3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(17):3321-3334. doi:10.7150/jca.88635 This issue Cite
Research Paper
High Expression of PSRC1 Predicts Poor Prognosis in Lung Adenocarcinoma
1. Department of respiratory and critical care medicine, the first affiliated hospital of Anhui medical university, Hefei 230022, China.
2. Department of infectious disease, Hefei second people's hospital, Hefei 230001, China.
3. Department of oncology, the first affiliated hospital of Anhui medical university, Hefei 230022, China.
4. Department of Occupational Disease, Hefei third clinical college of Anhui Medical University, Hefei 230022, China.
* These authors contributed equally to this work.
Received 2023-7-29; Accepted 2023-9-6; Published 2023-10-7
Abstract
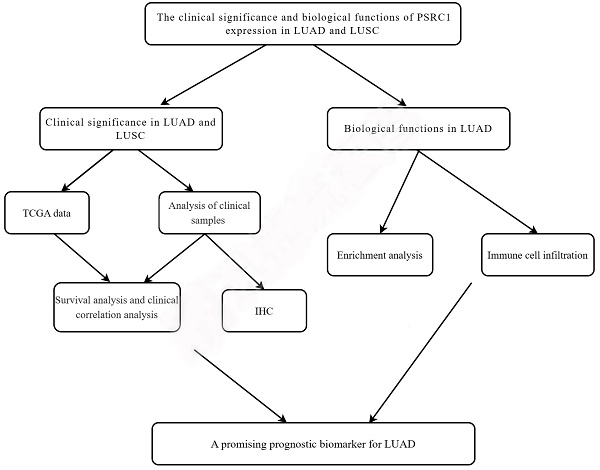
Background: The incidence of lung cancer is increasing annually, but the mechanism of its occurrence and development requires further study. This study aimed to investigate the biological function and prognostic value of proline- and serine-rich coiled-coil 1 (PSRC1) in lung cancer.
Methods: We used data from The Cancer Genome Atlas (TCGA) to analyze the association between clinical features and PSRC1 expression in non-small cell carcinoma. The relationship between PSRC1 expression and prognosis in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) was analyzed using Kaplan-Meier curves. The function of PSRC1 was identified using enrichment analysis, and the relationship between PSRC1 expression and immune cell infiltration was studied. In addition, the expression of PSRC1 in 150 patients with non-small cell carcinoma was detected using immunohistochemistry, and its clinical significance was analyzed.
Results: It was found that the expression level of PSRC1 was higher in LUAD and LUSC tumor tissues than in normal tissues, and the results were confirmed by immunohistochemistry in 150 patients. TCGA data showed that high PSRC1 expression in LUAD was associated with poorer overall survival (p = 0.003) and progression-free interval (p = 0.012). Multivariable analysis showed that PSRC1 was an independent risk factor for LUAD. Functional enrichment analysis showed that PSRC1 is related to tumor development.
Conclusion: High PSRC1 expression is significantly associated with LUAD survival and may be a promising prognostic biomarker.
Keywords: PSRC1, lung adenocarcinoma, prognosis, immunohistochemistry, bioinformatics
Introduction
Lung cancer is the most commonly diagnosed cancer worldwide and the leading cause of cancer-related deaths [1, 2]. Lung cancer includes small cell carcinoma and non-small cell carcinoma (NSCLC), which can be further divided include lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) [3]. Before the 1990s, LUSC accounted for a relatively high proportion of men with NSCLC; however, the incidence of adenocarcinoma gradually surpassed that of squamous cell carcinoma, with LUAD becoming the dominant histological subtype of NSCLC [4, 5]. According to statistics, lung cancer accounted for approximately one in nine cancers (11.4%) and one in six deaths (18.0%) in 2020, with an estimated 2.2 million new cancer cases and 1.8 million deaths worldwide[6]. Despite recent advances in the diagnosis and treatment of lung cancer, the prognosis of advanced lung cancer remains poor, with multiple targeted therapies and immune-checkpoint inhibitor-related drugs available [7, 8]. Studies have shown that lung cancer is a complex disease involving genetic and epigenetic changes [9], and molecular biological detection plays an important role in improving the early diagnosis rate and prognosis of NSCLC [10-13]. Therefore, revealing the intrinsic mechanism of NSCLC, seeking new potential targets, and identifying oncogenes associated with NSCLC prognosis are helpful in the development of new molecular biomarkers necessary for effective diagnostic and therapeutic strategies. With the development of gene chips and high-throughput second-generation sequencing technologies, an increasing amount of genetic data is being stored in public databases for researchers to mine. Therefore, a combination of gene expression data with bioinformatics methods can be used to determine the expression of differentially expressed genes (DEGs) in response to the occurrence and development of NSCLC, and to identify potential targets for treatment [14].
Proline- and serine-rich coiled-coil 1 (PSRC1) is also known as DDA3. PSRC1 is required for normal progression of mitosis, and is involved in a variety of signaling pathways [15, 16]. PSRC1 is a microtubule (MT) -associated cancer protein regulated by the downregulation of p53 transcription. Its promoter contains a putative p53-binding motif responsible for p53-mediated gene suppression [17]. Further experiments showed that PSRC1 has a unique domain. The C-terminal domain has the ability to bind to the mitotic spindle, while the regulatory N-terminal domain controls the C-terminal domain to bind to microtubules and determines the cellular activity of the PSRC1 protein [16]. Studies have shown that PSRC1 is overexpressed in a variety of cancers, including, colorectal cancer [18], hepatocellular carcinoma [19], and oral squamous cell carcinoma [20]; and is therefore a potential biomarker and therapeutic target. PSRC1 contains hot spot mutations in colorectal cancer and could potentially be used to develop personalized tumor analysis and therapy [18]. PSRC1 hypomethylation may lead to PSRC1 overexpression and tumor progression in hepatocellular carcinoma [19]. PSRC1 can be used as a prognostic predictor for oral squamous cell carcinoma without lymph node metastasis [20].
However, there are few studies on PSRC1 in NSCLC. Therefore, this study aimed to investigate the correlation between PSRC1 expression, and clinicopathological features and prognosis of NSCLC; and to clarify the biological role of PSRC1 in NSCLC.
