3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(5):852-860. doi:10.7150/jca.17845 This issue Cite
Research Paper
NR1H3 Expression is a Prognostic Factor of Overall Survival for Patients with Muscle-Invasive Bladder Cancer
1. Department of Urology, Fudan University Shanghai Cancer Center, Shanghai, 200032, China;
2. Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, China.
* These authors contributed equally to this work.
Received 2016-10-8; Accepted 2016-12-22; Published 2017-3-7
Abstract
Background: Nuclear receptors (NRs) are a class of transcription factors that regulate many cellular functions through manipulation of gene expression and also play important roles in tumorigenesis, proliferation, progression and prognosis in various kinds of cancers according to recent studies. This work aimed to determine the predictive ability of NRs in muscle-invasive bladder cancer (MIBC).
Patients and methods: A total of 308 MIBC patients with complete clinicopathological and RNASeq data from The Cancer Genome Atlas (TCGA) cohort were collected for filtration. Genes showed clear correlations with overall survival (OS) and recurrence free survival (RFS) were further validated in 123 MIBC patients recruited consecutively from 2008 to 2012 in Fudan University Shanghai Cancer Center (FUSCC) cohort. Cox proportional hazards regression model and Kaplan-Meier plot were used to assess the relative factors.
Results: In TCGA cohort, we found that high NR1H3 (HR=0.779, 95% CI: 0.634 - 0.957), NR2C1 (HR=0.673, 95% CI: 0.458 - 0.989) and NR2F6 (HR=0.750, 95% CI: 0.574 - 0.980) expressions were independent factors of favorable OS, while only low NR1H3 (log-rank test, P=0.0076) and NR2F6 (log-rank test, P=0.0395) expressions had the ability to predict poor prognosis for RFS. Further, in FUSCC validating cohort, we confirmed that low NR1H3 expression level was independent factor of poor OS (HR=1.295, 95% CI: 1.064 - 1.576) and it had the ability to predict poor RFS (log-rank test, P=0.0059).
Conclusions: Low NR1H3 expression level is an independent prognostic factor of poor OS, and can also predict worse RFS in MIBC patients. Our “TCGA filtrating and local database validating” model can help reveal more prognostic biomarkers and cast a new light in understanding certain gene function in MIBC.
Keywords: Nuclear receptor, NR1H3, muscle-invasive bladder cancer, overall survival
Introduction
Bladder cancer is the seventh most common cancer in the world [1] and is the most common urologic malignancy in China [2]. As newly estimated, there were 80,500 new cases in China in 2015, including 62,100 men and 18,400 women [3]. Bladder cancer is staged via the tumor-node-metastasis system, which describes the extent of invasion (Tis-T4) [4]. According to the 7th edition of the TNM classification published by the UICC, bladder cancer can be divided into non-muscle invasive bladder cancer and muscle invasive bladder cancer (stage T2 or above) [5]. Approximately 30% of patients suffer MIBC at the time of diagnosis and they have a less favorable prognosis with 5-year survival rate less than 50% [6, 7]. Despite improvements in surgical techniques and post-operative recovery, the complexity and high cost of procedure remains highly challenging [8], and the treatment has not been advanced for several decades [9]. Therefore, to further understand the prognosis and to develop new biological therapies, knowledge of novel biomarkers for MIBC is in an urgent need.
Nuclear receptors are a class of proteins that are responsible for sensing steroid and thyroid hormones and certain other molecules. These proteins can work with other molecules to regulate expression of specific genes. In humans, there are 48 kinds of NRs [10], which can be classified according to mechanism [11] or homolog [12]. Members of NR superfamily have the ability to regulate the development, homeostasis and metabolism, thus are correlated with a variety of diseases including cancer. Till today, many kinds of NRs are proved to be closely associated with the prognosis of cancer, including breast cancer [13, 14], gastric cancer [15], colon cancer [16], prostate cancer [17, 18], ovarian cancer [19, 20] and so many others. However, the potential roles of NRs in BCa, especially in MIBC, and their biological functions on tumorigenesis, recurrence and prognosis remain unknown. So in this study, we aim to find out if certain NR gene expression level is associated with patients' recurrence or prognosis with MIBC, and has the potential to serve as a novel biomarker.
Materials and methods
Patients and samples
Our study was carried out in accordance with the ethical standards of Helsinki Declaration II and approved by the Institution Review Board of Fudan University Shanghai Cancer Center. Written informed consent was obtained from each patient before any study-specific investigation was performed.
For TCGA cohort, patients' nuclear receptor gene expression and clinical information were downloaded from the website of Cancer Genomics Browser of University of California Santa Cruz (http://genome-cancer.ucsc.edu/). Forty-eight members of nuclear receptor gene family are included in the database as is presented in Supplementary Table 1. Originally, 436 lines' data was obtained. Then, we excluded patients with unclear event, age, gender, tumor subtype, tumor grade, stage and no RNASeq data, along with patients with a T stage of T0 or T1. After all, we included 308 patients of muscle-invasive bladder cancer from TCGA dataset.
