis resistance by forming a reciprocal feedback loop with Nrf2 to maintain redox homeostasis and stabilize Prdx1 in breast cancer. NPJ Breast Cancer. 2022;8:7
15. Feng T, Zhao R, Sun F, Lu Q, Wang X, Hu J. et al. TXNDC9 regulates oxidative stress-induced androgen receptor signaling to promote prostate cancer progression. Oncogene. 2020;39:356-67
16. Sun HN, Ren CX, Gong YX, Xie DP, Kwon T. Regulatory function of peroxiredoxin I on 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced lung cancer development. Oncol Lett. 2021;21:465
17. Liao W, Du J, Li L, Wu X, Chen X, Feng Q. et al. CircZNF215 promotes tumor growth and metastasis through inactivation of the PTEN/AKT pathway in intrahepatic cholangiocarcinoma. J Exp Clin Cancer Res. 2023;42:125
18. Lu E, Hu X, Pan C, Chen J, Xu Y, Zhu X. Up-regulation of peroxiredoxin-1 promotes cell proliferation and metastasis and inhibits apoptosis in cervical cancer. J Cancer. 2020;11:1170-81
19. Song C, Xiong G, Yang S, Wei X, Ye X, Huang W. et al. PRDX1 stimulates non-small-cell lung carcinoma to proliferate via the Wnt/beta-Catenin signaling. Panminerva Med. 2023;65:37-42
20. Jiang P, Sinha S, Aldape K, Hannenhalli S, Sahinalp C, Ruppin E. Big data in basic and translational cancer research. Nat Rev Cancer. 2022;22:625-39
21. Zhao S, Mo X, Wen Z, Ren L, Chen Z, Lin W. et al. Comprehensive bioinformatics analysis reveals the hub genes and pathways associated with multiple myeloma. Hematology. 2022;27:280-92
22. Su Y, Tian X, Gao R, Guo W, Chen C, Chen C. et al. Colon cancer diagnosis and staging classification based on machine learning and bioinformatics analysis. Comput Biol Med. 2022;145:105409
23. Sareen H, Ma Y, Becker TM, Roberts TL, de Souza P, Powter B. Molecular Biomarkers in Glioblastoma: A Systematic Review and Meta-Analysis. Int J Mol Sci. 2022 23
24. Kim K, Kim IH, Lee KY, Rhee SG, Stadtman ER. The isolation and purification of a specific "protector" protein which inhibits enzyme inactivation by a thiol/Fe(III)/O2 mixed-function oxidation system. J Biol Chem. 1988;263:4704-11
25. Troussicot L, Burmann BM, Molin M. Structural determinants of multimerization and dissociation in 2-Cys peroxiredoxin chaperone function. Structure. 2021;29:640-54
26. Ma P, Zhou Y, Fang P, Ke W, Xiao S, Fang L. Molecular cloning, prokaryotic expression and the anti-inflammatory activity of porcine PRDX5. Dev Comp Immunol. 2022;136:104515
27. Shau H, Gupta RK, Golub SH. Identification of a natural killer enhancing factor (NKEF) from human erythroid cells. Cell Immunol. 1993;147:1-11
28. Neumann CA, Cao J, Manevich Y. Peroxiredoxin 1 and its role in cell signaling. Cell Cycle. 2009;8:4072-8
29. Bjorklund G, Zou L, Wang J, Chasapis CT, Peana M. Thioredoxin reductase as a pharmacological target. Pharmacol Res. 2021;174:105854
30. Netto LE, Antunes F. The Roles of Peroxiredoxin and Thioredoxin in Hydrogen Peroxide Sensing and in Signal Transduction. Mol Cells. 2016;39:65-71
31. Lv C, Huang Y, Wang Q, Wang C, Hu H, Zhang H. et al. Ainsliadimer A induces ROS-mediated apoptosis in colorectal cancer cells via directly targeting peroxiredoxin 1 and 2. Cell Chem Biol. 2023;30:295-307 e5
