3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(11):1472-1480. doi:10.7150/jca.14844 This issue Cite
Research Paper
A Point Mutation in DNA Polymerase β (POLB) Gene Is Associated with Increased Progesterone Receptor (PR) Expression and Intraperitoneal Metastasis in Gastric Cancer
1. Laboratory of Molecular Oncology, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Peking University School of Oncology, Beijing Cancer Hospital & Institute, 52# Fu-Cheng-Lu, Hai-Dian District, Beijing, 100142, China;
2. Department of Gastroenterology, The Chengdu Military General Hospital, Chengdu, China;
3. Department of Sugary, Peking University School of Oncology, Beijing Cancer Hospital & Institute, 52# Fu-Cheng-Lu, Hai-Dian District, Beijing, 100142, China;
4. Department of Pathology, Peking University School of Oncology, Beijing Cancer Hospital & Institute, 52# Fu-Cheng-Lu, Hai-Dian District, Beijing, 100142, China;
5. Department of Genetics, Case Western Reserve University, Cleveland, OH 44106, USA;
6. Department of Medicine, The George Washington University School of Medicine and Health Sciences, 2300 Eye Street, N.W. Ross Hall 402C, Washington, DC 20037, USA.
*These authors contributed equally to this work.
Received 2015-12-29; Accepted 2016-3-15; Published 2016-7-5
Abstract
Increased expression of progesterone receptor (PR) has been reported in gastric cancer (GC). We have previously identified a functional T889C point mutation in DNA polymerase beta (POLB), a DNA repair gene in GC. To provide a detailed analysis of molecular changes associated with the mutation, human cDNA microarrays focusing on 18 signal transduction pathways were used to analyze differential gene expression profiles between GC tissues with T889C mutant in POLB gene and those with wild type. Among the differentially expressed genes, notably, PR was one of the significantly up-regulated genes in T889C mutant POLB tissues, which were subsequently confirmed in POLB gene transfected AGS cell line. Interestingly, patients with T889C mutation and PR positivity were associated with higher incidence of intraperitoneal metastasis (IM). In vitro studies indicate that PR expression was upregulated in AGS cell line when transfected with T889C mutant expression vector. Cotransfection of T889C mutant allele and PR gene induced cell migration in the cell line. These data demonstrated that T889C mutation-associated PR overexpression results in increased IM. Therefore, T889C mutation-associated PR overexpression may serve as a biomarker for an adverse prognosis for human GC.
Keywords: Gastric cancer, DNA polymerase beta, progesterone receptor, metastasis.
Introduction
Gastric cancer (GC) is the fourth most commonly diagnosed malignancy and the second leading cause of cancer-related mortality worldwide [1]. Influences of sex-hormones on gastric cancer development have been studied [2]. Progesterone is a critical regulator of normal female reproductive function, with diverse tissue-specific effects in humans. Progestogen has been classified by the International Agency for Research on Cancer (IARC) as possibly carcinogenic to humans and plays a role in cancer development and progression [3]. The effects of progesterone mediated by its nuclear receptor, progesterone receptors (PR), have been reported in adenocarcinoma of the stomach [4]. Further, primarily altering PR expression levels rather than impacting PR transcriptional activity has been reported to be involved in human cancer progression [5]. However, the roles of PR expression in GC are poorly defined.
Defective DNA repair can result in genomic instability and consequently induce oncogenic alterations. Base excision repair (BER) is a key pathway for maintaining genomic stability. A key enzyme in the BER pathway is DNA polymerase beta (POLB) [6-9]. POLB gene mutation and overexpression have been reported in various cancers including GC [10-13]. However, the detailed mechanism remains unclear.
In our previous studies, we identified a unique T889C missense mutation in POLB gene in human primary GCs [14]. Here, we investigate changes in gene expression associated with the T889C mutation in POLB gene using cDNA microarray technology. Interestingly, among the differentially expressed genes, notably, PR was one of the significantly up-regulated genes in T889C mutant POLB tissues. Patients with T889C mutation and PR positivity were associated with higher incidence of intraperitoneal metastasis (IM). Further functional relationship between PR expression and T889C mutation in GC cell line was investigated.
Materials and Methods
Cell lines and tissue samples
Human GC cell line AGS was obtained from the American Type Culture Collection (ATCC) and cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco) containing 10% fetal bovine serum (Gibco) and 10 μg/ml vancomycin. Primary GC specimens and their matched normal adjacent tissues were collected at the time of surgery. None of the patients had received any chemotherapy or radiotherapy before surgical resection. Tissue samples were snap-frozen in liquid nitrogen until further use. Clinical and histological features were evaluated (Table 1). Tumor tissues were histologically classified according to the Lauren's classification. The study was conducted according to the Helsinki Declaration, approved by the Institutional Ethical Standards Committee of Peking University and informed consents were obtained from all patients in this study.