We compared the expression of PSRC1 in LUAD and LUSC in both tumor and normal tissues using the dataset from The Cancer Genome Atlas (TCGA) database, and evaluated the correlation between PSRC1 expression levels and clinicopathological features. Immunohistochemical methods were used to detect differences in the expression of PSRC1 between tumor and normal tissues to verify the prognostic value of PSRC1 in NSCLC. Gene set enrichment analysis (GSEA) was used to identify the biological pathways associated with PSRC1. GSEA and immune-associated infiltration analysis revealed the biological influence of PSRC1 in LUAD and its potential mechanisms of action. Our results strongly support PSRC1 as a biomarker for predicting the prognosis and treatment outcomes of LUAD, but not LUSC. In summary, PSRC1 is a promising prognostic biomarker for LUAD.
Material and Methods
Bioinformatics Analysis Based on TCGA Database
We obtained gene expression data, corresponding clinical information, and survival data for TCGA-LUAD (594 samples, workflow type: HTSeq-TPM) and TCGA-LUSC (551 samples, workflow type: HTSeq-TPM) from TCGA. The LUAD dataset included 535 cases of tumors and 59 cases of normal tissue, whereas the LUSC dataset included 502 cases of tumors and 49 cases of normal tissue (https://cancergenome.nih.gov; Table S1). A total of 517 LUAD patients and 496 LUSC patients had complete survival data. We considered the median expression level as the critical value and divided the cases into two groups: the low and the high expression group for PSRC1. Gene expression data (HTSeq-Counts and HTSeq-FPKM), phenotypic data and detailed clinicopathological information of TCGA-LUAD were obtained by browser. Sequence data were retrieved using the Illumina HiSeq_RNA_Seq platform. HTSeq-FPKM gene expression data were converted to TPM (transcripts per million readings) for subsequent analysis. Since TPM produces results that are more similar to those of the microarray method, it is beneficial for comparison between samples [21].
Sample Collection
We collected 150 samples from patients who had been initially diagnosed with NSCLC. Two pathologists diagnosed lung cancer based on pathological findings, and determined the histological type and staging based on the eighth version of the tumor, node, and metastasis (TNM) staging system. There were 90 and 60 patients with LUAD and LUSC, respectively. Samples were collected from October 2019 to October 2021, and follow-up data were available until December 30, 2022, or the date of death. Tumor tissue samples and paired adjacent normal tissues (normal lung tissue > 5 cm from the tumor margin) were collected from 90 patients diagnosed with LUAD and 60 patients diagnosed with LUSC. Samples were formalin-fixed and paraffin-embedded for immunohistochemical analysis. None of the enrolled patients had a history of other malignant tumors, immune dysfunction, preoperative radiotherapy, or chemotherapy. The following clinical and pathological information was obtained from the enrolled patients: age, sex, surgical modalities, smoking history, tumor size, pathological type, pathological stage, T, N, and M stage; tumor differentiation, postoperative treatment, follow-up, tumor recurrence and progression, overall survival (OS), and progression-free interval (PFI) (Table S2). Sample exclusion criteria were as follows :(1) combined with other malignant tumors; (2) complicated with cardiovascular diseases; (3) the clinical baseline data missing.
Immunohistochemical Analysis of PSRC1 Expression
The procedure was similar to what was previously described [22]. Paraffin-embedded sections were dewaxed in xylene and ethanol, and antigen repair was performed in a citrate buffer solution (pH 6.0). Sections were incubated overnight with anti-PSRC1 antibody (GTX128047:GeneTex; 1:500 dilution) at 4°C and washed with phosphate-buffered saline (PBS). Then, the sections were incubated with HRP-conjugated goat anti-rabbit IgG (ab205718; Abcam) at 37°C for 2 h. Finally, 3, 3-diaminobenzidine tetra hydrochloride (DAB) staining, hematoxylin reverse staining, dehydration, neutral resin sealing, examination, and evaluation were performed. PSRC1 expression was analyzed by immunohistochemistry in tumor and normal tissue samples from 90 patients with LUAD and 60 patients with LUSC. Cytoplasmic staining of the tumor was positive for PSRC1 expression. Two independent pathologists evaluated PSRC1 expression under 400x optical microscopy using a semi-quantitative scoring system, while examining positive and negative controls in parallel [23]. The following categories were defined for the evaluation: (i) staining intensity: 0 (negative staining), 1 (mild staining), 2 (moderate staining), 3 (strong staining); (ii) percentage of immunopositive cells (5 fields were randomly selected at 400x magnification per slide): 0 (0%), 1 (1-25%), 2 (26-50%), 3 (51-75%), 4 (76-100%). The percentage of stained cells was multiplied by the fraction of stained cells to obtain the final score (score range: 0-12 points). We considered scores within the 0-2 range as indicating negative expression, within the 3-6 range as indicating low PSRC1 expression, and within the 7-12 range as indicating high PSRC1 expression.
Enrichment Analyses
We used a median Z-score to divide patients with LUAD into a low- and a high-expression group. DEGs were analyzed using R software with a log fold change (log FC) >1 and an adjusted p value <0.05. Volcano and heat maps were created to visualize the analysis results. The core modules and hub genes associated with PSRC1 were identified by weighted gene co-expression network analysis (WGCNA)[24]. We explored the biological function of PSRC1 in LUAD using the R clusterProfiler package for Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis [25]. GSEA has the advantage of grouping gene sets according to common biological functions, chromosomal regulatory mechanisms, or locations. GSEA was performed using the R package "clusterProfiler" with the following parameters: nPerm = 1000, minGSSize = 10, maxGSSize = 1000, p value cutoff = 0.05 [26]. The enrichment results were considered statistically significant when the false discovery rate (FDR) was <0.25 and the adjusted p value was <0.05.
Immune Infiltrate Analysis
Immunoinfiltration analysis of LUAD was performed using single-sample gene set enrichment analysis (ssGSEA, GSV A package). The ssGSEA algorithm is based on the expression information of specific marker genes in immune cells, which was derived from studies by Bindea et al. [27]. We also evaluated the relationship between PSRC1 expression and the abundance of 24 immune cell types. Spearman and Wilcoxon rank-sum tests were used, and p < 0.05 was considered statistically significant.
Statistical Analysis
Kaplan-Meier survival curves were used to estimate survival between patients with high and low gene expression levels and the log-rank test was used to compare differences in survival [28]. Univariable and multivariable cox regression analyses were used to determine prognostic factors for LUAD. Factors with statistical significance (p < 0.05) in the univariable analysis were included in the multivariable analysis to determine the independent risk factors. All statistical analyses and plots were performed using the R software (version 4.1.2). Combined with the expression value of PSRC1 and clinical variables, a nomogram was constructed to predict the 1-, 3-, and 5-year OS.