32. Leng Y, Wu Y, Lei S, Zhou B, Qiu Z, Wang K. et al. Inhibition of HDAC6 Activity Alleviates Myocardial Ischemia/Reperfusion Injury in Diabetic Rats: Potential Role of Peroxiredoxin 1 Acetylation and Redox Regulation. Oxid Med Cell Longev. 2018;2018:9494052
33. Rawat SJ, Creasy CL, Peterson JR, Chernoff J. The tumor suppressor Mst1 promotes changes in the cellular redox state by phosphorylation and inactivation of peroxiredoxin-1 protein. J Biol Chem. 2013;288:8762-71
34. Chang TS, Jeong W, Choi SY, Yu S, Kang SW, Rhee SG. Regulation of peroxiredoxin I activity by Cdc2-mediated phosphorylation. J Biol Chem. 2002;277:25370-6
35. Chu KL, Lew QJ, Rajasegaran V, Kung JT, Zheng L, Yang Q. et al. Regulation of PRDX1 peroxidase activity by Pin1. Cell Cycle. 2013;12:944-52
36. Mullen L, Hanschmann EM, Lillig CH, Herzenberg LA, Ghezzi P. Cysteine Oxidation Targets Peroxiredoxins 1 and 2 for Exosomal Release through a Novel Mechanism of Redox-Dependent Secretion. Mol Med. 2015;21:98-108
37. Fabbi M, Hagens ERC, van Berge Henegouwen MI, Gisbertz SS. Anastomotic leakage after esophagectomy for esophageal cancer: definitions, diagnostics, and treatment. Dis Esophagus. 2021 34
38. Gong F, Hou G, Liu H, Zhang M. Peroxiredoxin 1 promotes tumorigenesis through regulating the activity of mTOR/p70S6K pathway in esophageal squamous cell carcinoma. Med Oncol. 2015;32:455
39. Gong F, Liu H, Li J, Xue L, Zhang M. Peroxiredoxin 1 is involved in disassembly of flagella and cilia. Biochem Biophys Res Commun. 2014;444:420-6
40. Zhang J, Wang K, Zhang J, Liu SS, Dai L, Zhang JY. Using proteomic approach to identify tumor-associated proteins as biomarkers in human esophageal squamous cell carcinoma. J Proteome Res. 2011;10:2863-72
41. Ren P, Ye H, Dai L, Liu M, Liu X, Chai Y. et al. Peroxiredoxin 1 is a tumor-associated antigen in esophageal squamous cell carcinoma. Oncol Rep. 2013;30:2297-303
42. Chen Q, Li J, Yang X, Ma J, Gong F, Liu Y. Prdx1 promotes the loss of primary cilia in esophageal squamous cell carcinoma. BMC Cancer. 2020;20:372
43. Wu K, Liu F, Zhang T, Zhou Z, Yu S, Quan Y. et al. miR-375 suppresses the growth and metastasis of esophageal squamous cell carcinoma by targeting PRDX1. J Gastrointest Oncol. 2022;13:2154-68
44. Song Y, Liu H, Cui C, Peng X, Wang C, Tian X. et al. Silencing of Peroxiredoxin 1 Inhibits the Proliferation of Esophageal Cancer Cells and Promotes Apoptosis by Inhibiting the Activity of the PI3K/AKT Pathway. Cancer Manag Res. 2019;11:10883-90
45. Hoshino I, Matsubara H, Hanari N, Mori M, Nishimori T, Yoneyama Y. et al. Histone deacetylase inhibitor FK228 activates tumor suppressor Prdx1 with apoptosis induction in esophageal cancer cells. Clin Cancer Res. 2005;11:7945-52
46. Hoshino I, Matsubara H, Akutsu Y, Nishimori T, Yoneyama Y, Murakami K. et al. Tumor suppressor Prdx1 is a prognostic factor in esophageal squamous cell carcinoma patients. Oncol Rep. 2007;18:867-71
47. Machlowska J, Baj J, Sitarz M, Maciejewski R, Sitarz R. Gastric Cancer: Epidemiology, Risk Factors, Classification, Genomic Characteristics and Treatment Strategies. Int J Mol Sci. 2020 21