RNA extraction
Total RNA was isolated from the cells using the Trizol reagent (LifeTachnologies) following the manufacturer's instructions. Total RNA from excised tissue samples was isolated using the RNeasy Mini Kit (Qiagen) following the manufacturer's recommendations. The quality and quantity of the extracted total RNA was assessed by the spectrophotometric ultraviolet (UV) instrument.
Clinical and pathologic parameters of the 70 patient samples used in mutation analysis.
| Tissues parameters | Number |
|---|---|
| Tumor | 70 |
| Gender | |
| Male | 36 |
| Female | 34 |
| Age (years) | |
| Male | 31-71 |
| Female | 29-73 |
| Pathology | |
| Intestinal | 48 |
| Diffuse | 15 |
| Mixed | 3 |
| Adenosquamous | 4 |
| Tumor location | |
| GOJ a | 31 |
| Fundus | 6 |
| Greater curve-body | 10 |
| Lesser curve-body | 8 |
| Body nonspecified | 7 |
| Antrum | 8 |
| Differentiation | |
| Well | 14 |
| Moderate to poor | 33 |
| Poor | 23 |
| Depth of invasion | |
| Mucosa | 6 |
| Submucosa | 7 |
| Muscularis propria | 10 |
| Serosa/full thickness | 47 |
| Intraperitoneal metastasis | |
| + | 26 |
| - | 44 |
a Gastro-oesophageal junction.
cDNA expression array analysis
Microarray gene expression analysis was conducted using the nonradioactive signal transduction pathway finder GEArray Q series kit (GEArray Q Series HS-008-12/ HS-008N-12; SuperArray Inc., Bethesda, MD, USA), in accordance with the manufacturer's instructions. This array membrane is comprised of 96 signal transduction pathway genes, a plasmid pUC18 negative control, and four housekeeping genes, including glyceraldehyde-3-phosphate dehydrogenase (GAPDH), cyclophilin A, ribosomal protein L13a, and β-actin, each of which was printed with the Tetra Spots format. cDNA probes were synthesized from 5 μg of total RNA by gene-specific primers (supplied with the SuperArray kit) with biotin-16-dUTP. After 2 hours of pre-hybridization at 60°C, with GEAhyb Hybridization Solution (SuperArray) containing 100 μg of DNA/ml of denatured salmon sperm DNA (Invitrogen), the array membranes were hybridized overnight with denatured cDNA probes at 60°C. The membranes were then washed twice with 2x SSC, 1% SDS, and twice with 0.1x SSC, 0.5% SDS, for 15 minutes each at 60°C and the membrane was blocked for 40 minutes with GEAblocking Solution Q (SuperArray), then incubated with alkaline phosphatase-conjugated streptavidin for 10 minutes at room temperature. Chemiluminescent detection was conducted using the CDP-Star chemiluminescent substrate, and images of the arrays were acquired on X-ray films. The images were then scanned with a TouchToss SIS-3800 scanner (Samsung, Seoul, Korea). The resultant scanned images were converted to raw data files using the Scanalyse software. GEArray Analyzer software (SuperArray Inc.) was used for data analysis. The relative expression levels of the different genes were estimated via comparisons of their respective signal intensities with that of the internal controls. Genes were considered as not being expressed if their expression level was <20% of the control gene. The fold change of at least 1.5 fold is considered significant. The expression data was confirmed using the real-time PCR.
Semi-quantitative RT-PCR and real time PCR assays
First-strand cDNA was synthesized using the RT kit (SuperScript preamplification system; Life Technologies, Inc.-BRL). PR gene has two isoforms, A (PRA) and B (PRB) using two distinct promotors and translation start sites in the first exon. The two isoforms are indistinguishable by PCR primers we used for mRNA expression analysis. Primer sequences are available upon request. Total RNA (5 μg) was reverse-transcribed in the presence of 25 ng/μl of oligo (deoxythymidine) primer (Life Technologies, Inc.) Subsequently, 5 μl of the resulting cDNA were used as templates for PCR. The products were resolved on a 2.5% agarose ethidium bromide gel. Images were captured with Polaroid film under ultraviolet light. Products were quantified using the PhosphorImager and ImageQuant software (Molecular Dynamics, Inc.). Real-time PCR assays were carried out on a Bio-Rad iCycler iQ system (Bio-Rad, Hercules, CA, USA) using SYBR Green reagent [13].
