3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(18):3496-3507. doi:10.7150/jca.89360 This issue Cite
Research Paper
MUC15 is an independent prognostic factor that promotes metastases of MYCN non-amplified neuroblastoma
1. School of Medicine, South China University of Technology, Guangzhou, Guangdong, China.
2. Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, Guangzhou 510623, Guangdong, China.
3. Department of Pharmacology, School of Pharmacy, Binzhou Medical University, Yantai, 264003, China.
4. Laboratory Animal Center, School of Chemical Biology and Biotechnology, Peking University Shenzhen Graduate School, Shenzhen 518055, China.
*These authors contributed equally: Huiqin Guo, Wei-Xin Zhang and Qiu-yan Zhang
Received 2023-8-21; Accepted 2023-9-25; Published 2023-10-16
Abstract
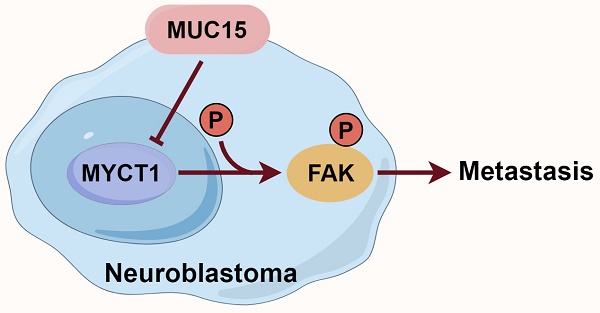
Background: Neuroblastoma (NB) is a cancer that arises from neural-crest-derived sympathoadrenal lineage. Less is known about the pathogenesis and molecular characteristics of MYCN non-amplified (MYCN-NA) NB.
Methods: We constructed a signature model targeting mucin family according to RNA sequencing data from GSE49710 dataset, and validated the prognostic performance. We also analyzed the gene expression matrix using DESeq2 R packages to screen the most differential mucin in high-risk NB samples. We further assessed its prognostic value, particularly in MYCN-NA NB samples. Moreover, we performed functional experiments to evaluate the impact of MUC15 overexpression on the migration of MYCN-NA NB cell lines.
Results: The 8-mucin signature model showed good prognostic performance in the GSE49710 dataset. Among the mucin genes, MUC15 was significantly upregulated in the high-risk NB cohort and was associated with poor prognosis, especially in MYCN-NA NB samples. Furthermore, MUC15 overexpression and exogenous MUC15 protein enhanced the migration of MYCN-NA NB cell lines. Mechanistically, MUC15 promoted the phosphorylation of focal adhesion kinase (FAK) by inhibiting the expression of MYCT1, a target of c-Myc.
Conclusions: Our findings suggested a potential network in controlling NB cell metastasis. Targeting MUC15 in MYCN-NA NB patients could be a promising therapeutic strategy.
Keywords: MUC15, neuroblastoma, MYCN non-amplified, metastases, MYCT1
1. Introduction
NB is the typical extracranial solid tumor that derives from sympathetic nervous system among childhood and represents roughly 15% in pediatric malignant tumor mortality [1]. The adrenal gland, abdomen and thorax are the frequent regions for NB [2]. Risk parameters have been established to determine the prognosis and treatment options for NB patients. Those with very low, low, and intermediate risks have a more favorable prognosis and treatment outcomes [3]. However, over 60% of NB patients are confirmed as high risk. Despite undergoing integrated treatment including radiotherapy, chemotherapy and immunotherapy, the 5-year event-free survival rate (EFS) of high-risk patients is still about 50% [4]. The heterogeneity of high-risk NB leads to poor outcomes with frequent recurrence and metastasis. Apart from age, the status of MYCN gene amplification is a principal factor in determining the risk grading. The EFS for NB patients with MYCN-amplified (MYCN-A) and MYCN-NA is 51.1% and 77.0% respectively [4]. Although many genetic abnormalities and molecular mechanisms have been discovered, the prospect of high-risk patients is still not optimistic [1, 5]. Compared to MYCN-A tumors, less is known about the pathogenesis and molecular characteristics of MYCN-NA NB. Therefore, there is an urgent need for molecular therapies targeting the MYCN-NA subgroup to address the treatment dilemma.
Mucins (MUCs) are highly glycosylated membrane proteins expressed broadly in epithelial cells. They have been characterized as transmembrane and secretory isoforms, which serve as protective barriers and transmit signals to the cell interior. Secretory mucins include MUC2, MUC5AC, MUC5B, MUC6 and MUC19, while transmembrane mucins include MUC1, MUC3, MUC4, MUC12, MUC13, MUC15, MUC16, MUC17, MUC21 and MUC22 [6, 7]. MUC1 and MUC16 have been broadly studied for their extensively aberrations in malignancy [8-13]. MUC1, also known as CA153 or CA199 in clinical diagnosis, interacts with various transcription factors such as p53, STAT3, NF-κB and β-catenin. These interactions activate multiple signaling pathways, including mitogen-activated protein kinase (MAPK), phosphatidylinositol 3-kinase (PI3K)/AKT and wingless type (Wnt), which contribute to malignant transformation in tumors [14]. On the other hand, MUC16, also refers to CA125, is highly expressed in breast, ovarian and pancreatic cancer. It is clinically used as a tumor marker for ovarian cancer [15]. MUC16 interacts with Janus kinase (JAK2) to induce STAT3 phosphorylation, thereby promoting the proliferation of breast and lung cancer cells [11, 12]. Both MUC1 and MUC16 play a role in mediating the epithelial-mesenchymal transition (EMT) process of tumor cells and facilitating metastasis [13]. Aberrant glycation in mucins is associated with tumor immune evasion and signal transduction issues, making them potential markers of tumor prognosis and treatment response [16].