Based on the optimal multivariate cox regression analysis to construct the nomogram, 1 year, 3 years and 5 years survival probability. R package rms (https://cran.r-project.org/web/packages/rms/index.html) is used to generate the column chart. Concordance index (C-index) and calibration plots are often used to evaluate the quality of nomogram models. Using Hmisc R package (https://cran.r-project.org/web/packages/Hmisc/index.html) evaluation C-index and calibration diagram. In this study, the C-index was used to determine the discriminative power of 1000 bootstrap replicates. With the increase of C-index, prediction accuracy increases. The calibration curve was evaluated visually by mapping the nomogram prediction to the observed probability, with the 45° line indicating the best predicted value.
Results
High PSRC1 Expression in LUAD and LUSC
From the TCGA dataset, 535 patients with LUAD and 502 patients with LUSC met the required clinical characteristics. To detect PSRC1 expression in LUAD and LUSC patients, we compared its expression level in tumor tissues with that in normal lung tissues. The results showed that the expression level of PSRC1 in LUAD and LUSC tissues was significantly higher than that in normal tissues (p < 0.001; Figure 1A and S1A), and that in the tumor tissue was higher than that in the paired normal tissue (p < 0.001; Figure 1B and S1B). To verify the differences in PSRC1 expression, immunohistochemical analyses were performed on normal and tumor tissues from 90 patients with LUAD and 60 patients with LUSC. The results were validated in LUAD, LUSC, and paired normal lung tissues (p < 0.001; Figure 1C and S1C). Representative immunohistochemical images are shown in Figure 1D, 1E, S1D and S1E.
PSRC1 expression in LUAD. (A)PSRC1 expression levels in LUAD and normal tissues in TCGA. (B)PSRC1 expression levels in LUAD and matched normal tissues in TCGA. (C)PSRC1 expression of IHC on 90 patients with LUAD tissues and 90 adjacent normal lung tissues. Representative IHC images of PSRC1 expression in normal tissues (D) and LUAD tissues (E). Abbreviations: PSRC1, Proline and serine rich coiled-coil 1; LUAD, lung adenocarcinoma; TCGA, The Cancer Genome Atlas; IHC, immunohistochemistry.

Correlation between PSRC1 expression and clinicopathologic features in patients with LUAD in TCGA database (p < 0.05). The correlation between PSRC1 expression and pathologic T stage (A), pathologic stage (B), primary therapy outcome (C), age (D), gender (E) and number pack year smoked (F) of patients with LUAD. Abbreviations: PSRC1, Proline and serine rich coiled-coil 1; LUAD, lung adenocarcinoma; TCGA, The Cancer Genome Atlas.

Expression of PSRC1 in relation to clinicopathological features of LUAD and LUSC from the TCGA dataset
The expression of PSRC1 was different according to age, sex, smoking history, T stage, and pathological stage group in LUAD patients (Figure 2). The analysis showed that the expression of PSRC1 in LUAD patients at T2/T3/T4 was higher than that in patients at T1 (p = 0.003; Figure 2A), the expression of PSRC1 in LUAD patients with a high pathological stage was higher than in those with a low pathological stage (p = 0.023; Figure 2B), and the expression of PSRC1 in LUAD patients with improved or stable primary therapy outcome was lower than that in those with tumor progression (p = 0.002; Figure 2C). Older LUAD patients exhibited lower expression compared to patients under 65 years (p < 0.001; Figure 2D), and male LUAD patients showed higher expression than female LUAD patients (p < 0.001; Figure 2E). PSRC1 expression in LUAD patients with long smoking duration was higher than that in LUAD patients with short smoking duration (p = 0.018; Figure 2F).
In LUSC, high PSRC1 expression was not correlated with clinical features, as shown in Figure S2A-H.
High expression of PSRC1 was an independent risk factor for poor prognosis in LUAD
Kaplan-Meier analysis was used to estimate survival rates in LUAD and LUSC patients. Figure 3A, 3B, S3A and S3B included patient death and disease recurrence as the end point events, respectively, and the two survival curves were compared using the log-rank test. The cox proportional hazards regression model was used to analyze the influencing factors of OS and PFI in figure 3C, 3D, S3C and S3D. The results showed that the survival time of LUAD tumor patients with high PSRC1 expression was shorter than that of patients with low PSRC1 expression (OS, p = 0.003, Figure 3A; and PFI, p = 0.013, Figure 3B). This conclusion is consistent with the results obtained in 90 patients with LUAD (OS, Figure 3C; PFI, Figure 3D; p < 0.001). However, no significant correlation was observed in the TCGA LUSC dataset (Figure S3A and B; p > 0.05) or in the 60 LUSC patients (Figure S3C and D; p > 0.05).
We performed a further subgroup analysis and found that high expression of PSRC1 significantly decreased OS at the LUAD T1/T2 stage (p = 0.013), N0 stage (p = 0.03), M0 stage (p = 0.012), and pathological stages I/II/III (p = 0.005); in patients aged > 65 years (p = 0.003), and in female patients (p = 0.006; Figure S4A-F). These results confirmed that high PSRC1 expression is associated with poor prognosis in patients with LUAD.
Since no correlation between PSRC1 expression and LUSC prognosis was observed in the TCGA dataset and the clinical data of 60 LUSC cases, univariable and multivariable cox regression and enrichment analyses were conducted only for LUAD cases. In TCGA, univariable cox regression analysis showed that the expression of PSRC1, TNM stage, pathological stage, and primary therapy outcome were correlated with OS, while PSRC1 expression, T stage, N stage, and primary therapy outcome were correlated with PFI. Multivariable analysis showed that PSRC1 expression, T stage, N stage, and primary therapy outcome were independent risk factors for OS in LUAD patients, while PSRC1 expression, T stage, and primary therapy outcome were independent risk factors for PFI (Table 1). Univariable and multivariable cox analyses were performed on a cohort of 90 patients with LUAD. Univariable analysis revealed that N stage, differentiation, and PSRC1 expression were prognostic factors for OS. The univariable analysis results for PFI were similar to those for OS (Table 2). Multivariable analysis was conducted using variables with p values <0.05 in univariable analysis, and the results showed that high expression of PSRC1 was an independent risk factor for OS and PFI in patients with LUAD. The results are shown in Table 2.