For the FUSCC cohort, 123 muscle-invasive bladder cancer patients from 2008-2012 who underwent radical cystectomy were enrolled retrospectively and consecutively. All these tissue samples were collected during surgeries and stored at -70℃ in the tissue bank of FUSCC. Clinicopathological characteristics were obtained from our electronic records. Patients were regularly followed up by telephone or in the clinic once every 3 months. Events, including death and recurrence, were recorded.
Patient demographic and clinical characteristics
| Characteristics | TCGA cohort (N=308) | FUSCC cohort (N=123) | P value | ||
|---|---|---|---|---|---|
| N | % | N | % | ||
| Age, median (range) | 70.0 | 38 - 90 | 61.0 | 25 - 78 | 0.010a |
| Gender | 0.016b | ||||
| Male | 224 | 72.7 | 103 | 83.7 | |
| Female | 84 | 27.3 | 20 | 16.3 | |
| Tumor subtype | |||||
| Papillary | 93 | 30.2 | - | - | |
| Non-papillary | 215 | 69.8 | - | - | |
| pT stage | <0.001b | ||||
| T2 | 101 | 32.8 | 76 | 61.8 | |
| T3 | 159 | 51.6 | 34 | 27.6 | |
| T4 | 48 | 15.6 | 13 | 10.6 | |
| pN stage | <0.001c | ||||
| N0 | 188 | 61.0 | 100 | 81.3 | |
| N1 | 35 | 11.4 | 10 | 8.1 | |
| N2 | 63 | 20.5 | 12 | 9.8 | |
| N3 | 6 | 1.9 | 1 | 0.8 | |
| Nx | 16 | 5.2 | 0 | 0 | |
| pM stage | |||||
| M0 | 146 | 47.4 | 123 | 100 | |
| M1 | 4 | 1.3 | - | - | |
| Mx | 158 | 51.3 | - | - | |
| Pathological stage | |||||
| Stage II | 87 | 28.2 | - | - | |
| Stage III | 115 | 37.3 | - | - | |
| Stage IV | 106 | 34.4 | - | - | |
| Histological grade | 0.020b | ||||
| Low grade | 14 | 4.5 | 13 | 10.6 | |
| High grade | 294 | 95.5 | 110 | 89.4 | |
| Family history of cancer | |||||
| Yes | 107 | 34.7 | - | - | |
| No | 201 | 65.3 | - | - | |
| Smoking status | |||||
| Yes | 212 | 68.8 | - | - | |
| No | 86 | 27.9 | - | - | |
| Not mentioned | 10 | 3.2 | - | - | |
| Adjuvant chemotherapy | |||||
| Yes | - | - | 46 | 37.4 | |
| No | - | - | 77 | 62.6 | |
| Patient group | |||||
| Initial case | - | - | 90 | 73.2 | |
| Relapse case | - | - | 33 | 26.8 | |
a. Student's t test.
b. Chi-square test.
c. Chi-square test, N stage as a binary variable: No lymph node involvement versus otherwise.
RNA extraction, reverse transcription and qRT PCR analysis
In the FUSCC cohort, total RNA of tumor tissue was isolated from all 123 patients' samples using TRIzol reagent (15596-026, Invitrogen). A PrimeScript RT reagent kit (K1622, Thermo Scientific) was used to synthesize first-strand cDNA from total RNA. After that, we performed SYBR Green real-time PCR analysis using ABI 7900HT machine (Applied Biosystems, USA). We used β-actin mRNA expression level as a reference. NR1H3 and NR2F6 mRNA expression level was normalized to β-actin [21]. The primers for qRT-PCR analysis were synthesized by Sangon (Shanghai, P.R. China), sequences of which are shown as following:
NR1H3: Forward primer: 5'-CCTTCAGAACCCACAGAGATCC-3'; Reverse primer: 5'-ACGCTGCATAGCTCGTTCC-3'
NR2F6: Forward primer: 5'-GAGCGGCAAGCATTACGGT-3'; Reverse primer: 5'-GGCAGGTGTAGCTGAGGTT-3'
β-actin: Forward primer: 5'-CATGTACGTTGCTATCCAGGC-3'; Reverse primer: 5'-CTCCTTAATGTCACGCACGAT-3'
Statistical analysis
Overall survival was calculated from the date of surgery to the date of death or last follow-up. Recurrence-free survival was defined as the time from the date of surgery to the date of first recurrence or last follow-up. Patients without events or death were recorded as censored at the time of last follow-up. Gender, tumor subtype (papillary vs non-papillary), histological grade (high vs low), family history, adjuvant chemotherapy and patient group (initial vs relapse) were considered binary variables and presented as proportions. Pathological stage, pT stage, pN stage, pM stage and smoking status were considered multiple categorical variables and shown as proportions. Age was deemed to be continuous variable and reported in the form of a median (range). Gene expression level from TCGA database, NR1H3 △Ct, NR2F6 △Ct were also deemed to be continuous variables. The higher the △Ct value, the lower the gene expression level. NR1H3 △Ct and NR2F6 △Ct were calculated using equations below [22]:
NR1H3 △Ct = Ct (NR1H3)-Ct (β-actin) (1)
NR2F6 △Ct = Ct (NR2F6)-Ct (β-actin) (2)
We used Kaplan-Meier method to construct survival curves and used log-rank tests to assess differences between groups. For continuous variables, we used X-tile 3.6.1 (Yale University, New Haven, CT, USA) to determine the best cutoff value for subgroups. Adjusted hazard ratio (HR) was calculated using Cox proportional hazards models. Univariate and multivariate Cox analysis for patients with muscle-invasive bladder cancer in TCGA database and FUSCC cohort were performed. Normally distributed data were compared using Student's t test. The Chi-square test was used to compare the distribution of categorical data between groups. All tests were two-tailed and P values less than 0.05 were considered to be statistically significant. SPSS software, version 22.0 (SPSS Inc., Chicago, IL, USA) were used for data analysis.