In our study, we identified an 8-mucin signature model that showed good prognostic value in the GSE49710 dataset. Specifically, MUC15 was found to be upregulated in the high-risk cohort, particularly in MYCN-NA NB samples. MUC15 belongs to the family of transmembrane mucins. Its molecular structure consists of an N-terminal region and a C-terminal region. The N-terminal region contains a signal peptide and an extracellular domain formed by 10 N-glycosylated and 14 O-glycosylated modifications. The C-terminal region includes a transmembrane domain and a cytoplasmic tail [17]. Abnormal expression of MUC15 plays a crucial role in malignant tumors, affecting cell growth, adhesion, invasion and metastasis. However, the role of MUC15 in cancer is context-dependent and even opposite among different tumors. In some cases, MUC15 activates the ERK pathway, promoting the progression of glioma, thyroid and pancreatic cancer, leading to poor prognosis [18-20]. Additionally, high expression of MUC15 activates the Wnt/β-catenin signaling pathway, supporting invasion and migration in osteosarcoma [21]. Conversely, an elevated level of MUC15 has been shown to inhibit metastasis in renal cell carcinoma, suppress tumorigenicity and increase chemoresistance in liver cancer by inhibiting the PI3K/AKT signaling pathway [22-24]. Furthermore, aberrant downregulation of MUC15 has been linked to GSK3β phosphorylation, triggering epithelial-mesenchymal transition (EMT) and promoting stemness in prostate cancer [25]. MUC15 has garnered increasing attention in the realm of tumor development. However, the expression and effects of MUC15 in NB remain comparatively unknown.
Here, we demonstrated that MUC15 promoted migration of MYCN-NA NB cell lines by suppressing MYCT1/FAK axis. Targeting the metastatic ability of MUC15 in MYCN-NA NB could therefore be a promising and novel therapeutic option.
2. Materials and methods
2.1 GEO datasets acquisition
The microarray data of NB samples from GSE45547 and GSE49710 were downloaded from the Gene Expression Omnibus (GEO) database. GSE45547 included 608 tumor samples (550 MYCN-NA, 93 MYCN-A, and 6 not applicable), while GSE49710 contained 498 tumor samples (401 MYCN-NA, 92 MYCN-A, and 5 not applicable). The corresponding clinical characteristics for these samples were also acquired.
2.2 Construction and validation of mucin-related prognostic signature
We constructed a prognostic gene signature focusing on the mucin family genes (MFGs) within the 401 MYCN-NA samples from the GSE49710 cohort. To minimize the risk of overfitting, we employed LASSO-penalized Cox regression analysis with the 'glmnet' package to create a prognostic model, implementing a 10-fold cross-validation. The independent variable was the normalized expression matrix of MFGs, while the dependent variables were the overall survival (OS) and status of the patients. The penalty parameter (λ) for the model was determined through tenfold cross-validation, selecting the value that resulted in the lowest partial likelihood deviance as the minimum criteria. Then we computed each gene's risk score in this model along with its corresponding regression coefficients, categorized the patients into mucins-high-risk and mucins-low-risk groups based on the median value. The risk score was determined according to the following formula:
score= esum (each gene's expression × corresponding coefficient)
Principal component analysis (PCA) and t-SNE analysis were performed using the "stats" and "Rtsne" packages, respectively. For survival analysis, we established the optimal cut-off expression value using the "survminer" package and evaluated the predictive efficacy of the gene signature via time-dependent ROC curve analyses with the "survivalROC" package.
Utilizing the "limma" package in R, we identified differentially expressed genes (DEGs) between high-risk (83 samples) and low-risk (318 samples) NB groups among the 401 MYCN-NA samples in the GSE49710 cohort. DEGs were selected based on their absolute log2-fold change (|log2FC| > 1) and a false discovery rate (FDR) less than 0.05. The DEGs were visually represented using the "EnhancedVolcano" package in R. Finally, to identify MFGs with prognostic significance, we conducted a univariate Cox regression analysis on the overall survival data, setting a p-value threshold of less than 0.05.
2.3 Functional enrichment analysis
For pathway analysis, the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Set Enrichment Analysis (GESA) methods were used to explore significant pathways related to differentially expressed proteins.
2.4 Cell lines and cell culture
Human neuroblastoma cell lines (SK-N-SH, SK-N-AS, IMR-32) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). The SK-N-SH and IMR32 cell lines were cultured in Eagle's minimum essential medium (EMEM) and SK-N-AS was cultured in Roswell Park Memorial Institute (RPMI-1640) medium. All the mediums containing 10% fetal bovine serum (FBS), 1% Pen/Strep and maintained at 37 °C in 5% CO2 incubator.