Construction of a Nomogram
We constructed a prognostic nomogram by weighing the TNM stage, primary treatment outcome, age, sex, and PSRC1 expression. Figure 4A shows a forest map based on the survival analysis of different clinical subgroups of LUAD patients with PSRC1 expression. The prognosis map includes clinical features independently associated with survival in the multivariable analysis (i.e., high PSRC1 expression, T stage, N stage, and primary treatment outcomes; Figure 4B). These results also confirm that high PSRC1 expression is associated with poor prognosis. Nomogram discrimination was evaluated using c-index. In our prognosis chart, LUAD's c-index of LUAD was 0.815(95%CI:0.771-0.860). As shown in the calibration diagram (Figure S5), the predicted values were in good agreement with the observed values.
Kaplan-Meier curve for OS in LUAD. (A) Overall survival and (B) progress free interval. Kaplan-Meier survival curves of 90 LUAD patients with high and low PSRC1-expressing tumors (C) Overall survival and (D) progression free interval.

Correlation and Enrichment Analyses
We used TCGA data to analyze the correlation between PSRC1 and other genes in LUAD to predict the function of PSRC1. We used R (| logFC | > 1, adjusted p values < 0.05) analysis of the TCGA dataset (Figure 5). PSRC1 between the high- and low-expression groups identified 3703 DEGs (2200 upregulated and 1503 downregulated). Volcano and heat maps were created to visualize the results (Figure 5A and B). The brown core module associated with PSRC1 was identified by WGCNA, so we performed enrichment analysis with the brown module using the clusterProfiler R package (Figure S6). Figure 5C shows the most important GO terms for biological processes, cellular components, and molecular functions. In LUAD, GO analysis revealed 326 GO terms associated with the biological processes, indicating that PSRC1 is involved in many biological processes such as organelle fission, nuclear division, and chromosome segregation. 44 GO terms associated with the cell component category "chromosome". And there are 21 GO terms related to molecular functions, such as tubulin binding, ATP hydrolysis activity and microtubule binding. According to KEGG functional enrichment analysis, related genes were involved in cell cycle, biosynthesis of amino acids and glycolysis/gluconeogenesis.
We performed GSEA to determine the signaling pathway activated in LUAD, and adjusted p values < 0.05 and FDR < 0.25 were considered significant. The results showed that the following eight pathways were significantly different between the two groups: G-protein couple receptor (GPCR) ligand binding, signaling by RHO GTPases, M phase, neuronal system, transcriptional regulation by Tp53, DNA repair, signaling by WNT, and metabolism of amino acids and derivatives (Figure 6A-H).
Relationship between PSRC1 expression and different clinicopathological factors and overall survival. (A) Analysis of PSRC1 expression on survival in different clinical subgroups of LUAD patients. (B) Nomogram for predicting probability of patients with 1-, 3- and 5-year OS.

Correlations Between PSRC1 Expression and Overall Survival in Patients with Lung Adenocarcinoma
| Characteristics | Total (N) | OS | PFI | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Univariable analysis | Multivariable analysis | Univariable analysis | Multivariable analysis | ||||||
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| T stage | 527 | 2.352 (1.614 - 3.426) | < 0.001 | 1.801 (1.056 - 3.073) | < 0.05 | 1.840 (1.268 - 2.670) | 0.001 | 1.909 (1.286 - 2.833) | 0.001 |
| T1&T2 | 461 | ||||||||
| T3&T4 | 66 | ||||||||
| N stage | 514 | 2.547 (1.904 - 3.407) | < 0.001 | 1.904 (1.278 - 2.836) | < 0.01 | 1.484 (1.131 - 1.948) | 0.004 | 1.117 (0.832 - 1.501) | 0.461 |
| N0 | 345 | ||||||||
| N1&N2&N3 | 169 | ||||||||
| M stage | 381 | 2.176 (1.272 - 3.722) | < 0.01 | 1.473 (0.692 - 3.138) | 0.315 | 1.542 (0.872 - 2.728) | 0.137 | ||
| M0 | 356 | ||||||||
| M1 | 25 | ||||||||
| Pathologic stage | 522 | 2.338 (1.397 - 3.912) | < 0.01 | ||||||
| Stage I & Stage II & Stage III | 496 | ||||||||
| Stage IV | 26 | ||||||||
| Primary therapy outcome | 442 | 0.272 (0.191 - 0.388) | < 0.001 | 0.246 (0.158 - 0.383) | < 0.001 | 0.165 (0.122 - 0.223) | < 0.001 | 0.176 (0.129 - 0.241) | < 0.001 |
| PD | 71 | ||||||||
| SD&PR&CR | 371 | ||||||||
| Age | 520 | 1.216 (0.910 - 1.625) | 0.186 | 1.019 (0.781 - 1.330) | 0.889 | ||||
| <=65 | 257 | ||||||||
| >65 | 263 | ||||||||
| Gender | 530 | 1.087 (0.816 - 1.448) | 0.569 | 1.189 (0.914 - 1.547) | 0.197 | ||||
| Female | 283 | ||||||||
| Male | 247 | ||||||||
| Race | 472 | 1.493 (0.913 - 2.440) | 0.110 | 1.411 (0.670 - 2.970) | 0.365 | 1.188 (0.792 - 1.781) | 0.406 | ||
| Asian&Black or African American | 63 | ||||||||
| White | 409 | ||||||||
| PSRC1 | 530 | 1.545 (1.155 - 2.065) | < 0.01 | 1.770 (1.174 - 2.667) | < 0.01 | 1.400 (1.075 - 1.823) | 0.013 | 1.334 (1.005 - 1.770) | 0.046 |
| Low | 266 | ||||||||
| High | 264 | ||||||||
Differentially expressed genes in LUAD patients with high and low PSRC1 expression. (A) Volcano map of differentially expressed genes based on PSRC1 expression. (B) Heat maps of the first five up-regulated and the first five down-regulated genes were selected according to the expression status of PSRC1. (Pearson). (C) GO/KEGG enrichment analysis of PSRC1 expression-related genes. *p < 0.05; ** p < 0.01; *** p < 0.001. Abbreviations: PSRC1, Proline and serine rich coiled-coil 1; LUAD, lung adenocarcinoma; GO, Gene Ontology.