Results
Clinical characteristics of patients in TCGA and FUSCC database
In TCGA cohort, the median age of patients with muscle-invasive bladder cancer was 70.0 years old, ranging from 38 to 90 years old. In this database, 224 patients were male (72.7%) while 84 patients were female (27.3%). Information of TNM stage, tumor subtype, family history, smoking status and so on was recorded in Table 1. The median follow-up time was 14.5 months and 115 patients died during follow-up.
In FUSCC database, the median age of 123 patients who underwent radical cystectomy was 61.0 years old, with a range from 25 to 78 years old. One hundred and three patients were men (83.7%) while twenty patients were women (16.3%). Records of TNM stage, adjuvant chemotherapy information, histological grade and patient catalog were also presented in Table 1, along with comparison of clinical characteristics between TCGA and FUSCC cohort. The median follow-up time in FUSCC cohort was 53.1 months and 42 people died.
NR1H3, NR2C1 and NR2F6 expression were independent prognostic factors of OS in TCGA cohort
To examine which nuclear receptor genes have prognostic value for predicting overall survival in muscle-invasive bladder cancer patients, we used TCGA cohort to do the filtration. Clinical pathological data and 48 nuclear receptor genes' expression profile were included in Cox proportion hazard ratio analysis. In univariate analysis, we found that age, pT stage, pN stage, pathological stage, ESRRA, NR1H3, NR1H4, NR2C1, NR2F6 and THRB expression level were significantly associated with overall survival in patients with bladder cancer in TCGA cohort (Table 2). Then, we performed reduced model of multivariate Cox analysis, only including variables with a statistical P value less than 0.1 in univariate analysis, to find independent prognostic factors for predicting OS. After adjustment, multivariate Cox regression for potential prognostic factors indicated that age (HR=1.043, 95% CI: 1.022 - 1.065), pT stage (HR=1.433, 95% CI: 1.061 - 1.936), pN stage (HR=2.007, 95% CI: 1.365 - 2.950), NR1H3 expression (HR=0.779, 95% CI: 0.634 - 0.957), NR2C1 expression (HR=0.673, 95% CI: 0.458 - 0.989) and NR2F6 expression (HR=0.750, 95% CI: 0.574 - 0.980) were independent prognostic factors for OS of patients with muscle-invasive bladder cancer in TCGA cohort (All P<0.05, shown in Table 2).
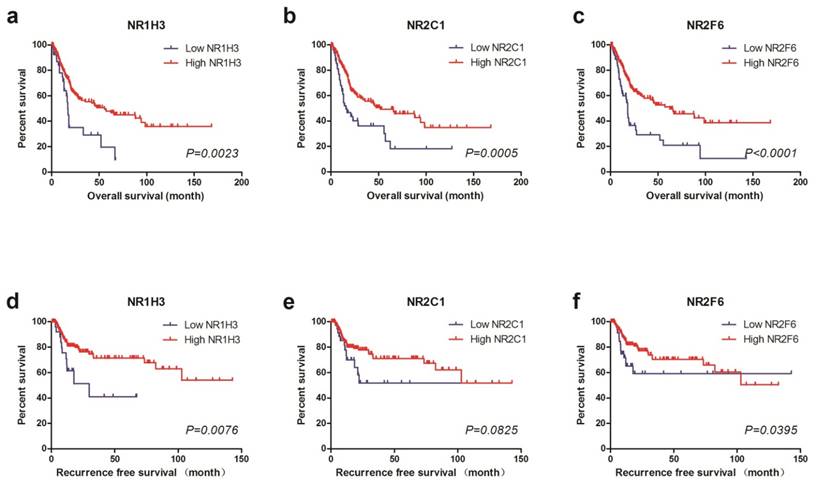
For further study, we divided TCGA cohort into low expression and high expression groups by using X-tile to find out the best cutoff values for OS of NR1H3, NR2C1 and NR2F6 expression (Supplementary Figure 1). We also used the same cutoff value of OS as the cutoff of RFS. Thus, we found that low NR1H3 expression (log-rank test, P=0.0023), low NR2C1 expression (log-rank test, P=0.0005) and low NR2F6 expression (log-rank test, P<0.0001) were all significantly associated with poor prognosis for OS. However, by performing log-rank test for RFS, we found that only low NR1H3 expression (log-rank test, P=0.0076) and low NR2F6 expression (log-rank test, P=0.0395) were associated with poor prognosis for RFS. There's no obvious difference of RFS between high and low NR2C1 expression groups (log-rank test, P=0.0825) (Figure 1).