48. Zhuang K, Tang H, Guo H, Yuan S. Geraniol prevents Helicobacterium pylori-induced human gastric cancer signalling by enhancing peroxiredoxin-1 expression in GES-1 cells. Microb Pathog. 2023;174:105937
49. Tan BB, Li Y, Li SJ, Zhao Q, Fan LQ, Liu QW. et al. [Effect and mechanism of PRDX1 in epithelial mesenchymal transformationin of gastric cancer cells]. Zhonghua Zhong Liu Za Zhi. 2020;42:919-24
50. Xu R, Pan J, Mei J, Zhang Q. Systematic Characterization of Prognostic Values of Peroxiredoxin Family in Gastric Cancer. Biomed Res Int. 2020;2020:3948183
51. Zhang Z, Dai DQ. MicroRNA-596 acts as a tumor suppressor in gastric cancer and is upregulated by promotor demethylation. World J Gastroenterol. 2019;25:1224-37
52. Tan B, Li Y, Zhao Q, Fan L, Zhang M. The impact of Harmine hydrochloride on growth, apoptosis and migration, invasion of gastric cancer cells. Pathol Res Pract. 2020;216:152995
53. Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S. et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6
54. Vogel A, Meyer T, Sapisochin G, Salem R, Saborowski A. Hepatocellular carcinoma. Lancet. 2022;400:1345-62
55. Anwanwan D, Singh SK, Singh S, Saikam V, Singh R. Challenges in liver cancer and possible treatment approaches. Biochim Biophys Acta Rev Cancer. 2020;1873:188314
56. Huang A, Yang XR, Chung WY, Dennison AR, Zhou J. Targeted therapy for hepatocellular carcinoma. Signal Transduct Target Ther. 2020;5:146
57. An Y, Jiang J, Zhou L, Shi J, Jin P, Li L. et al. Peroxiredoxin 1 is essential for natamycin-triggered apoptosis and protective autophagy in hepatocellular carcinoma. Cancer Lett. 2021;521:210-23
58. Ma Q, Hui Y, Huang BR, Yang BF, Li JX, Fan TT. et al. Ferroptosis and cuproptosis prognostic signature for prediction of prognosis, immunotherapy and drug sensitivity in hepatocellular carcinoma: development and validation based on TCGA and ICGC databases. Transl Cancer Res. 2023;12:46-64
59. He Y, Li S, Tang D, Peng Y, Meng J, Peng S. et al. Circulating Peroxiredoxin-1 is a novel damage-associated molecular pattern and aggravates acute liver injury via promoting inflammation. Free Radic Biol Med. 2019;137:24-36
60. Sun QK, Zhu JY, Wang W, Lv Y, Zhou HC, Yu JH. et al. Diagnostic and prognostic significance of peroxiredoxin 1 expression in human hepatocellular carcinoma. Med Oncol. 2014;31:786
61. Cheng ML, Lu YF, Chen H, Shen ZY, Liu J. Liver expression of Nrf2-related genes in different liver diseases. Hepatobiliary Pancreat Dis Int. 2015;14:485-91
62. Luo L, Yao X, Xiang J, Huang F, Luo H. Identification of ferroptosis-related genes for overall survival prediction in hepatocellular carcinoma. Sci Rep. 2022;12:10007
63. Aguilar-Melero P, Prieto-Alamo MJ, Jurado J, Holmgren A, Pueyo C. Proteomics in HepG2 hepatocarcinoma cells with stably silenced expression of PRDX1. J Proteomics. 2013;79:161-71
64. Kang MA, Jeon YK, Nam MJ. Auricularia auricula increases an apoptosis in human hepatocellular carcinoma cells via a regulation of the peroxiredoxin1. J Food Biochem. 2020: e13373.