Univariable and Multivariable Analyses of OS and PFI in 90 Patients with Lung Adenocarcinoma
| Characteristics | Total(N) | OS | PFI | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Univariable analysis | Multivariable analysis | Univariable analysis | Multivariable analysis | ||||||
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| Age | 90 | 0.795 (0.373-1.695) | 0.553 | 0.832 (0.390-1.773) | 0.634 | ||||
| <=65 | 49 | ||||||||
| >65 | 41 | ||||||||
| Gender | 90 | 1.254 (0.588-2.672) | 0.558 | 1.366 (0.641-2.912) | 0.419 | ||||
| Male | 50 | ||||||||
| Female | 40 | ||||||||
| Smoker | 90 | 1.113 (0.520-2.384) | 0.783 | 1.089 (0.509-2.332) | 0.826 | ||||
| Yes | 50 | ||||||||
| No | 40 | ||||||||
| Pathologic stage | 90 | 0.000 (0.000-Inf) | 0.997 | 0.000 (0.000-Inf) | 0.997 | ||||
| I & II & III | 87 | ||||||||
| IV | 3 | ||||||||
| T stage | 90 | 1.049 (0.361-3.053) | 0.930 | 0.935 (0.320-2.730) | 0.903 | ||||
| T1&T2 | 78 | ||||||||
| T3&T4 | 12 | ||||||||
| N stage | 90 | 0.209 (0.091-0.480) | <0.001 | 0.065 (0.008-0.537) | 0.011 | 0.203 (0.088-0.466) | <0.001 | 0.195 (0.082-0.469) | <0.001 |
| N0 | 54 | ||||||||
| N1&N2 | 36 | ||||||||
| M stage | 90 | 0.000 (0.000-Inf) | 0.997 | 0.000 (0.000-Inf) | 0.997 | ||||
| M0 | 87 | ||||||||
| M1 | 3 | ||||||||
| Differentiation | 90 | 5.383 (2.123-13.647) | <0.001 | 3.262 (1.083-9.831) | 0.036 | 4.927 (1.952-12.437) | <0.001 | 2.182 (0.745-6.396) | 0.155 |
| Well | 41 | ||||||||
| Moderate& Poor | 49 | ||||||||
| PSRC1 Expression | 90 | 6.920 (2.877-16.645) | <0.001 | 4.264 (1.626-11.181) | 0.003 | 7.752 (3.271-18.372) | <0.001 | 6.292 (2.279-17.370) | <0.001 |
| Low expression | 54 | ||||||||
| High expression | 36 | ||||||||
Gene set enrichment analysis (GSEA) enrichment plot. (A) GPCR ligand binding, (B) signaling by RHO GTPases, (C) M phase, (D) neuronal system, (E) transcriptional regulation by Tp53, (F) DNA repair, (G) signaling by WNT, (H) metabolism of amino acids and derivatives.

Correlation between immune cell infiltration and PSRC1 in LUAD. (A) Correlation between PSRC1 expression level in LUAD and infiltration of 24 immune cells. Relationship between PSRC1 expression and infiltration of (B) mast cells, (C) eosinophils, (D) dendritic cells (DC), (E) immature dendritic cells (iDC), (F) type 2 T-helper (Th2) cells and (G) γδ T (Tgd) cells.

Correlation between immune cell infiltration and PSRC1 in LUAD
The survival of cancer patients is associated with immune cell infiltration. Spearman's r was used to analyze the correlation between PSRC1 expression levels in LUAD and infiltration of 24 immune cell types (Figure 7A). The results showed that the expression of PSRC1 was negatively correlated with mast cell, eosinophil, dendritic cell (DC), and immature dendritic cell (iDC) infiltration (p < 0.001; Figure 7B-E), and positively correlated with type 2 T-helper (Th2) and γδ T (Tgd) cell infiltration (p < 0.001; Figure 7F-G).
Discussion
Lung cancer, a complex disease involving genetic and epigenetic changes, is the leading cause of cancer-related deaths worldwide. Lung adenocarcinoma is the most important subtype of lung cancer, and its incidence is on the rise [5]. Personalized, biomarker-driven therapy and screening in high-risk groups have led to improvements in LUAD's survival rate [28]. Lung cancer has a 5-year survival rate of 10-20%, depending on the stage and region [6]. Treatment for LUAD usually involves a combination of surgery, chemotherapy, and/or radiation; and in recent years the development of various chemotherapies, advances in targeted therapy, and immunotherapy have provided hope for the treatment of lung cancer [29-31]. However, owing to the lack of early diagnosis, most lung cancers are only detected in advanced stages of local tumor invasion or distant metastasis. A comprehensive study by the Tyrol Registry has shown that most patients with NSCLC are already in an advanced stage at the time of initial diagnosis and have multiple comorbidities, which limits treatment options [32]. Therefore, it is critical to identify biomarkers for tumorigenesis and poor prognosis in these patients.
PSRC1 is a novel microtubule-associated protein that encodes a serine- and proline-rich protein containing a coiled-coil region and six putative SH3-domain-binding motifs, PXXP [33]. PSRC1 interacts with the MT depolymerase Kif2a in a microtubule-dependent manner, recruiting Kif2a and ANKRD53 to the mitotic spindle and spindle pole [34]. It was reported that PSRC1, as a p53 target gene, interacts physically and functionally with microtubules and their associated proteins EB3 and APC2, and enhances the expression of β-catenin-dependent genes, which provides the basis for the functional connection between the p53 and the β-catenin pathways, and may be involved in the role of p53 in microtubule-dependent activities [33]. PSRC1 may also be involved in regulating neurite formation and elongation [35]. Proper regulation of mitotic spindle dynamics is key to successful cell division, and PSRC1 plays a role in this regulation. It was found that the suppressor of Cullin 5-interacting suppressor of the cytokine signaling box protein ASB7 ubiquitinates the regulator PSRC1 for proteasomal degradation. ASB7 may play a critical role in regulating spindle dynamics and genome integrity by controlling PSRC1 expression [36].
Previous studies have demonstrated that PSRC1 is a biomarker of cardiovascular disease [37-39], and there is increasing evidence that PSRC1 is involved in the occurrence and progression of multiple cancers. PSRC1 is regulated by the downregulation of p53 transcription and is responsible for p53-mediated gene inhibition, which plays an important role in human tumors [17]. As reported by Wei et al., PSRC1 is involved in the prognosis of patients with hepatocellular carcinoma [40].