Tissue microarray construction, immunohistochemical staining and evaluation
Tissue microarrays were constructed as described previously [15]. Briefly, the GC tissues and matched normal gastric mucosa were ethanol fixed, paraffin embedded for creating tissue microarrays. H&E-stained sections were used for histological verification. An affinity purified goat polyclonal antibody POLB (sc-5927, Santa Cruz) was used for immunohistochemical staining. All the slides were stained by the avidin-biotin-peroxidase (ABC) protocol. The labeling index (LI) of POLB gene was determined by counting tumor and normal cells stained with the antibody. For evaluation, the total fields were counted microscopically under high power (×200). The results of POLB expression were graded as: negative (-), <5% of cell stained; positive (+), >5% of cells stained.
Construction of eukaryotic expression vector and transfection
The entire coding region of cDNA containing wild type POLB was amplified by RT-PCR, and the T889C mutant POLB was obtained by site-directed mutagenesis before they were cloned into pcDNA3.1 vector. The pCMV6-XL4/PR containing PRA and PRB cDNA sequence was purchased from Origene Company. AGS cells were transfected with either POLB wild type or T889C mutant plasmids, along with PR expression plasmid. 5 X 105 cells were plated in 10 cm2 culture dishes, and 2.5 μg of plasmid DNA/dish was transfected using the calcium phosphate precipitation method.
Immunofluorescence assays
Cells were seeded at 2 × 104 cells per well on glass coverslips in six-well plates and fixed in 2% paraformaldehyde as described previously [16] . Confocal images were obtained using a LSM 510 Confocal microscope (Carl Zeiss). The number of nuclei containing at least one localized area of immunofluorescence was determined by examination of the confocal images. Antibodies for immunofluorescence were as follows: goat polyclonal antibody POLB (sc-5927, Santa Cruz) at final concentration (1:600), Rabbit anti-progesterone receptor Polyclonal antibody (NBP1-61401, Novus Biologicals, 1:500), Alexa Flour 568 goat anti-mouse IgG (Invitrogen), 1:500 and Alexa Flour 568 goat anti-mouse IgG (Invitrogen), 1:500.
Western blot analysis
Protein extraction and Western blot analysis with chemiluminescent detection were as described [16]. The following antibodies and dilution factors were used: affinity purified goat polyclonal antibody POLB (sc-5927, Santa Cruz, 1:800), Rabbit anti-progesterone receptor polyclonal antibody (NBP1-61401, Novus Biologicals, 1:500), rabbit anti-actin (H-196, Santa Cruz, 1:1000), donkey anti-goat IgG-HRP (sc-2020, Santa Cruz, 1:2000) and goat anti-rabbit IgG-HRP (sc-2030, Santa Cruz, 1:2000).
Transwell matrigel invasion assay
Matrigel invasion assays were performed using the BD BioCoat™ Matrigel™ Invasion Chamber (BD Biosciences). BD BioCoat Matrigel Invasion Chambers provide cells with the conditions that allow assessment of their invasive property in vitro. It consists of a BD Falcon™ TC Companion Plate with Falcon Cell Culture Inserts containing an 8 micron pore size PET membrane with a thin layer of matrigel basement membrane matrix. Briefly, prior to the start of each experiment, 500 μl of warm (37°C) serum-free DMEM medium was added to the upper and lower chambers and allowed to rehydrate for 2h in a 37°C cell culture incubator. After 2h rehydration, the medium was removed from the upper and lower chambers, 750 μl of DMEM with 10% fetal bovine serum was added to lower chambers. Then 2.5 x 105 AGS cells transfected with PR, wild-type or mutant POLB and negative control for 24h were seeded onto the top chamber cultured in 500 μl serum-free DMEM with 0.1% BSA in the top chamber. Cells were incubated in a matrigel chamber in a 37°C humidified incubator with 5% CO2 for 24 h. The non-invasive cells were removed from the upper surface of the membrane by scrubbing with a cotton swab and the invasive cells present on the bottom of the membrane were fixed, stained with the Diff-Quick staining solution and counted (five microscope fields under the 10X len). Experiments were done in duplicates for each cell line at least four times. Cell counts were performed on five nonoverlapping random fields for each chamber and four chambers were counted for each experimental point, and the number of invasive cells was compared to the corresponding control [17].
Statistical analysis
Means ± SD were calculated, and statistical analysis was performed using one-way ANOVA, followed by the nonparametric Kruskal-Wallis test; P < 0.05 was considered statistically significant.