65. Ciardiello F, Ciardiello D, Martini G, Napolitano S, Tabernero J, Cervantes A. Clinical management of metastatic colorectal cancer in the era of precision medicine. CA Cancer J Clin. 2022;72:372-401
66. Dharwadkar P, Zaki TA, Murphy CC. Colorectal Cancer in Younger Adults. Hematol Oncol Clin North Am. 2022;36:449-70
67. Islam MR, Akash S, Rahman MM, Nowrin FT, Akter T, Shohag S. et al. Colon cancer and colorectal cancer: Prevention and treatment by potential natural products. Chem Biol Interact. 2022;368:110170
68. Li HX, Sun XY, Yang SM, Wang Q, Wang ZY. Peroxiredoxin 1 promoted tumor metastasis and angiogenesis in colorectal cancer. Pathol Res Pract. 2018;214:655-60
69. Xu S, Ma Y, Tong Q, Yang J, Liu J, Wang Y. et al. Cullin-5 neddylation-mediated NOXA degradation is enhanced by PRDX1 oligomers in colorectal cancer. Cell Death Dis. 2021;12:265
70. Rho JH, Qin S, Wang JY, Roehrl MH. Proteomic expression analysis of surgical human colorectal cancer tissues: up-regulation of PSB7, PRDX1, and SRP9 and hypoxic adaptation in cancer. J Proteome Res. 2008;7:2959-72
71. Chu G, Li J, Zhao Y, Liu N, Zhu X, Liu Q. et al. Identification and verification of PRDX1 as an inflammation marker for colorectal cancer progression. Am J Transl Res. 2016;8:842-59
72. Xu H, Zhao H, Ding C, Jiang D, Zhao Z, Li Y. et al. Celastrol suppresses colorectal cancer via covalent targeting peroxiredoxin 1. Signal Transduct Target Ther. 2023;8:51
73. Tai WC, Wong WY, Lee MM, Chan BD, Lu C, Hsiao WL. Mechanistic study of the anti-cancer effect of Gynostemma pentaphyllum saponins in the Apc(Min/+) mouse model. Proteomics. 2016;16:1557-69
74. Dahou H, Minati MA, Jacquemin P, Assi M. Genetic Inactivation of Peroxiredoxin-I Impairs the Growth of Human Pancreatic Cancer Cells. Antioxidants (Basel). 2021 10
75. Zhou J, Shen W, He X, Qian J, Liu S, Yu G. Overexpression of Prdx1 in hilar cholangiocarcinoma: a predictor for recurrence and prognosis. Int J Clin Exp Pathol. 2015;8:9863-74
76. Shen Y, Xu H, Li L, Lu Y, Zhang M, Huang X. et al. Assessment of Potential Prognostic Value of Peroxiredoxin 1 in Oral Squamous Cell Carcinoma. Cancer Manag Res. 2021;13:5725-37
77. Park HJ, Choi YJ, Lee JH, Nam MJ. Naringenin causes ASK1-induced apoptosis via reactive oxygen species in human pancreatic cancer cells. Food Chem Toxicol. 2017;99:1-8
78. Lau HCH, Kranenburg O, Xiao H, Yu J. Organoid models of gastrointestinal cancers in basic and translational research. Nat Rev Gastroenterol Hepatol. 2020;17:203-22
79. Guo W, Liu X, Li J, Shen Y, Zhou Z, Wang M. et al. Prdx1 alleviates cardiomyocyte apoptosis through ROS-activated MAPK pathway during myocardial ischemia/reperfusion injury. Int J Biol Macromol. 2018;112:608-15
80. Cao J, Schulte J, Knight A, Leslie NR, Zagozdzon A, Bronson R. et al. Prdx1 inhibits tumorigenesis via regulating PTEN/AKT activity. EMBO J. 2009;28:1505-17
81. Gao L, Meng J, Yue C, Wu X, Su Q, Wu H. et al. Integrative analysis the characterization of peroxiredoxins in pan-cancer. Cancer Cell Int. 2021;21:366
82. Xu M, Xu J, Zhu D, Su R, Zhuang B, Xu R. et al. Expression and prognostic roles of PRDXs gene family in hepatocellular carcinoma. J Transl Med. 2021;19:126
Author contact
![]() Corresponding author: Dr Bing Pei, The Affiliated Suqian First People's Hospital of Nanjing Medical University, No. 120 Suzhi Road, Suqian, P.R. China, 223800 E-mail: m15996557561com.
Corresponding author: Dr Bing Pei, The Affiliated Suqian First People's Hospital of Nanjing Medical University, No. 120 Suzhi Road, Suqian, P.R. China, 223800 E-mail: m15996557561com.

 Global reach, higher impact
Global reach, higher impact