Our study suggests that PSRC1 is a potential therapeutic target and prognostic predictor. Bioinformatics analysis was performed using RNA-seq data from TCGA to assess the prognostic value of PSRC1 in LUAD and LUSC. Survival analysis revealed the prognostic value of PSRC1 in patients with LUAD. High PSRC1 expression was associated with poor OS and PFI in patients with LUAD, but this was not the case for LUSC. The results were confirmed by immunohistochemistry of the clinical samples. In addition, we also found significant differences in the expression levels of PSRC1 at different TNM stages (T1/T2, N0, M0), pathological stages (I/II/III), in advanced age (> 65 years), and in female patients, which may influence the prognosis of LUAD patients. These results suggest that PSRC1 acts as an oncogene in LUAD. To date, no studies have reported the role of PSRC1 in the progression of LUAD, and therefore these new results may provide a new avenue for exploring new treatment options for this particular type of cancer.
Since our results showed that PSRC1 expression had no significant effect on the prognosis of LUSC patients, functional enrichment and immunoinfiltration analyses were performed only for LUAD patients. Campbell et al. examined the exome sequences and copy number profiles of 1,144 types of lung cancer genes and found that mutated genes and somatic cell copy number changes were significantly different between the two types of lung cancer [41]. This is consistent with gene expression studies showing that oncogenic drivers may differ to a large extent between lung adenocarcinoma and lung squamous cell carcinoma, and somatic alterations may have different oncogenic potential in different cellular environments [42]. These differences may explain the different sensitivities of LUAD and LUSC to immunotherapy and targeted therapy [43].
Enrichment analysis indicated that PSRC1 expression is related to various biological processes, such as organelle fission, nuclear division, and chromosome segregation, and plays an important role in the cell cycle. Downregulation of PSRC1 controls mitosis by increasing the frequency of misaligned chromosomes in metaphase cells and the metaphase delay, and decreasing the metaphase intercentromere tension and the anaphase chromosome separation rate [15]. Our results suggest that high PSRC1 expression may accelerate mitosis, promote cell migration and proliferation, and lead to tumor development, which is consistent with previous reports in other types of cancer [19, 44]. Tumor genomes are usually abnormal due to mutagenesis damage and incomplete DNA repair. DNA repair has recently received considerable attention as a therapeutic target [45]. GSEA showed enrichment in the expression of the PSRC1 gene, which also suggests PSRC1 as a new target for anticancer therapy in lung cancer.
It is well known that the immune system represents an important barrier to the formation and development of tumors, and escaping immune destruction is one of the hallmarks of cancer [46]. Tumors use tumor-induced damage to antigen presentation, activation of negative co-stimulatory signals, and immunosuppression to thwart immune responses, and cell populations such as regulatory T cells, natural killer T cells, and different subpopulations of immature and mature dendritic cells may contribute to this immunosuppressive network [47]. The ability of of NSCLC to escape immune vigilance is determined by the level of immune invasion and the existence of an immune escape mechanism; the invasiveness of tumors is related to the immune regulatory function of the body [48]. Immunotherapy aims to promote the activity of cytotoxic T lymphocytes in tumor, T cell immune infiltrating level is related to the curative effect of immunotherapy, so is the current focus in the study of cancer immunotherapy [49]. Studies have shown that high T cell expression is associated with better OS in many types of cancer, including breast cancer, lung cancer, and melanoma [50]. Dendritic cells are professional antigen-presenting cells that influence anti-tumor immune responses by displaying a wide range of dysfunctional states in the tumor microenvironment. DC-based therapy can restore the function of DC in the tumor microenvironment, thus showing promise in tumor therapy [51]. Our results showed that PSRC1 expression level was correlated with immune infiltration, including levels of T cells, mast cells and dendritic cells. These results suggest that the expression level of PSRC1 may indicate the level of immune infiltration and provide a reference for the application of immunotherapy in LUAD.
We further investigated the relationship between PSRC1 expression and immune cell infiltration in LUAD patients. The results showed that PSRC1 expression was negatively correlated with mast cell, eosinophil, DC, and iDC infiltration, and positively correlated with Th2 and Tgd cell infiltration. Studies have shown that mast cells and eosinophils may have therapeutic targets and may also improve cancer immunotherapy [52, 53]. In fact, NSCLC tumors are infiltrated by T cells [54], and human primary NSCLC is Th2-skewed and contains many tumor-promoting Treg cells [55]. Some symbiotic microbiota can promote lung cancer development through Tgd cells [56]. In summary, PSRC1 may alter the prognosis of patients with LUAD by influencing immune cell infiltration.
Our study has some limitations. First, our dataset is derived from a single medical institution, and the number of clinical samples is relatively small. Therefore, studies involving a larger number of samples are still needed to verify the results. Second, our results mainly focused on LUAD. Bioinformatics analysis of the online dataset revealed the relationship between PSRC1 expression and immune cell infiltration, but no significant correlation was found for LUSC. Therefore, more experiments with larger sample sizes are needed to study the specific mechanism of PSRC1 in the occurrence and development of NSCLC.
Conclusion
Through bioinformatics analysis based on online datasets, our study showed that PSRC1 expression in tumor tissues was higher than that in normal tissues in both LUAD and LUSC. By collecting clinical samples, we focused on immunohistochemical and survival analyses of NSCLC. The results showed that high PSRC1 expression was significantly associated with poor prognosis in LUAD, but no correlation was observed in the case of LUSC. Therefore, PSRC1 may be a promising prognostic biomarker that could serve as a valuable therapeutic target for LUAD patients.
Abbreviations
DEGs: differentially expressed genes; DC: dendritic cell; GSEA: Gene set enrichment analysis; GPCR: G-protein couple receptor; iDC: immature dendritic cell; KEGG: Kyoto Encyclopedia of Genes and Genomes; NSCLC: non-small cell carcinoma; LUAD: lung adenocarcinoma; LUSC: lung squamous cell carcinoma; MT: microtubule; PBS: phosphate-buffered saline; PFI: progression-free interval; PSRC1: Proline- and serine-rich coiled-coil 1; OS: overall survival; ssGSEA: single-sample gene set enrichment analysis; TCGA: The Cancer Genome Atlas; Th2: type 2 T-helper; TNM: tumor, node, and metastasis.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
We acknowledge the TCGA database for providing their platforms and contributors for uploading their meaningful datasets.