Results
POLB T889C mutation altered gene expression profile in GC
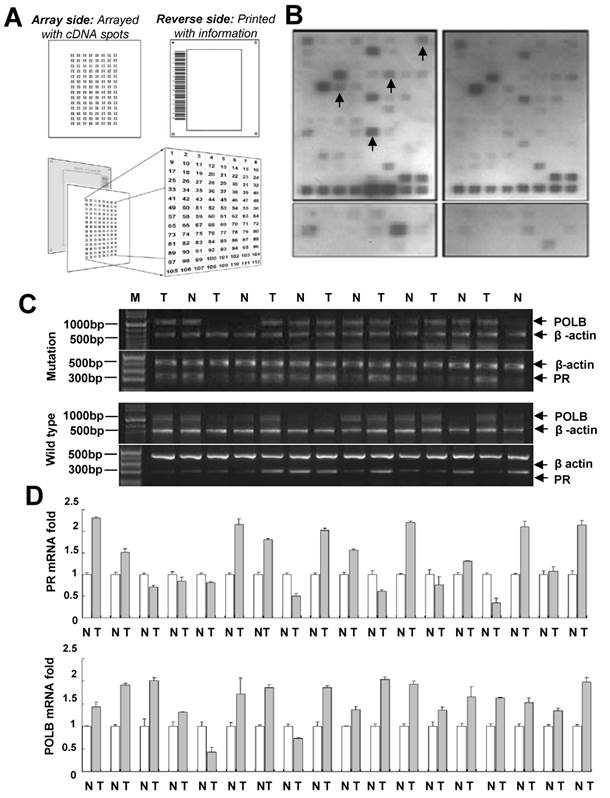
In order to characterize the signal transduction pathways that might be modulated by the T889C mutation in POLB gene, we conducted a series of microarrays. The GEArray Signal Transduction Pathway Finder Gene Array membrane contains 96 marker genes, which are associated with 18 different signal transduction pathways (Mitogenic Pathway, Wnt Pathway, Hedgehog Pathway, TGF-β Pathway, Survival Pathway, p53 Pathway, Stress Pathway, NF-κB Pathway, NFAT Pathway, CREB Pathway, Jak-Stat Pathway, Estrogen Pathway, Androgen Pathway, Calcium and Protein Kinase C Pathway, Phospholipase C Pathway, Insulin Pathway, LDL Pathway, and Retinoic Acid Pathway) and four housekeeping genes, including GAPDH, cyclophilin A, ribosomal protein L13a, and β-actin. Patient cohorts included paired GC and their adjacent normal samples from 10 patients with wild-type and 10 patients with T889C mutant POLB gene. The expression profiles of GC samples with wild-type and T889C mutant POLB gene were analyzed by the Superarray platform. These data were evaluated using the GEArray Analyzer. Upregulated PR expression was detected in 8 of 10 tumor samples with mutant T889C POLB, while only in 3 of 10 wild-type tumor samples compared with matched normal tissues (P < 0.05). In addition, upregulated CIAP2 (Homo sapiens baculoviral IAP repeat-containing 2) and Krox-24/EGR-1 (Early growth response 1) expression was found in 7 of 10 tumor samples with T889C mutation, while in 3 of 10 for CIAP2 and 4 of 10 for EGR-1 in wild-type tumor samples, compared with matched normal tissues. Upregulated c-Fos gene was identified in 6 of 10 tumor samples with T889C mutation while only in 2 of 10 wild-type tumor samples compared with matched normal tissues (P < 0.05) (Figure 1A, 1B and Supplementary Table 1). Since upregulated PR expression was frequently associated with T889C mutation, we determined to focus on the clinical significance of this association in GC prognosis.
POLB and PR co-expression was expressed more frequently in GC tissue with POLB T889C mutation compared to that without the mutation
To confirm the cDNA array results, 70 GC samples (Table 1) including 18 GC tissues with the mutation and 52 without were subjected to real-time PCR analysis. POLB overexpression was detected in 16 of 18 (88.9%) GC tissues with T889C mutation, and 31 of 52 (59.6 %) GC tissues without the mutation. PR overexpression was observed in 13 of 18 (72.2%) GCs with the mutation whereas in 14 of 52 (26.9 %) GCs without the mutation.
There was a significant positive correlation between T889C point mutation and PR overexpression (P < 0.05). Co-expression of POLB and PR was observed in 13 of 18 (72.2%) GCs with the mutation (including 11 co-overexpression and 2 co-downexpression) whereas in 9 of 52 (17.3%) GCs without the mutation (Figure 1C and 1D).
Co-expression of POLB and PR associated with T889C mutation. A. Superarray Q Series HS-008-12/ HS-008N-12. B. A representative cDNA array results to determine T889C mutation related genes in human GC tissues with and without T889C mutation. Gene expression was compared with matched normal adjacent tissues. Detectable upregulated genes upon normalization were marked in arrows. C. A representative gel-based RT-PCR for POLB and PR mRNA expression in tumor and the matched normal tissue from representative GCs. D. Real-time PCR detection of 11 co-overexpression and 2 co-downexpression of POLB and PR mRNA in GCs with the mutation in tumor (T) vs. matching normal (N).