2.5 Plasmid transfection
MUC15 short hairpin RNA (shRNA) was applied to knock-down MUC15 in NB cell lines. pCMV3-MUC15 and pCMV3-MYCT1 vectors were used to overexpress MUC15 and MYCT1, respectively. Ctrl was transfected the empty vector (pcDNA3.1) plasmid. Transient transfection was performed with Lipofectamine 3000 kit (Invitrogen, USA). The quantitative real‐time PCR (RT-PCR) assay was used to evaluate the transfection.
2.6 RT-PCR
Total RNA was extracted using TRIzol reagent according to the manufacturer's protocol. Each group of RNA was quantified and inverted into cDNAs using Evo M-MLV RT Master Mix (Accurate Biology, China) according to the manufacturer's instruction. The RT-PCR assay (ABI Q6 System, Applied Biosystems) was used to perform the relative expressions of identified genes.
2.7 Transwell assay
2-4×104 tumor cells were harvested and supplemented with 200µl serum-free RIPA-1640 into upper chamber of transwell insert (24 well 8.0-µm pore size, BD Falcon, USA). The lower chamber was added 600µl medium with 10% FBS. Following incubation for 24 h, all the transwell apparatus were fixed with carbinol for 10 min and subsequently stained with 0.1% crystal violet for 15 min at room temperature (RT). Then, the cell count was performed in five randomly selected fields under a light microscope at a magnification of 100×.
2.8 Western blot analysis
Cell lysates were obtained by treating with lysis buffer (Thermo Scientific, USA) supplemented with phosphatase inhibitor (MedChemExpress, USA) and protease inhibitor cocktail (Abcam, MA) for 10 min on ice, and centrifuged at 12,000 g for 15 min at 4°C. The resulting supernatants were added into 5× loading buffer and boiled at 95°C for 10 min. For western blot assay, equal concentration of samples was separated by 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS/PAGE), then transferred onto 0.45µm Immobilon-P PVDF membranes (Merck Millipore, Ireland) and blocked in western blocking buffer (Beyotime Biotechnology, China) for 2h at RT. After incubating primary antibodies (MUC15, Aanta Cruz Biotechnology, USA; MYCT1 and β-actin, Proteintech, USA, FAK Antibody Sample Kit, Cell Signaling Technology, USA) at 4 °C overnight, the membranes were washed for 3 times with TBST buffer. Subsequently, the membranes were incubated with HRP-conjugated anti-mouse/rabbit secondary antibodies (dilution of 1:20000) and the protein bands were visualized using WB ultrasensitive ECL luminescent liquid (Invigentech, USA).
2.9 Cell counting kit-8 (CCK8) assay
The tumor cells were seeded in 96-well cell culture plates. Following transfection as indicated, CCK-8 (Invigentech, USA) was added and incubating at 37°C for 2h. Finally, absorbance was recorded at 450 nm using an enzyme-labeled instrument.
2.10 Immunofluorescence staining
Paraffin sections of human NB tissues were subjected to dewaxing and repairing with EDTA antigen retrieval solution (PH=8.0, ZSGB-BIO, China). Blocking was performed using goat serum. The sections were then incubated overnight at 4°C with anti-MUC15 antibody (dilution of 1:100, Santa Cruz Biotechnology, USA). After removing catalase with 0.3% hydrogen peroxide, the sections were labeled with Alexa Fluor 555 Conjugate anti-mouse IgG secondary antibodies (Cell Signaling Technology, USA). Eventually, sections were sealed and nuclei stained by antifade mounting medium containing DAPI (Beyotime Biotechnology, China). Fluorescence images were captured using a Leica TCS SP8 confocal laser scanning microscope.
2.11 Cytoskeletal staining
For immunofluorescence imaging, cells were seeded in confocal plates prior to transfection. After treatment, the cells were fixed in 4% paraformaldehyde for 30 minutes. Subsequently, the cells were permeabilized with 0.1% Triton X-100 for 10 minutes. To stain F-actin filaments beneath the cell membranes, FITC-labeled phalloidin (dilution of 1:200, Yeasen Biotechnology, China) was applied and incubated for 90 minutes at RT protected from light The nucleus was stained by antifade mounting medium with DAPI (Beyotime Biotechnology, China). Cell morphology in each group was observed using a fluorescence inverted microscope (Leica DMi8).
2.12 Statistical analysis
All the western blot bands presented in this study are representative images obtained from two independent biological replicates. For immunofluorescence, ten images of each stage were randomly selected for statistics. For transwell assay, each group was randomly photographed in five different locations for statistics. Pearson's correlation analysis was employed to calculate the correlations between identified genes. The bar graphs display the means ± standard deviations (SDs). All the data were derived from a minimum of three independent experiments and analyzed using Student's t-test (unpaired, two-tailed) or one-way ANOVA with GraphPad Prism 9.5.1 software (GraphPad, La Jolla, CA, USA). A p-value of less than 0.05 was considered statistically significant.