Kaplan-Meier plots of survival in TCGA cohort are shown according to NR1H3, NR2C1 and NR2F6 expression. a, b, c Kaplan-Meier estimates of overall survival (OS) are shown according to NR1H3, NR2C1 and NR2F6 expression. d, e, f Kaplan-Meier estimates of recurrence free survival (RFS) are shown according to NR1H3, NR2C1 and NR2F6 expression.
Univariate and multivariate Cox proportional hazards analysis of nuclear receptor gene expression and overall survival for patients with bladder cancer in TCGA cohort
| Variable | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Age | 1.041 | 1.020 - 1.062 | <0.001 | 1.043 | 1.022 - 1.065 | <0.001 |
| Gender | 1.067 | 0.715 - 1.594 | 0.750 | |||
| Tumor subtype | 0.661 | 0.415 -1.054 | 0.082 | 0.933 | 0.571 - 1.527 | 0.784 |
| pT stage | 1.652 | 1.252 - 2.179 | <0.001 | 1.433 | 1.061 - 1.936 | 0.019 |
| pN stagea | 2.437 | 1.679 - 3.537 | <0.001 | 2.007 | 1.365 - 2.950 | <0.001 |
| pM stagea | 1.443 | 0.995 - 2.092 | 0.053 | 1.290 | 0.882 - 1.887 | 0.189 |
| Pathological stage | 1.972 | 1.524 - 2.550 | <0.001 | 1.221 | 0.748 - 1.991 | 0.424 |
| Histological grade | 21.658 | 0.146 - 3216.531 | 0.228 | |||
| Family history | 0.780 | 0.531 - 1.147 | 0.207 | |||
| Smoking statusa | 1.265 | 0.833 - 1.921 | 0.270 | |||
| AR | 0.984 | 0.912 - 1.062 | 0.679 | |||
| ESR1 | 1.047 | 0.952 - 1.152 | 0.346 | |||
| ESR2 | 0.853 | 0.717 - 1.015 | 0.074 | 0.860 | 0.715 - 1.035 | 0.110 |
| ESRRA | 0.709 | 0.519 - 0.968 | 0.030 | 0.906 | 0.627 - 1.310 | 0.600 |
| ESRRB | 0.875 | 0.735 - 1.040 | 0.131 | |||
| ESRRG | 0.945 | 0.867 - 1.029 | 0.191 | |||
| HNF4A | 0.989 | 0.884 - 1.105 | 0.840 | |||
| HNF4G | 0.939 | 0.873 - 1.009 | 0.088 | 0.988 | 0.912 - 1.070 | 0.760 |
| NR0B1 | 1.057 | 0.967 - 1.156 | 0.220 | |||
| NR0B2 | 0.966 | 0.778 - 1.200 | 0.758 | |||
| NR1D1 | 1.123 | 0.933 - 1.352 | 0.218 | |||
| NR1D2 | 0.883 | 0.677 - 1.152 | 0.360 | |||
| NR1H2 | 0.723 | 0.472 - 1.109 | 0.137 | |||
| NR1H3 | 0.784 | 0.641 - 0.958 | 0.018 | 0.779 | 0.634 - 0.957 | 0.018 |
| NR1H4 | 0.926 | 0.860 - 0.997 | 0.041 | 0.962 | 0.879 - 1.053 | 0.405 |
| NR1I2 | 0.956 | 0.825 - 1.108 | 0.549 | |||
| NR1I3 | 0.928 | 0.790 - 1.090 | 0.362 | |||
| NR2C1 | 0.522 | 0.365 - 0.745 | <0.001 | 0.673 | 0.458 - 0.989 | 0.044 |
| NR2C2 | 0.924 | 0.758 - 1.125 | 0.429 | |||
| NR2E1 | 1.020 | 0.925 - 1.124 | 0.694 | |||
| NR2E3 | 0.874 | 0.709 - 1.077 | 0.205 | |||
| NR2F1 | 1.111 | 0.980 - 1.259 | 0.099 | 1.085 | 0.963 - 1.223 | 0.181 |
| NR2F2 | 1.036 | 0.859 - 1.249 | 0.713 | |||
| NR2F6 | 0.713 | 0.565 - 0.900 | 0.004 | 0.750 | 0.574 - 0.980 | 0.035 |
| NR3C1 | 1.115 | 0.952 - 1.307 | 0.178 | |||
| NR3C2 | 1.028 | 0.933 - 1.133 | 0.581 | |||
| NR4A1 | 1.012 | 0.903 - 1.135 | 0.834 | |||
| NR4A2 | 0.982 | 0.857 - 1.124 | 0.789 | |||
| NR4A3 | 1.024 | 0.929 - 1.128 | 0.634 | |||
| NR5A1 | 1.067 | 0.971 - 1.174 | 0.177 | |||
| NR5A2 | 0.917 | 0.775 - 1.085 | 0.314 | |||
| NR6A1 | 1.063 | 0.892 - 1.266 | 0.497 | |||