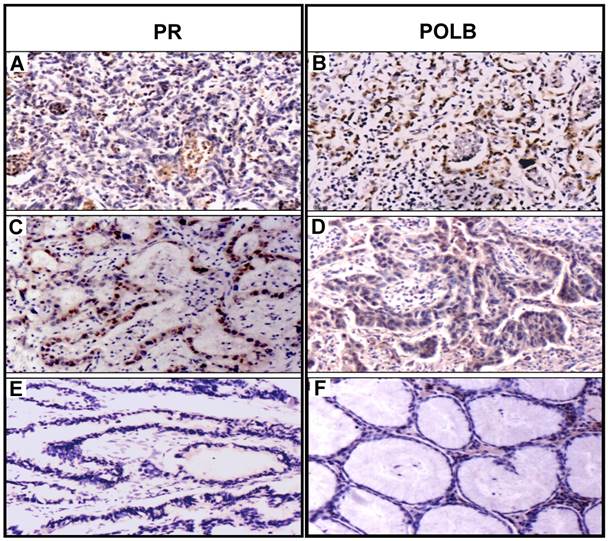
To examine the expression in protein level, we constructed tissue microarrays consisting of GC samples from the 70 patients (Table 1) and 20 normal adjacent tissues (5 matched adjacent normal tissues with the T889C mutation and 15 matched adjacent normal tissues without the mutation). Expression of POLB and PR status was analyzed by immunohistochemistry. The positive hybridization signals of POLB protein were observed in 47 (67.1%) of 70 GCs, including 16 of 18 (88.9%) GC tissues with T889C mutation, of which one was from the 5 matched samples, and 31 of 52 (59.6 %) GC tissues without the mutation, of which four were from the 15 matched samples (Table 2). Positive PR proteins were detected in 30 of 70 (42.9%) GCs, including 13 of 18 (72.2%) GC tissues with T889C mutation and 17 (32.7 %) of 52 GC tissues without the mutation. Co-expression of POLB and PR protein was detected in 10 of 18 (55.6%) GCs with the mutation, including 9 co-positive signals (POLB+/PR+) and one co-negative signal (POLB-/PR-). Co-expression of two proteins was detected in 7 of 52 (13.5%) GCs without the mutation, including 4 POLB+/PR+ and 3 POLB-/PR-, in addition to 3 of 20 (15%) matched adjacent normal tissues (Figure 2 and Table 2). The co-expression of two proteins was significantly different between GCs with- and without- the T889C mutation (p<0.01). Our data suggests that T889C mutation significantly correlated with the co-expression of POLB and PR genes.
Co-expression of POLB and PR genes is associated with intraperitoneal metastasis (IM)
We next investigated whether the co-expression of POLB and PR correlates with clinicopathological parameters, such as gender, age, tumor location, depth of invasion, differentiation and metastasis in the 70 samples from primary GC patients. Of the 70 samples from GC patients, IM are present in 26 (37.1%) of the patients (Table 1). In 17 patients with T889C mutation, IM was presents in 7 of 9 (77.8%) with POLB+/PR+ signals, and in 3 of 8 (37.5%) with POLB+/ PR- or POLB-/ PR+ signals. In 49 patients with wild type POLB, IM was present in 2 of 4 (50%) with POLB+/PR+ signals, 14 of 45 (31.1%) patients with non- coexpression signals. There were significant differences between the patients with coexpression and non-coexpression of POLB and PR signals in regard to IM (p = 0.002, Table 3) in patients with T889C mutation. Thus, co-expression of POLB and PR was more likely associated with increased susceptibility of IM in patients with T889C mutation. Our results suggest that co-expression of POLB and PR plays a role during metastasis of GC with T889C mutation.
Correlation of POLB and PR expression between GC samples with mutant T889C and wild type POLB on tissue arrays.
| Samples | POLB | PR | Co-expression | Total |
|---|---|---|---|---|
| + - | + - | Co+* Co-* +/- | ||
| Tumor | ||||
| Mutation | 16(88.9%) 2 | 13 (72.2%) 5 | 9(50%) 1 8 | 18 |
| Wild type | 31(59.6%) 21 | 17(26.9%) 35 | 4(7.7%) 3 45 | 52 |
| Total | 47 23 | 30 40 | 13 4 53 | 70 |
| Normal | ||||
| Mutation matched | 1(25%) 4 | 1(20%) 4 | 1(20%) 0 4 | 5 |
| Wild type matched | 4(36.3%) 11 | 1(6.7%) 14 | 1(6.7%) 1 13 | 15 |
| Total | 5 15 | 2 18 | 2 1 17 | 20 |
*p < 0.01.
Coexpression of PR and POLB protein in representative GC tissues with T889C mutation. Panels A and B show protein signals stained with PR and POLB antibodies in same GC sample, so do panels C and D for another GC case. Panels E and F show protein signals stained with PR and POLB antibodies in the same matched normal sample.