Results
3.1 Establishment and Validation of a prognostic gene model targeting mucin family
In GSE49710 dataset, we took advantage of the LASSO algorithm's variable selection and shrinkage properties, to establish an 8-mucin signature model including MUC12, MUC13, MUC15, MUC20, MUC3A, MUC5AC, MUC7, and MUC8 (Figure 1A). The correlation network of mucin family genes was shown in Figure S1A. Meanwhile, seven of the 8-mucin signature model were significantly associated with the OS of NB patients, especially for MUC15 (Figure 1B). We subsequently stratified the patients into mucins-high-risk and mucins-low-risk groups based on the median risk score. The distribution of two risk groups was well clustered and discernible (Figure 1C, Figure S1B). Additionally, the risk and status of patients also showed reasonable clustering by mucins-high-risk and mucins-low-risk groups (Figure S1C, D). Kaplan-Meier survival analysis demonstrated that NB patients in the mucins-low-risk group had a superior prognosis (Figure 1D). To validate the prognostic accuracy of our model, we created a time-dependent ROC curve, which demonstrated its ability to accurate predict 1-, 3-, and 5-year OS rates (Figure 1E).
Notably, Cox univariate regression and multivariate Cox regression analyses demonstrated that our 8-mucin signature model could act as an independent prognostic factor for predicting the risk, stage, and age of NB patients (Figure 1F, G). Furthermore, we identified differentially expressed mucins between high-risk and low-risk NB groups, with MUC15 markedly upregulated in the high-risk cohort (Figure 1H, Figure S1E). The above data revealed that MUC15 emerged as a unique intersection, underscoring its significant role in the prognostic model.
Establishment and validation of a prognostic gene signature targeting mucin family base on LASSO Cox regression model in GSE49710. A. Left: Ten times cross-validation for tuning parameter selection in the LASSO model. Right: The LASSO coefficient profiles for clinicopathologic variables. B. Forest plots showed the results of the univariate Cox regression analysis of the relationship between gene expression and OS. C. PCA plot of mucin-high-risk and mucin-low-risk group in GSE49710. D. Kaplan-Meier curves were generated for the OS of patients in the mucin-high-risk and mucin-low-risk groups in GSE49710. E. AUC of time-dependent ROC curves were verified the prognostic performance of the risk score in GSE49710. F-G. Results of the univariate and multivariate Cox regression analyses regarding OS in the GSE49710 cohort. H. Volcano plot visualizing the differentially expressed mucin genes between high-risk and low-risk NB groups in GSE49710.

3.2 MUC15 is identified as a potential oncogene in MYCN-NA NB
Transcriptome analyses of online GEO datasets (GSE45547 and GSE49710) screened out highly expressed MUC15 in advanced NB (INSS Ⅳ stage) (Figure 2A). We were divided into MUC15-high and MUC15-low groups based on the median count of MUC15. In the subset of GSE49710, MUC15 preferred to exist in high-risk patients (Figure 2B) and led to shorter EFS and OS (Figure 2C). Remarkably, MUC15 was obvious upregulated in MYCN-NA cohort compared with MYCN-A (Figure 2D). The expression difference in advanced stage and high-risk of NB was more significant under MYCN-NA cohort (Figure 2E, F). In particular, increased MUC15 was more related to the shorter EFS and OS in MYCN-NA cohort (Figure 2G, H), indicating MUC15 may serve as a potential oncogene to predict prognosis in MYCN-NA patients. Together, these observations demonstrated that MUC15 is involved in carcinogenesis of MYCN-NA NB.
MUC15 is associated with poor prognosis in MYCN-NA NB from GEO datasets. A. The expression level of MUC15 across different INSS stages of NB (GSE45547 and GSE49710). B. Comparison of MUC15 expression between high-risk and non-high-risk NB cohorts (GSE49710). C. Differences in EFS and OS were evaluated in NB samples from GSE49710. D. Analysis of MUC15 expression in MYCN-amplified (MYCN-A) and MYCN-non-amplified (MYCN-NA) NB samples (GSE45547 and GSE49710). E. The expression of MUC15 across different INSS stages of MYCN-NA NB cohorts (GSE45547 and GSE49710). F. Comparison of MUC15 expression between high-risk and non-high-risk MYCN-NA NB cohorts (GSE49710). G-H. The EFS and OS of MUC15 in MYCN-NA group of GSE45547 and GSE49710.

MUC15 is highly expressed in MYCN-NA NB. A. Expression of MUC15 in tissues with different stages. Blue and green fluorescence represent DAPI (nucleus) and MUC15, respectively. The histogram showed the relative fluorescence area (n=10). B. RT-PCR showed the MUC15 expression in patients with different stages (I+II, n=15; Ⅲ+Ⅳ, n=20). C. Western blotting revealed the ratio of MUC15 to internal reference protein in three different neuroblastoma cell lines. *p < 0.05, **p < 0.01.

3.3 MUC15 is highly expressed in MYCN-NA NB
We performed immunofluorescence staining and RT-PCR assay to investigate the express situation of MUC15 in MYCN-NA NB slices. As was shown in Figure 3A and B, MUC15 was enriched in Ⅲ, Ⅳ stages compared to Ⅰ, Ⅱ stages. Interestingly, we observed MUC15 content in IMR-32 (MYCN-A cell line) had lower expression than SK-N-SH and SK-N-AS (MYCN-NA cell lines) (Figure 3C). Thus, we confirmed that MUC15 is highly expressed in MYCN-NA NB with poor prognosis.