| PGR | 1.079 | 0.959 - 1.213 | 0.208 | |||
| PPARA | 1.013 | 0.811 - 1.264 | 0.911 | |||
| PPARD | 0.918 | 0.685 - 1.231 | 0.568 | |||
| PPARG | 0.928 | 0.861 - 1.001 | 0.053 | 1.013 | 0.891 - 1.151 | 0.847 |
| RARA | 0.994 | 0.741 - 1.335 | 0.970 | |||
| RARB | 1.022 | 0.916 - 1.141 | 0.694 | |||
| RARG | 0.855 | 0.714 - 1.024 | 0.089 | 0.897 | 0.739 - 1.089 | 0.273 |
| RORA | 1.033 | 0.898 - 1.190 | 0.647 | |||
| RORB | 1.014 | 0.892 - 1.153 | 0.826 | |||
| RORC | 0.946 | 0.881 - 1.016 | 0.126 | |||
| RXRA | 0.907 | 0.723 - 1.137 | 0.397 | |||
| RXRB | 0.879 | 0.564 - 1.371 | 0.571 | |||
| RXRG | 1.024 | 0.883 - 1.188 | 0.750 | |||
| THRA | 1.160 | 0.958 - 1.405 | 0.127 | |||
| THRB | 0.897 | 0.805 - 0.998 | 0.046 | 0.987 | 0.864 - 1.127 | 0.843 |
| VDR | 0.914 | 0.778 - 1.074 | 0.274 | |||
Binary variables: No lymph node involvement versus otherwise; Non-metastasis versus otherwise; Non-smoker versus otherwise.
NR1H3 expression was an independent prognostic factor of OS in FUSCC cohort
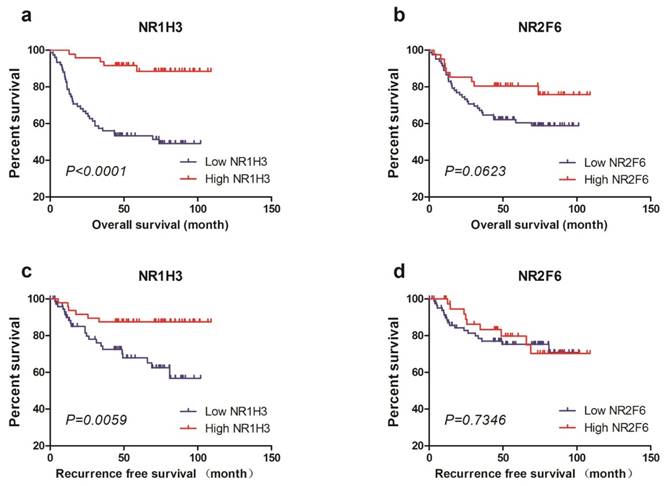
To validate the predictive value of NR1H3 and NR2F6 expression for OS and RFS in muscle-invasive bladder cancer patients, we tested tumor NR1H3 and NR2F6 expression levels in FUSCC cohort and performed statistical analysis. We used X-tile to find out the best cutoff values for OS of NR1H3 and NR2F6 expression level (△Ct) and divided patients into low expression and high expression groups (Supplementary Figure 2). Also we used the same cutoff value of OS as the cutoff of RFS. Firstly, we used Kaplan-Meier method to perform survival analysis and found that low NR1H3 expression was significantly associated with poor prognosis for both OS (log-rank test, P<0.0001) and RFS (log-rank test, P=0.0059). However, low NR2F6 expression level was relatively associated with poor prognosis for OS with a borderline significance (log-rank test, P=0.0623). There was no difference of RFS between high NR2F6 and low NR2F6 expression groups (Figure 2).
Furthermore, we performed univariate and multivariate Cox analysis to investigate if NR1H3 or NR2F6 gene expression was an independent prognostic factor of OS in FUSCC cohort. In univariate Cox model, patients who received adjuvant chemotherapy (HR=1.957, 95% CI: 1.067 - 3.587) and patients with low NR1H3 expression level (HR=1.312, 95% CI: 1.090 - 1.580) had significantly poor prognosis. However, in multivariate Cox analysis, only low NR1H3 expression level remained the significance (HR=1.295, 95% CI: 1.064 - 1.576) (Table 3). The same regression analysis was performed to determine whether gene expression was independent factor of RFS in muscle-invasive bladder cancer patients. It showed that only low NR1H3 expression level was significantly associated with poor prognosis of RFS in univariate Cox analysis (HR=1.259, 95% CI: 1.008 - 1.572). The statistical significance vanished in multivariate Cox regression model (Table 4).