Coexpression of POLB and PR associated with increased IM.
| Protein signals | POLB+/ PR+ | POLB-/PR- | POLB+/PR- and POLB-/PR+ | Total |
|---|---|---|---|---|
| POLB Mutation | 9 | 1 | 8 | 18 |
| IM | 7 (77.8%)* | 0 | 3 (37.5%) | 10 |
| No IM | 2 (22.2%)* | 1 | 5 (62.5%) | 8 |
| POLB Wild type | 4 | 3 | 45 | 52 |
| IM | 2 (50%) | 0 | 14 (31.1%) | 16 |
| No IM | 2 (50%) | 3 | 31 (68.9%) | 36 |
*p < 0.01.
Transfected T889C mutation elevates PR expression in human GC cell lines
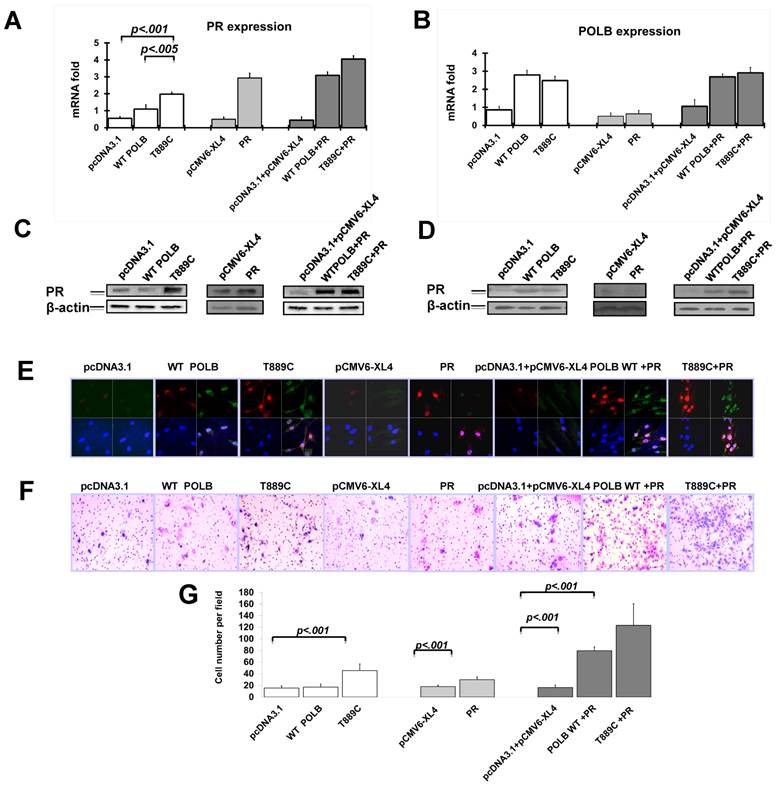
Having established the relationship between T889C mutant POLB and PR gene expression in GC tissues, we tested whether the T889C mutant POLB affects PR levels in vitro. We analyzed PR expression on mRNA level by real time PCR and protein level by Western blot and Immunofluorescence after transfecting plasmids containing T889C mutant POLB or wild type POLB into AGS cells. PR expression was significantly increased in T889C mutant POLB transfected cell lines compared to the cells expressing wild-type POLB (Figure 3A, 3C and 3E). There was no significant change of POLB expression when transfected pCMV6-XL4/PRB PR expression plasmid to AGS cells (Figure 3B, 3D and 3E). These results indicate that the T889C mutant POLB have somehow increased PR expression, though the exact mechanism that is yet to be elucidated.
Co-transfection of T889C mutant POLB and PR promotes cell invasion in human GC cell lines
We have observed the relationship between co-expression of POLB and PR, and increased IM. Since cell migration is a critical property for cancer cell metastasis, we further investigated the cell invasiveness using in vitro invasion assays. Cells co-transfected with T889C mutant POLB and PR showed a significant increase in the number of migrated cells, compared to those co-transfected with wild type POLB and PR, or T889C mutant POLB, or wild type POLB or PR alone (Figure 3F). We found that AGS cell line transfected with mutant POLB plasmid exhibited significant increase of invasion ability compared to the wild-type POLB as well as the empty vector control (p<0.01) (Figure 3G). Moreover, AGS cell line transfected with mutant POLB plasmid along with PR expression plasmid showed more significant invasion compared to the wild-type POLB plasmid with PR expression plasmid (p<0.01) (Figure 3G). These results indicate that T889C mutation alone, or with enhanced PR expression promotes invasion properties of GC cells.