3.4 MUC15 promotes NB cell migration in vitro
In order to elucidate the mechanism of MUC15, NB cells subjected to MUC15 knocking-down or full-length overexpression were first validated by RT-PCR and immunoblotting (Figure S2A, B). We then carried out whether MUC15 influence the proliferation and chemoresistance of NB. As shown in Figure S2C and D, endogenous MUC15 alteration or exogeneous MUC15 exerted trivial effects on proliferation of SK-N-AS and SK-N-SH cells with or without cisplatin treatment.
Considering migration-related pathways such as focal adhesion and ECM-receptor interaction were positive enriched in GSE45447 set (Figure S2E), migration abilities were further explored. We noticed that overexpression MUC15 vastly increased the migration of both SK-N-AS and SK-N-SH cells (Figure 4A). Knocking-down MUC15 decreased the migration of SK-N-AS, but seldom affected SK-N-SH. In addition, the enrichment of ECM-receptor interaction and focal adhesion pathway in MYCN-NA cohort supported the conclusion that migration-related genes were highly expressed (Figure 4B). Furthermore, western blotting results suggested that the level of p-FAK (925), p-FAK (397) and p-FAK (576) were increased under MUC15 overexpression and exogeneous MUC15 in SK-N-SH (Figure 4C). Cytoskeletal fluorescence staining in SK-N-AS showed increased and irregular skeletal proteins in MUC15 overexpression and exogeneous MUC15 groups, the appearance of filopodia indicated enhanced migration ability of above two groups (Figure 4D). To sum up, overexpression MUC15 promoted migration of MYCN-NA NB cells in vitro.
MUC15 promotes NB cell migration. A. Left: Transwell images of migration for NB cells randomly selected from each treatment group. Right: The bar chart showed the statistics of migration results of SK-N-AS and SK-N-SH. B. GESA analysis of migration-related pathways in the MYCN-NA subgroup of GSE45547. C. Western blotting showed increased expressions of three phosphorylated FAKs exposing to MUC15 overexpression and exogeneous MUC15 (20ng/ml) in SK-N-SH. D. Cytoskeletal protein staining in SK-N-AS. The green fluorescence for phalloidin combined with filamentous actin (F-actin) and excited with 488nm photoexcitation. The blue staining is DAPI for nuclear staining and MERGE as an overlapping image of two channels. SH#1&2 MUC15, knockdown of MUC15; OE-MUC15, overexpression of MUC15; MUC15, exogeneous MUC15 protein (20ng/ml). *p < 0.05, **p < 0.01, ****P<0.0001.

3.5 MUC15 facilitates migration via MYCT1
To figure out the downstream mechanism of MUC15, we compared the RNA sequences of overexpressed MUC15 and control groups in IMR-32. As shown in Figure 5A, we found MYCT1 reduced after overexpressing MUC15. RT-PCR as well as western blotting of SK-N-SH also validated the consequence (Figure 5B, C). Furthermore, correlation analysis revealed that MYCT1 was negatively associated with MUC15, whether in total samples or MYCN-NA subgroups (GSE45547 and GSE49710) (Figure 5D).
MUC15 facilitates migration via MYCT1. A. Transcriptome analysis of MUC15 overexpression and control groups. B. MYCT1 expression situation after overexpression of MUC15 by RT-PCR in SK-N-SH (n=6). C. The content of MYCT1 after MUC15 overexpression in SK-N-SH was detected by western blotting. D. The correlation analysis between MUC15 and MYCT1 in GSE45547 and GSE49710. OE-MUC15, overexpression of MUC15; MUC15, exogeneous MUC15 protein (20ng/ml). *p < 0.05.

MYCT1 rescues the MUC15-induced migration of NB. A. Transwell images of migration for NB cells were randomly selected from each group treated as indicated. B. The statistics analysis of SK-N-AS and SK-N-SH in A. C. Western blotting showed the expressions of three FAK phosphorylation in SK-N-SH treated as indicated. OE-MUC15, overexpression of MUC15; OE-(MUC15+MYCT1), overexpression of MUC15 and MYCT1; MUC15, exogeneous MUC15 protein (20ng/ml); MUC15+OE-MYCT1, exogeneous MUC15 protein (20ng/ml) and overexpression of MYCT1; OE-MYCT1, overexpression of MYCT1. ****P<0.0001.

3.6 MYCT1 rescues the MUC15-induced migration of NB
Later, the full length MYCT1 plasmid was constructed and transfected into MUC15-overexpressing or exogeneous MUC15-treated cell lines. Our results showed that MUC15-induced migration was depressed by MYCT1 in both overexpression and exogeneous MUC15-treated groups among SK-N-AS and SK-N-SH (Figure 6A, B). At the same time, western blotting in SK-N-SH also showed that phosphorylation of FAK were all inhibited, especially in the exogeneous MUC15-treated subgroup (Figure 6C), indicating that the phosphorylation of FAK induced by MUC15 could be inhibited by MYCT1. Totally, our study demonstrated MUC15 as an independent prognostic factor to predict the prognosis of NB patients with MYCN-NA. MYCT1 acted as an inhibitory factor in the process of MUC15 promoting migration of NB.