Funding
This research was supported by the fund for Natural Science Foundation of China (No.81970051), Excellent Top Talent Cultivation Project of Anhui Higher Education Institutions (gxgwfx2021014), A sub-project of the Anhui Medical University National first-class undergraduate specialty construction program (clinical medicine), Construction projects of key disciplines in Hefei (Occupational medicine), and the Applied Medical Research Project of Hefei Health Commission (Hwk2021zd008, Hwk2022zd013).
Ethics approval and consent to participate
This research had been approved by the Institutional Review Boards of First Affiliated Hospital of Anhui Medical University (Quick-PJ 2023-02-28). This project was following the principles of the Helsinki Declaration.
Consent for publication
All patients signed an informed consent approved by the Institutional Review Board.
Availability of data and materials
The dataset supporting the conclusions of this article is included within the article and its additional files. The data of this manuscript can be downloaded from The Cancer Genome Atlas database (https://portal.gdc.cancer.gov/). The original contributions presented in the study are included in the article and supplements. Further inquiries can be directed to the corresponding authors.
Author contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7-33
2. Cao M, Li H, Sun D, Chen W. Cancer burden of major cancers in China: A need for sustainable actions. Cancer Commun (Lond). 2020;40:205-10
3. Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB. et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol. 2015;10:1243-60
4. Lortet-Tieulent J, Soerjomataram I, Ferlay J, Rutherford M, Weiderpass E, Bray F. International trends in lung cancer incidence by histological subtype: adenocarcinoma stabilizing in men but still increasing in women. Lung Cancer. 2014;84:13-22
5. Shukla S, Evans JR, Malik R, Feng FY, Dhanasekaran SM, Cao X. et al. Development of a RNA-Seq Based Prognostic Signature in Lung Adenocarcinoma. J Natl Cancer Inst. 2017 109
6. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-49
7. Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung cancer. Lancet. 2021;398:535-54
8. Liu Y, Li HM, Wang R. Effectiveness and Safety of Adding Bevacizumab to Platinum-Based Chemotherapy as First-Line Treatment for Advanced Non-Small-Cell Lung Cancer: A Meta-Analysis. Front Med (Lausanne). 2021;8:616380
9. Zhu K, Xu A, Xia W, Li P, Zhang B, Jiang H. et al. Association Between NAT2 Polymorphism and Lung Cancer Risk: A Systematic Review and Meta-Analysis. Front Oncol. 2021;11:567762
10. Vargas AJ, Harris CC. Biomarker development in the precision medicine era: lung cancer as a case study. #N/A. 2016 16: 525-37
11. Zhang B, Xu A, Wu D, Xia W, Li P, Wang E. et al. ARL14 as a Prognostic Biomarker in Non-Small Cell Lung Cancer. J Inflamm Res. 2021;14:6557-74
12. Li P, Chen X, Zhou S, Xia X, Wang E, Han R. et al. High Expression of DEPDC1B Predicts Poor Prognosis in Lung Adenocarcinoma. J Inflamm Res. 2022;15:4171-84
13. Arbour KC, Riely GJ. Systemic Therapy for Locally Advanced and Metastatic Non-Small Cell Lung Cancer: A Review. Jama. 2019;322:764-74
14. Yu Y, Tian X. Analysis of genes associated with prognosis of lung adenocarcinoma based on GEO and TCGA databases. Medicine (Baltimore). 2020;99:e20183
15. Jang CY, Wong J, Coppinger JA, Seki A, Yates JR 3rd, Fang G. DDA3 recruits microtubule depolymerase Kif2a to spindle poles and controls spindle dynamics and mitotic chromosome movement. J Cell Biol. 2008;181:255-67
16. Jang CY, Fang G. The N-terminal domain of DDA3 regulates the spindle-association of the microtubule depolymerase Kif2a and controls the mitotic function of DDA3. #N/A. 2009 8: 3165-71
17. Hsieh WJ, Hsieh SC, Chen CC, Wang FF. Human DDA3 is an oncoprotein down-regulated by p53 and DNA damage. Biochem Biophys Res Commun. 2008;369:567-72
18. Gylfe AE, Kondelin J, Turunen M, Ristolainen H, Katainen R, Pitkänen E. et al. Identification of candidate oncogenes in human colorectal cancers with microsatellite instability. Gastroenterology. 2013;145:540-3.e22
19. Yang JD, Seol SY, Leem SH, Kim YH, Sun Z, Lee JS. et al. Genes associated with recurrence of hepatocellular carcinoma: integrated analysis by gene expression and methylation profiling. J Korean Med Sci. 2011;26:1428-38
20. Wang W, Lim WK, Leong HS, Chong FT, Lim TK, Tan DS. et al. An eleven gene molecular signature for extra-capsular spread in oral squamous cell carcinoma serves as a prognosticator of outcome in patients without nodal metastases. Oral Oncol. 2015;51:355-62
21. Zhao S, Ye Z, Stanton R. Misuse of RPKM or TPM normalization when comparing across samples and sequencing protocols. RNA. 2020;26:903-9
22. Wang R, Zhou S, Wu P, Li M, Ding X, Sun L. et al. Identifying Involvement of H19-miR-675-3p-IGF1R and H19-miR-200a-PDCD4 in Treating Pulmonary Hypertension with Melatonin. Mol Ther Nucleic Acids. 2018;13:44-54
23. Endo K, Terada T. Protein expression of CD44 (standard and variant isoforms) in hepatocellular carcinoma: relationships with tumor grade, clinicopathologic parameters, p53 expression, and patient survival. J Hepatol. 2000;32:78-84
24. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559
25. Consortium GO. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049-56
26. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. #N/A. 2012 16: 284-7
27. Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC. et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782-95
28. The L. Lung cancer: some progress, but still a lot more to do. Lancet. 2019;394:1880
29. Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL. et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299-311