Discussion
Based on our previous studies [14, 18], we predicted that the T889C mutation in POLB gene mediates GC progression by affecting the global gene expression and associated molecular pathways. Thus, our goal was to identify the differentially expressed genes and understand the mechanisms of their differential regulation by comparing wild type and T889C mutant POLB gene in GC tissue and cell line. In this study, we performed targeted gene expression profiling to understand how the T889C mutation in POLB gene altered molecular pathways associated with GC. We identified dramatic gene expression differences at the transcriptional level between wild type and mutant T889C POLB gene carrying GCs. The results were further verified by RT-PCR, real-time PCR and immunofluorescence assays. Interestingly, we identified a T889C mutation-driven gene signature that is associated with significant PR overexpression in human GCs, resulting in increased IM. However, we did not observe a correlation between overexpression of PR and POLB with either age or sex [18]. Additionally, transfection of GC cells expressing only mutant T889C POLB increased PR expression, which further implicated that the T889C mutation upregulated PR expression. Our studies support a mechanistic link between mutant T889C POLB in PR specific action and GC cell metastasis.
Existing data suggest that sex hormonal factors are involved in a variety of cancers including gastric cancer [19]. Progesterone has been found to increase the migration of breast cancer cells [20]. The role of sex hormones in the pathogenesis of cancer is still not clear. Our data suggest that the increased IM in GC associated with mutant T889C POLB may be due to the modulation of PR overexpression, which is related to tumor metastatic pathways. Further studies may shed light on the exact mechanisms of these important findings.
Conclusions
The T889C point mutation in POLB gene is strongly associated with increased PR expression in both GC tissue and cell line. The T889C mutation-associated PR overexpression increased IM in GC. Therefore, detection of T889C mutation-associated PR overexpression may serve as a biomarker for an adverse prognosis of human GC. The exact mechanism of how POLB mutation affects PR expression is yet to be elucidated.
The T889C mutant POLB increase PR expression in human GC cell line, AGS. Cells were transfected with pcDNA3.1, wild type POLB, T889C mutant POLB, PR plasmids and co-transfection of these plasmids into AGS cell line. The expression of PR and POLB was analyzed by real time PCR (A and B), Western blot (C and D) and immunofluorescence (E, red signals are PR, and green signals are POLB). F. The effect on cell invasion by co-transfection of T889C mutant expression vector and PR expression plasmid in human GC cell line, AGS. The Transwell assay with matrigel was performed for the invasion activity of AGS cells transfected with pcDNA3.1, WT POLB, T889C mutant POLB, PR plasmids and co-transfection of these plasmids. Cells co-transfected T889C mutant POLB with PR show a significant increase in the number of migrated cells. Co-transfected wild type POLB and PR show a slightly weak migration, while single transfection of WT POLB, T889C mutant POLB, and PR presents a decreased migration. G. Invasion ability of the cells is displayed as an absolute cell numbers. Results are displayed as mean data ± SE. (**P <0.01). Five fields of unit area on each membrane or whole membrane were counted for cell numbers, and the experiments were repeated at least four times in duplicate.
Supplementary Material
Supplementary table 1.
Abbreviations
GC, Gastric cancer; POLB, DNA polymerase beta; PR, progesterone receptors; IM, intraperitoneal metastasis.
Acknowledgements
We would like to thank individual patients for participating in this study. This work was supported by grants from the National Key Basic Research Program Project of China (Grant# 2004CB518708), the National Natural Science Foundation of China (NSFC) (Grant# 39625016), and the National Bio-Tech 863 Program (Grant# 2012-AA02A203) to YL, and the Elaine H. Snyder Cancer Research Award to SWF.
Authors' contributions
XT contributed to conception and design, development of methodology, acquisition of data, analysis and interpretation of data, statistical analyses, writing of the manuscript. XW performed microarray data analysis and interpretation, and edited the manuscript. SR performed qRT-PCR and matrigel invasion assays and helped write/revise the manuscript. HY was involved in patient's accrual and performed surgery. ZL identified pathological cases with clinical information. WA performed array data analysis. WL and JC involved in cell culture and matrigel invasion assays. GL designed and interpreted data and helped write the manuscript. RSS and SWF analyzed data, and revised the manuscript. YL designed and interpreted data, and wrote and revised the manuscript. All authors read and approved the final version of this manuscript.
Competing interest
The authors declare that they have no competing interests.