As low NR1H3 expression was found to be tightly correlated with overall survival in MIBC patients, we also evaluated if patients with low NR1H3 expression can benefit from adjuvant chemotherapy. However, it turned out that there was no significant benefit from adjuvant chemotherapy in patients with low NR1H3 expression (P=0.7627, Supplementary Figure 3).
Kaplan-Meier plots of survival in FUSCC cohort are shown according to NR1H3 and NR2F6 expression. a, b Kaplan-Meier estimates of overall survival (OS) are shown according to NR1H3 and NR2F6 expression. c, d Kaplan-Meier estimates of recurrence free survival (RFS) are shown according to NR1H3 and NR2F6 expression.
Univariate and multivariate Cox proportional hazards analysis of overall survival for patients with bladder cancer in FUSCC cohort.
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Age | 1.028 | 0.990 - 1.067 | 0.146 | 1.029 | 0.990 - 1.070 | 0.147 |
| Gender | 0.474 | 0.169 - 1.331 | 0.156 | 0.499 | 0.177 - 1.407 | 0.189 |
| pT stage | 1.452 | 0.967 - 2.181 | 0.072 | 1.125 | 0.702 - 1.803 | 0.624 |
| pN stagea | 1.575 | 0.774 - 3.204 | 0.210 | 1.783 | 0.849 - 3.742 | 0.126 |
| Histological grade | 1.648 | 0.509 - 5.336 | 0.404 | 1.376 | 0.393 - 4.812 | 0.617 |
| Adjuvant chemotherapy | 1.957 | 1.067 - 3.587 | 0.030 | 1.754 | 0.955 - 3.222 | 0.070 |
| Patient group | 1.069 | 0.547 - 2.088 | 0.846 | 1.006 | 0.474 - 2.135 | 0.987 |
| NR1H3 △Ct | 1.312 | 1.090 - 1.580 | 0.004 | 1.295 | 1.064 - 1.576 | 0.010 |
| NR2F6 △Ct | 0.921 | 0.766 - 1.107 | 0.381 | 0.876 | 0.720 - 1.067 | 0.189 |
a. Binary variable: No lymph node involvement versus otherwise
Univariate and multivariate Cox proportional hazards analysis of recurrence-free survival for patients with bladder cancer in FUSCC cohort.
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Age | 1.002 | 0.962 - 1.044 | 0.917 | 0.996 | 0.951 - 1.044 | 0.877 |
| Gender | 0.157 | 0.021 - 1.160 | 0.070 | 0.144 | 0.020 - 1.068 | 0.058 |
| pT stage | 1.275 | 0.775- 2.098 | 0.339 | 0.966 | 0.527 - 1.771 | 0.910 |
| pN stagea | 1.423 | 0.576 - 3.511 | 0.445 | 1.912 | 0.756 - 4.833 | 0.171 |
| Histological grade | 1.720 | 0.408 - 7.253 | 0.460 | 1.133 | 0.250 - 5.141 | 0.871 |
| Adjuvant chemotherapy | 1.808 | 0.861 - 3.795 | 0.117 | 1.967 | 0.936 - 4.133 | 0.074 |
| Patient group | 0.631 | 0.256 - 1.557 | 0.318 | 0.643 | 0.242 - 1.706 | 0.375 |
| NR1H3 △Ct | 1.259 | 1.008 - 1.572 | 0.042 | 1.218 | 0.944 - 1.572 | 0.129 |
| NR2F6 △Ct | 0.924 | 0.775 - 1.101 | 0.375 | 0.909 | 0.750 - 1.103 | 0.333 |
Discussion
In our study, we were the first to demonstrate that NR superfamily gene expression was correlated with prognosis of patients with MIBC, in both OS and RFS. In TCGA filtrating cohort, we found that low NR1H3, NR2C1 and NR2F6 expressions were independent factors of poor OS, while only low NR1H3 and NR2F6 expressions had the ability to predict poor prognosis for RFS. Further, in our FUSCC validating cohort, we confirmed that low NR1H3 expression level was independent factor of poor OS and it had the ability to predict poor RFS. However, NR2F6 failed to maintain statistical significance in predicting prognosis of both OS and RFS.
Nuclear receptors are one of the largest classes of transcription factors and regulate many cellular functions through manipulation of gene expression [23]. They play important roles in many physiological and pathological processes [24]. Liver X receptor alpha (LXRα) is a nuclear receptor encoded by NR1H3 gene, and forms a subfamily of the NR superfamily along with liver X receptor β. Previously understood, LXRα was a key regulator of macrophage function [25] and lipid homeostasis [26-28]. But recent researches revealed that LXRα was associated with various cancer types. Savic D et al found that activation of LXRα could inhibit colorectal cancer cell proliferation and decrease glutathione levels, consistent with increased cellular oxidative stress [29, 30]. In breast cancer, some researchers declared that NR1H3 was likely to be an onco-suppressor gene [31, 32]. Similar results were also achieved in prostate cancer [33], gallbladder cancer [34], ovarian carcinoma [35] and so on. All the publications mentioned above illustrated that NR1H3 might be an onco-suppressor gene in various cancer types, which was in line with the unfavorable prognosis in MIBC patients with low NR1H3 expression level found in our investigation.