30. Jiang H, Xu A, Xia W, Xia X, Li P, Zhang B. et al. Correction to: Nivolumab monotherapy or combination therapy with ipilimumab for lung cancer: a systemic review and meta-analysis. Cancer Cell Int. 2021;21:520
31. Zhang B, Liu Y, Zhou S, Jiang H, Zhu K, Wang R. Predictive effect of PD-L1 expression for immune checkpoint inhibitor (PD-1/PD-L1 inhibitors) treatment for non-small cell lung cancer: A meta-analysis. Int Immunopharmacol. 2020;80:106214
32. Kocher F, Hilbe W, Seeber A, Pircher A, Schmid T, Greil R. et al. Longitudinal analysis of 2293 NSCLC patients: a comprehensive study from the TYROL registry. Lung Cancer. 2015;87:193-200
33. Hsieh PC, Chang JC, Sun WT, Hsieh SC, Wang MC, Wang FF. p53 downstream target DDA3 is a novel microtubule-associated protein that interacts with end-binding protein EB3 and activates beta-catenin pathway. Oncogene. 2007;26:4928-40
34. Kim S, Jang CY. ANKRD53 interacts with DDA3 and regulates chromosome integrity during mitosis. Biochem Biophys Res Commun. 2016;470:484-91
35. Hsieh PC, Chiang ML, Chang JC, Yan YT, Wang FF, Chou YC. DDA3 stabilizes microtubules and suppresses neurite formation. J Cell Sci. 2012;125:3402-11
36. Uematsu K, Okumura F, Tonogai S, Joo-Okumura A, Alemayehu DH, Nishikimi A. et al. ASB7 regulates spindle dynamics and genome integrity by targeting DDA3 for proteasomal degradation. J Cell Biol. 2016;215:95-106
37. Wallace C, Newhouse SJ, Braund P, Zhang F, Tobin M, Falchi M. et al. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am J Hum Genet. 2008;82:139-49
38. Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ. et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189-97
39. Luo T, Guo Z, Liu D, Guo Z, Wu Q, Li Q. et al. Deficiency of PSRC1 accelerates atherosclerosis by increasing TMAO production via manipulating gut microbiota and flavin monooxygenase 3. #N/A. 2022 14: 2077602
40. Wei J, Wang B, Gao X, Sun D. Prognostic Value of a Novel Signature With Nine Hepatitis C Virus-Induced Genes in Hepatic Cancer by Mining GEO and TCGA Databases. Front Cell Dev Biol. 2021;9:648279
41. Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS. et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48:607-16
42. Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S. et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158:929-44
43. Cardarella S, Johnson BE. The impact of genomic changes on treatment of lung cancer. Am J Respir Crit Care Med. 2013;188:770-5
44. Long J, Wu XD, Liu Z, Xu YH, Ge CL. Integrated regulatory network involving differently expressed genes and protein-protein interaction on pancreatic cancer. Eur Rev Med Pharmacol Sci. 2015;19:2423-8
45. Postel-Vinay S, Vanhecke E, Olaussen KA, Lord CJ, Ashworth A, Soria JC. The potential of exploiting DNA-repair defects for optimizing lung cancer treatment. Nat Rev Clin Oncol. 2012;9:144-55
46. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-74
47. Rabinovich GA, Gabrilovich D, Sotomayor EM. Immunosuppressive strategies that are mediated by tumor cells. Annu Rev Immunol. 2007;25:267-96
48. Rosenthal R, Cadieux EL, Salgado R, Bakir MA, Moore DA, Hiley CT. et al. Neoantigen-directed immune escape in lung cancer evolution. Nature. 2019;567:479-85
49. Borst J, Ahrends T, Bąbała N, Melief CJM, Kastenmüller W. CD4(+) T cell help in cancer immunology and immunotherapy. #N/A. 2018 18: 635-47
50. Iglesia MD, Parker JS, Hoadley KA, Serody JS, Perou CM, Vincent BG. Genomic Analysis of Immune Cell Infiltrates Across 11 Tumor Types. J Natl Cancer Inst. 2016 108
51. Zhu S, Yang N, Wu J, Wang X, Wang W, Liu YJ. et al. Tumor microenvironment-related dendritic cell deficiency: a target to enhance tumor immunotherapy. Pharmacol Res. 2020;159:104980
52. Lichterman JN, Reddy SM. Mast Cells: A New Frontier for Cancer Immunotherapy. #N/A. 2021 10: 1270
53. Grisaru-Tal S, Rothenberg ME, Munitz A. Eosinophil-lymphocyte interactions in the tumor microenvironment and cancer immunotherapy. Nat Immunol. 2022;23:1309-16
54. Stankovic B, Bjørhovde HAK, Skarshaug R, Aamodt H, Frafjord A, Müller E. et al. Immune Cell Composition in Human Non-small Cell Lung Cancer. Front Immunol. 2018;9:3101
55. Frafjord A, Buer L, Hammarström C, Aamodt H, Woldbæk PR, Brustugun OT. et al. The Immune Landscape of Human Primary Lung Tumors Is Th2 Skewed. Front Immunol. 2021;12:764596
56. Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B. et al. Commensal Microbiota Promote Lung Cancer Development via γδ T Cells. Cell. 2019;176:998-1013.e16
Author contact
![]() Corresponding authors: Guanghe Fei Ph.D., Department of Respiratory and Critical Care Medicine, The First Affiliated Hospital of Anhui Medical University, Hefei 230022, China. Phone: 86-551-62922913, Fax: 86-551-62922913, E-mail: guanghefeicom. Sijing Zhou, Department of Occupational Diseases, Hefei Third Clinical College of Anhui Medical University, Hefei 230022, China. Phone: 0551-63511309, Fax: 0551-63511309, E-mail: zhousijingnet. Ran Wang Ph.D., Department of Respiratory and Critical Care Medicine, The First Affiliated Hospital of Anhui Medical University, Hefei 230022, China. Phone: 86-551-62922913, Fax: 86-551-62922913, E-mail: wangranedu.cn.
Corresponding authors: Guanghe Fei Ph.D., Department of Respiratory and Critical Care Medicine, The First Affiliated Hospital of Anhui Medical University, Hefei 230022, China. Phone: 86-551-62922913, Fax: 86-551-62922913, E-mail: guanghefeicom. Sijing Zhou, Department of Occupational Diseases, Hefei Third Clinical College of Anhui Medical University, Hefei 230022, China. Phone: 0551-63511309, Fax: 0551-63511309, E-mail: zhousijingnet. Ran Wang Ph.D., Department of Respiratory and Critical Care Medicine, The First Affiliated Hospital of Anhui Medical University, Hefei 230022, China. Phone: 86-551-62922913, Fax: 86-551-62922913, E-mail: wangranedu.cn.

 Global reach, higher impact
Global reach, higher impact