4. Discussion
Investigating the role of membrane proteins in neuroblastoma is critical in tackling the challenge of discovering novel drug targets and precisely targeting the microenvironment. Transmembrane mucins expressed in tumors play a crucial role in promoting proliferation, metastasis, and chemoresistance. In this study, we conducted a comprehensive analysis of expression matrix of mucin family genes using the online GEO database (GSE49710). From this analysis, we developed an 8-mucin signature model that specifically targeted mucin family genes. By categorizing NB patients into mucins-low-risk and mucins-high-risk groups based on this model, we were able to demonstrate that the model had superior prognostic predictive capabilities for NB patients, including risk, stage and age. Notably, we observed significant upregulation of MUC15 in the high-risk NB group.
The extracellular domain of MUC15 contains N- and O-glycosylated modifications, alterations in N-glycan had been shown to decrease FAK phosphorylation and were associated with tumor migration [26]. FAK, the intersection of multiple signaling pathways for tumors, is a crucial tyrosine kinase protein that regulates cell migration and adhesion. FAK exists in an inactive form in the cytoplasm, but after phosphorylation activation, it interacts with membrane receptors and nucleus transcription factors to transmit diverse signals [27]. FAK promotes tumor development. In fact, several FAK phosphorylation inhibitors have entered clinical trials, highlighting the therapeutic potential of targeting this protein in cancer treatment [28].
The MYC family, which includes c-Myc, N-Myc and L-Myc transcription factors, plays a crucial role in tumorigenesis, maintenance and progression in various cancers. Each subtype of MYC exhibits unique features, leading to distinct oncogenic mechanisms in tumor development [29]. In NB, N-Myc (MYCN) amplification is a common variation occurring in approximately 25% of cases, which contributes to malignant proliferation and metastasis [30]. With the deepening of research, the mechanism of c-Myc affects NB has attracted increasing attention in recent years [31-34]. However, the exact mechanism of c-Myc in NB remains largely unclear.
As a direct downstream target of c-Myc, MYCT1 has two E-boxes in front of its promoter, which can bind c-Myc transcription complex [35, 36]. MYCT1, also known as MT-MC1 or MTCL, was initially discovered in myeloid cells [37]. MYCT1 plays inhibitory roles in controlling the growth, adhesion, transformation, apoptosis and migration in various cancers [38-43]. However, not all evidence supports MYCT1 as a tumor suppressor gene. Kabir et al. reported that MYCT1 regulated actin cytoskeleton in endothelial motility to promote tumor angiogenesis. Targeting MYCT1 enhanced antitumor immunity in the microenvironment and improved the effectiveness of anti-PD1 treatment [44]. Based on our transcriptome data and mechanistic work, we have identified MYCT1 as a factor that dampened MUC15-induced FAK phosphorylation and NB cell migration, which is consistent with its migration-inhibiting activity.
In summary, we have discovered that MUC15 served as a potential target and novel biomarker for NB prognosis assessment. Overexpression of MUC15 significantly enhanced NB cell migration. Mechanically, MYCT1 served as a negative regulator of elevated MUC15, thereby reducing the MUC15-induced migration of NB cells. Further investigation is needed to clarify the underlying mechanism of how MUC15 influences MYCT1 and to establish MUC15 as a candidate for assessing prognosis in NB patients with MYCN-NA. Our study is the first to identify molecular marker and signaling pathway for MYCN-NA NB membrane surface protein. Our findings complemented the understanding of NB pathogenesis, providing a foundation for future precision treatments.
Abbreviations
EFS: event-free survival; FAK: focal adhesion kinase; MYCN-A: MYCN amplificated; MYCN-NA: MYCN non-amplificated; MUC: mucin; MYCT1: MYC target 1; NB: neuroblastoma; OS: overall survival.
Supplementary Material
Supplementary figures.
Acknowledgements
The graphical abstract was created by Figdraw (www.figdraw.com).
Funding
This study was supported by grants from National Natural Science Foundation of China (82002635, 82002636, 82173593).
Data availability
All the data supporting of this work are available within the article and its Supplementary files.