LXRs were closely related to cholesterol metabolism in cancer. LXR activation was absolutely a hot-spot in cancer research and was a potential bona fide therapeutic strategy. As reported, tumor-related proliferation status was associated with enhanced cholesterol requirement which is satisfied not only by up regulating de novo biosynthesis but also by increasing cholesterol uptake via LDLR. Furthermore, in cancer cells, despite the intracellular cholesterol abundance, LXR transcriptome was also down regulated [36]. In most studies, LXR activation promoted cholesterol removal from cells by increasing expression of membrane transporters ABCA1, ABCG5, ABCG8 and ABCG1 [37], and inhibited uptake via LDLR by inducing IDOL-mediated LDLR degradation [38], to inhibit cancer cell proliferation. Besides, some groups reported that activation of LXR can play an anti-tumor role through other mechanisms, like altering microenvironment by activating marcophages [39], disrupting key growth pathways by suppressing β-catenin signaling [40] or activating apoptotic processes through caspase pathway [41]. Overall, LXRs play important role in tumor suppression through various mechanisms. Our study was the first to link LXRα to prognosis of MIBC and casted a new light in therapeutic potential of LXRα in MIBC.
Although adjuvant chemotherapy in MIBC remains controversial and many powerful guidelines, like EAU [42] and NCCN [43] guidelines, are very restrictive with regard to its recommendation, adjuvant chemotherapy in a multimodal strategy will be probably used frequently in the future in bladder cancer treatment due to recent findings [44]. In our study, we found that MIBC patients with low NR1H3 expression not only had worse prognosis, but showed resistance to adjuvant chemotherapy as well. It indicated that maybe we should utilize other managing strategies instead of chemotherapy in those patients and a target therapy for those patients was in an urgent need. Indeed, subsequent well-designed large-cohort studies should be performed to confirm our finding. Response to neoadjuvant chemotherapy should also be evaluated in the future if possible.
A major strength of this study is that our data was got from two large populations with a relatively long follow-up, but there are certainly some limitations. Firstly, the limitations include the retrospective nature of this study and the selected bias of patients in FUSCC cohort, which from a single cancer center. Secondly, the prognosis of MIBC is affected by many factors, such as tumor stage, surgical performance, response to adjuvant chemotherapy and genetic background, so a single biological biomarker is not enough. Thirdly, qRT-PCR technique only measures the mRNA expression level of certain gene, not referring to protein level. So in the future, we will expand our cohort to multicenter cohorts for further validation, and perform immunohistochemistry or western blotting to measure protein expression level in tumor tissues. Furthermore, research in cell lines and deep molecular mechanism should be explored to make a step forward to potential application.
In conclusion, low NR1H3 expression level is an independent prognostic factor of poor OS, and can also predict worse RFS in MIBC patients. Our “TCGA filtrating and local database validating” model can help reveal more prognostic biomarkers of MIBC and cast a new light in understanding certain gene function in MIBC.
Abbreviations
NR: nuclear receptor; MIBC: muscle-invasive bladder cancer; TCGA: The Cancer Genome Atlas; OS: overall survival; RFS: recurrence free survival; FUSCC: Fudan University Shanghai Cancer Center; HR: hazard ratio; LXRα: Liver X receptor alpha.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
We wish to thank all our colleagues in the Department of Urology, Shanghai Cancer Center, without you, we could not have completed the work. This work is supported by Project 81502192 by the National Natural Science Foundation of China.
Author contributions
Junlong Wu and Fangning Wan acquired the data and drafted the manuscript, Haoyue Sheng and Guowen Lin analyzed and interpreted the data, Guohai Shi prepared all figures, Yijun Shen and Bo Dai edited all tables. Dingwei Ye and Yiping Zhu designed the study. All authors reviewed and approved the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Abdollah F, Gandaglia G, Thuret R, Schmitges J, Tian Z, Jeldres C. et al. Incidence, survival and mortality rates of stage-specific bladder cancer in United States: a trend analysis. Cancer epidemiology. 2013;37:219-25
2. Chen W, Zheng R, Zeng H, Zhang S, He J. Annual report on status of cancer in China, 2011. Chinese journal of cancer research = Chung-kuo yen cheng yen chiu. 2015;27:2-12
3. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA: a cancer journal for clinicians. 2016;66:115-32
4. Rocken C, Behrens HM. Validating the prognostic and discriminating value of the TNM-classification for gastric cancer - a critical appraisal. European journal of cancer (Oxford, England: 1990). 2015;51:577-86