References
1. Al-Marzoqee FY, Khoder G, Al-Awadhi H, John R, Beg A, Vincze A. et al. Upregulation and inhibition of the nuclear translocation of Oct4 during multistep gastric carcinogenesis. Int J Oncol. 2012;41:1733-43 doi:10.3892/ijo.2012.1608
2. Tokunaga A NK, Matsukura N, Tanaka N, Onda M, Shirota A, Asano G, Hayashi K. Estrogen and progesterone receptors in gastric cancer. Cancer. 1986;57:1376-9
3. Brambilla G MA. Are some progestins genotoxic liver carcinogens. Mutat Res. 2002;12:155-63
4. Oshima CT WD, Catarino RM, Mattos D, Forones NM. Estrogen and progesterone receptors in gastric and colorectal cancer. Hepatogastroenterology. 1999;46:3155-8
5. Knutson TP, Daniel AR, Fan D, Silverstein KA, Covington KR, Fuqua SA. et al. Phosphorylated and sumoylation-deficient progesterone receptors drive proliferative gene signatures during breast cancer progression. Breast cancer research: BCR. 2012;14:R95. doi:10.1186/bcr3211
6. Masaoka A, Horton JK, Beard WA, Wilson SH. DNA polymerase beta and PARP activities in base excision repair in living cells. DNA Repair (Amst). 2009;8:1290-9 doi:10.1016/j.dnarep.2009.08.004
7. Cağlayan M BV, Sassa A, Prasad R, Wilson SH. Role of polymerase β in complementing aprataxin deficiency during abasic-site base excision repair. Nat Struct Mol Biol. 2014;21:497-9
8. Prasad R WJ, Hou EW, Wilson SH. Pol β associated complex and base excision repair factors in mouse fibroblasts. Nucleic Acids Res. 2012;40:11571-82
9. Mark WY, Liao JC, Lu Y, Ayed A, Laister R, Szymczyna B. et al. Characterization of segments from the central region of BRCA1: an intrinsically disordered scaffold for multiple protein-protein and protein-DNA interactions? J Mol Biol. 2005;345:275-87 doi:10.1016/j.jmb.2004.10.045
10. Starcevic D, Dalal S, Sweasy JB. Is there a link between DNA polymerase beta and cancer? Cell Cycle. 2004;3:998-1001 doi:1062 [pii]
11. Khanra K, Panda K, Bhattacharya C, Mitra AK, Sarkar R, Banerjee S. et al. Association between newly identified variant form of DNA polymerase beta(Delta 208-304) and ovarian cancer. Cancer Biomark. 2012;11:155-60 doi:Doi 10.3233/Cbm-2012-0275
12. Sweasy JB LT, Starcevic D, Sun KW, Lai CC, Dimaio D, Dalal S. Expression of DNA polymerase β cancer-associated variants in mouse cells results in cellular transformation. Proc Natl Acad Sci U S A. 2005;102:14350-5
13. Donigan KA HS, Eckert KA, Sweasy JB. The human gastric cancer-associated DNA polymerase β variant D160N is a mutator that induces cellular transformation. DNA repair. 2012;11:381-90
14. Tan XH, Zhao M, Pan KF, Dong Y, Dong B, Feng GJ. et al. Frequent mutation related with overexpression of DNA polymerase beta in primary tumors and precancerous lesions of human stomach. Cancer letters. 2005;220:101-14 doi:10.1016/j.canlet.2004.07.049
15. Yang YH, Deng H, Li WM, Zhang QY, Hu XT, Xiao B. et al. Identification of matrix metalloproteinase 11 as a predictive tumor marker in serum based on gene expression profiling. Clin Cancer Res. 2008;14:74-81 doi:Doi 10.1158/1078-0432.Ccr-07-1179
16. Tan X, Anzick SL, Khan SG, Ueda T, Stone G, Digiovanna JJ. et al. Chimeric negative regulation of p14ARF and TBX1 by a t(9;22) translocation associated with melanoma, deafness, and DNA repair deficiency. Hum Mutat. 2013;34:1250-9 doi:10.1002/humu.22354
17. Tan X, Peng J, Fu Y, An S, Rezaei K, Tabbara S. et al. miR-638 mediated regulation of BRCA1 affects DNA repair and sensitivity to UV and cisplatin in triple negative breast cancer. Breast cancer research: BCR. 2014;16:435. doi:10.1186/s13058-014-0435-5
18. Tan X, Wang H, Luo G, Ren S, Li W, Cui J. et al. Clinical significance of a point mutation in DNA polymerase beta (POLB) gene in gastric cancer. Int J Biol Sci. 2015;11:144-55 doi:10.7150/ijbs.10692
19. Camargo MC, Goto Y, Zabaleta J, Morgan DR, Correa P, Rabkin CS. Sex hormones, hormonal interventions, and gastric cancer risk: a meta-analysis. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012;21:20-38 doi:10.1158/1055-9965.EPI-11-0834
20. Diaz J, Aranda E, Henriquez S, Quezada M, Espinoza E, Bravo ML. et al. Progesterone promotes focal adhesion formation and migration in breast cancer cells through induction of protease-activated receptor-1. J Endocrinol. 2012;214:165-75 doi:Doi 10.1530/Joe-11-0310
Author contact
![]() Corresponding authors: Sidney W. Fu, sfuedu or Youyong Lu , youyonglucom.
Corresponding authors: Sidney W. Fu, sfuedu or Youyong Lu , youyonglucom.

 Global reach, higher impact
Global reach, higher impact