Ethics approval statement
This work was approved by the Ethics Committee of Guangzhou Women and Children's Medical Center.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Zafar A, Wang W, Liu G, Wang X, Xian W, McKeon F. et al. Molecular targeting therapies for neuroblastoma: Progress and challenges. Med Res Rev. 2021;41:961-1021
2. Tsubota S, Kadomatsu K. Origin and initiation mechanisms of neuroblastoma. Cell Tissue Res. 2018;372:211-21
3. Monclair T, Brodeur GM, Ambros PF, Brisse HJ, Cecchetto G, Holmes K. et al. The International Neuroblastoma Risk Group (INRG) staging system: an INRG Task Force report. J Clin Oncol. 2009;27:298-303
4. Irwin MS, Naranjo A, Zhang FF, Cohn SL, London WB, Gastier-Foster JM. et al. Revised Neuroblastoma Risk Classification System: A Report From the Children's Oncology Group. J Clin Oncol. 2021;39:3229-41
5. Matthay KK, Maris JM, Schleiermacher G, Nakagawara A, Mackall CL, Diller L. et al. Neuroblastoma. Nat Rev Dis Primers. 2016;2:16078
6. Kufe DW. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer. 2009;9:874-85
7. Singh PK, Hollingsworth MA. Cell surface-associated mucins in signal transduction. Trends Cell Biol. 2006;16:467-76
8. Pochampalli MR, el Bejjani RM, Schroeder JA. MUC1 is a novel regulator of ErbB1 receptor trafficking. Oncogene. 2007;26:1693-701
9. Wei X, Xu H, Kufe D. Human MUC1 oncoprotein regulates p53-responsive gene transcription in the genotoxic stress response. Cancer Cell. 2005;7:167-78
10. Raina D, Ahmad R, Kumar S, Ren J, Yoshida K, Kharbanda S. et al. MUC1 oncoprotein blocks nuclear targeting of c-Abl in the apoptotic response to DNA damage. EMBO J. 2006;25:3774-83
11. Lakshmanan I, Salfity S, Seshacharyulu P, Rachagani S, Thomas A, Das S. et al. MUC16 Regulates TSPYL5 for Lung Cancer Cell Growth and Chemoresistance by Suppressing p53. Clin Cancer Res. 2017;23:3906-17
12. Lakshmanan I, Ponnusamy MP, Das S, Chakraborty S, Haridas D, Mukhopadhyay P. et al. MUC16 induced rapid G2/M transition via interactions with JAK2 for increased proliferation and anti-apoptosis in breast cancer cells. Oncogene. 2012;31:805-17
13. Lee DH, Choi S, Park Y, Jin HS. Mucin1 and Mucin16: Therapeutic Targets for Cancer Therapy. Pharmaceuticals (Basel). 2021 14
14. Nath S, Mukherjee P. MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med. 2014;20:332-42
15. Aithal A, Rauth S, Kshirsagar P, Shah A, Lakshmanan I, Junker WM. et al. MUC16 as a novel target for cancer therapy. Expert Opin Ther Targets. 2018;22:675-86
16. Bhatia R, Gautam SK, Cannon A, Thompson C, Hall BR, Aithal A. et al. Cancer-associated mucins: role in immune modulation and metastasis. Cancer Metastasis Rev. 2019;38:223-36
17. Zhang S, Zhang W, Xiao Y, Qin T, Yue Y, Qian W. et al. Targeting MUC15 Protein in Cancer: Molecular Mechanisms and Therapeutic Perspectives. Curr Cancer Drug Targets. 2020;20:647-53
18. Cheng M, Liu L. MUC15 promotes growth and invasion of glioma cells by activating Raf/MEK/ERK pathway. Clin Exp Pharmacol Physiol. 2020;47:1041-8
19. Choi C, Thi Thao Tran N, Van Ngu T, Park SW, Song MS, Kim SH. et al. Promotion of tumor progression and cancer stemness by MUC15 in thyroid cancer via the GPCR/ERK and integrin-FAK signaling pathways. Oncogenesis. 2018;7:85
20. Wang S, Li J, You L, Dai M, Zhao Y. High Expression of MUC15 Is Correlated with Poor Prognosis of Pancreatic Cancer and Promotes Migration, Invasion, and Chemo-Resistance In Vitro. Med Sci Monit. 2020;26:e926432
21. Chen T, Chen Z, Lian X, Wu W, Chu L, Zhang S. et al. MUC 15 Promotes Osteosarcoma Cell Proliferation, Migration and Invasion through Livin, MMP-2/MMP-9 and Wnt/beta-Catenin Signal Pathway. J Cancer. 2021;12:467-73
22. Yue Y, Hui K, Wu S, Zhang M, Que T, Gu Y. et al. MUC15 inhibits cancer metastasis via PI3K/AKT signaling in renal cell carcinoma. Cell Death Dis. 2020;11:336
23. Wang RY, Chen L, Chen HY, Hu L, Li L, Sun HY. et al. MUC15 inhibits dimerization of EGFR and PI3K-AKT signaling and is associated with aggressive hepatocellular carcinomas in patients. Gastroenterology. 2013;145:1436-48 e1-12
24. Han T, Zheng H, Zhang J, Yang P, Li H, Cheng Z. et al. Downregulation of MUC15 by miR-183-5p.1 promotes liver tumor-initiating cells properties and tumorigenesis via regulating c-MET/PI3K/AKT/SOX2 axis. Cell Death Dis. 2022;13:200
25. Wu S, Yue Y, Gu Y, Wang Q, Liu T, Li L. et al. MUC15 loss facilitates epithelial-mesenchymal transition and cancer stemness for prostate cancer metastasis through GSK3beta/beta-catenin signaling. Cell Signal. 2021;84:110015
26. Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15:540-55
27. Sulzmaier FJ, Jean C, Schlaepfer DD. FAK in cancer: mechanistic findings and clinical applications. Nat Rev Cancer. 2014;14:598-610
28. Dawson JC, Serrels A, Stupack DG, Schlaepfer DD, Frame MC. Targeting FAK in anticancer combination therapies. Nat Rev Cancer. 2021;21:313-24
29. Baluapuri A, Wolf E, Eilers M. Target gene-independent functions of MYC oncoproteins. Nat Rev Mol Cell Biol. 2020;21:255-67
30. Bartolucci D, Montemurro L, Raieli S, Lampis S, Pession A, Hrelia P. et al. MYCN Impact on High-Risk Neuroblastoma: From Diagnosis and Prognosis to Targeted Treatment. Cancers (Basel). 2022 14
31. Le Grand M, Mukha A, Puschel J, Valli E, Kamili A, Vittorio O. et al. Interplay between MycN and c-Myc regulates radioresistance and cancer stem cell phenotype in neuroblastoma upon glutamine deprivation. Theranostics. 2020;10:6411-29
32. Yang XH, Tang F, Shin J, Cunningham JM. A c-Myc-regulated stem cell-like signature in high-risk neuroblastoma: A systematic discovery (Target neuroblastoma ESC-like signature). Sci Rep. 2017;7:41
33. Zimmerman MW, Liu Y, He S, Durbin AD, Abraham BJ, Easton J. et al. MYC Drives a Subset of High-Risk Pediatric Neuroblastomas and Is Activated through Mechanisms Including Enhancer Hijacking and Focal Enhancer Amplification. Cancer Discov. 2018;8:320-35
34. Wei SJ, Nguyen TH, Yang IH, Mook DG, Makena MR, Verlekar D. et al. MYC transcription activation mediated by OCT4 as a mechanism of resistance to 13-cisRA-mediated differentiation in neuroblastoma. Cell Death Dis. 2020;11:368
35. Fu S, Guo Y, Chen H, Xu ZM, Qiu GB, Zhong M. et al. MYCT1-TV, a novel MYCT1 transcript, is regulated by c-Myc and may participate in laryngeal carcinogenesis. PLoS One. 2011;6:e25648
36. Abou Ziki R, Teinturier R, Luo Y, Cerutti C, Vanacker JM, Poulard C. et al. MEN1 silencing triggers the dysregulation of mTORC1 and MYC pathways in ER+ breast cancer cells. Endocr Relat Cancer. 2022;29:451-65
37. Yin X, Grove L, Rogulski K, Prochownik EV. Myc target in myeloid cells-1, a novel c-Myc target, recapitulates multiple c-Myc phenotypes. J Biol Chem. 2002;277:19998-20010
38. Yue PJ, Sun YY, Li YH, Xu ZM, Fu WN. MYCT1 inhibits the EMT and migration of laryngeal cancer cells via the SP1/miR-629-3p/ESRP2 pathway. Cell Signal. 2020;74:109709
39. Wang HT, Tong X, Zhang ZX, Sun YY, Yan W, Xu ZM. et al. MYCT1 represses apoptosis of laryngeal cancerous cells through the MAX/miR-181a/NPM1 pathway. FEBS J. 2019;286:3892-908
40. Zhang ZX, Zhang WN, Sun YY, Li YH, Xu ZM, Fu WN. CREB promotes laryngeal cancer cell migration via MYCT1/NAT10 axis. Onco Targets Ther. 2018;11:1323-31
41. Pu J, Wang J, Xu Z, Lu Y, Wu X, Wu Y. et al. miR-632 Functions as Oncogene in Hepatocellular Carcinoma via Targeting MYCT1. Hum Gene Ther Clin Dev. 2019;30:67-73
42. Xu XP, Peng XQ, Yin XM, Liu Y, Shi ZY. miR-34a-5p suppresses the invasion and metastasis of liver cancer by targeting the transcription factor YY1 to mediate MYCT1 upregulation. Acta Histochem. 2020;122:151576
43. Qiu GB, Gong LG, Hao DM, Zhen ZH, Sun KL. Expression of MTLC gene in gastric carcinoma. World J Gastroenterol. 2003;9:2160-3
44. Kabir AU, Subramanian M, Lee DH, Wang X, Krchma K, Wu J. et al. Dual role of endothelial Myct1 in tumor angiogenesis and tumor immunity. Sci Transl Med. 2021 13
Author contact
![]() Corresponding authors: Huimin Xia, Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, 9 Jinsui Road, Guangzhou 510623, Guangdong, China. Tel.: (+86-020) 38076001, Fax: (+86-020) 38076020; E-mail: xia-huimincom. Or Lei Miao, Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, 9 Jinsui Road, Guangzhou 510623, Guangdong, China, Email: miaoleiedu.cn. Or Jing He, Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, 9 Jinsui Road, Guangzhou 510623, Guangdong, China, Email: hejing198374com.
Corresponding authors: Huimin Xia, Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, 9 Jinsui Road, Guangzhou 510623, Guangdong, China. Tel.: (+86-020) 38076001, Fax: (+86-020) 38076020; E-mail: xia-huimincom. Or Lei Miao, Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, 9 Jinsui Road, Guangzhou 510623, Guangdong, China, Email: miaoleiedu.cn. Or Jing He, Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, 9 Jinsui Road, Guangzhou 510623, Guangdong, China, Email: hejing198374com.

 Global reach, higher impact
Global reach, higher impact