5. Sobin L H GM, Wittekind C. TNM classification of malignant tumours (Uicc international union against cancer). 7th ed ed. 2009
6. Soloway MS. Bladder cancer: Lack of progress in bladder cancer-what are the obstacles? Nature reviews Urology. 2013;10:5-6
7. Chamie K, Litwin MS. Quality of bladder cancer care in the USA. Expert review of pharmacoeconomics & outcomes research. 2011;11:619-21
8. Johnson DC, Greene PS, Nielsen ME. Surgical advances in bladder cancer: at what cost? The Urologic clinics of North America. 2015;42:235-52 ix
9. Corcoran AT, Handorf E, Canter D, Tomaszewski JJ, Bekelman JE, Kim SP. et al. Variation in performance of candidate surgical quality measures for muscle-invasive bladder cancer by hospital type. BJU international. 2015;115:230-7
10. Zhang Z, Burch PE, Cooney AJ, Lanz RB, Pereira FA, Wu J. et al. Genomic analysis of the nuclear receptor family: new insights into structure, regulation, and evolution from the rat genome. Genome research. 2004;14:580-90
11. Novac N, Heinzel T. Nuclear receptors: overview and classification. Current drug targets Inflammation and allergy. 2004;3:335-46
12. Nuclear Receptors Nomenclature Committee. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161-3
13. Doan TB, Eriksson NA, Graham D, Funder JW, Simpson ER, Kuczek ES. et al. Breast cancer prognosis predicted by nuclear receptor-coregulator networks. Molecular oncology. 2014;8:998-1013
14. Chang LY, Liu LY, Roth DA, Kuo WH, Hwa HL, Chang KJ. et al. The Major Prognostic Features of Nuclear Receptor NR5A2 in Infiltrating Ductal Breast Carcinomas. International journal of genomics. 2015;2015:403576
15. Han Y, Cai H, Ma L, Ding Y, Tan X, Chang W. et al. Expression of orphan nuclear receptor NR4A2 in gastric cancer cells confers chemoresistance and predicts an unfavorable postoperative survival of gastric cancer patients with chemotherapy. Cancer. 2013;119:3436-45
16. Han YF, Cao GW. Role of nuclear receptor NR4A2 in gastrointestinal inflammation and cancers. World journal of gastroenterology. 2012;18:6865-73
17. Seton-Rogers S. Prostate cancer: Connecting androgen receptor and immunity. Nature reviews Cancer. 2016;16:273
18. Visakorpi T, Hyytinen E, Koivisto P, Tanner M, Keinanen R, Palmberg C. et al. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nature genetics. 1995;9:401-6
19. Hawkins SM, Loomans HA, Wan YW, Ghosh-Choudhury T, Coffey D, Xiao W. et al. Expression and functional pathway analysis of nuclear receptor NR2F2 in ovarian cancer. The Journal of clinical endocrinology and metabolism. 2013;98:E1152-62
20. Masuyama H, Nakamura K, Nobumoto E, Hiramatsu Y. Inhibition of pregnane X receptor pathway contributes to the cell growth inhibition and apoptosis of anticancer agents in ovarian cancer cells. International journal of oncology. 2016;49:1211-20
21. Song B, Wang Y, Xi Y, Kudo K, Bruheim S, Botchkina GI. et al. Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene. 2009;28:4065-74
22. Chandra S, Narang R, Sreenivas V, Bhatia J, Saluja D, Srivastava K. Association of angiotensin II type 1 receptor (A1166C) gene polymorphism and its increased expression in essential hypertension: a case-control study. PloS one. 2014;9:e101502
23. Vogeler S, Bean TP, Lyons BP, Galloway TS. Dynamics of nuclear receptor gene expression during Pacific oyster development. BMC developmental biology. 2016;16:33
24. Itaba N, Wairagu PM, Aramaki N, Yasui T, Matsumi Y, Kono Y. et al. Nuclear receptor gene alteration in human induced pluripotent stem cells with hepatic differentiation propensity. Hepatology research: the official journal of the Japan Society of Hepatology. 2014;44:E408-19
25. Joseph SB, Bradley MN, Castrillo A, Bruhn KW, Mak PA, Pei L. et al. LXR-dependent gene expression is important for macrophage survival and the innate immune response. Cell. 2004;119:299-309
26. Bensinger SJ, Bradley MN, Joseph SB, Zelcer N, Janssen EM, Hausner MA. et al. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell. 2008;134:97-111
27. Mitro N, Mak PA, Vargas L, Godio C, Hampton E, Molteni V. et al. The nuclear receptor LXR is a glucose sensor. Nature. 2007;445:219-23
28. Zelcer N, Hong C, Boyadjian R, Tontonoz P. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science (New York, NY). 2009;325:100-4
29. Savic D, Ramaker RC, Roberts BS, Dean EC, Burwell TC, Meadows SK. et al. Distinct gene regulatory programs define the inhibitory effects of liver X receptors and PPARG on cancer cell proliferation. Genome medicine. 2016;8:74
30. Vedin LL, Gustafsson JA, Steffensen KR. The oxysterol receptors LXRalpha and LXRbeta suppress proliferation in the colon. Molecular carcinogenesis. 2013;52:835-44
31. Garattini E, Bolis M, Gianni M, Paroni G, Fratelli M, Terao M. Lipid-sensors, enigmatic-orphan and orphan nuclear receptors as therapeutic targets in breast-cancer. Oncotarget. 2016
32. Vedin LL, Lewandowski SA, Parini P, Gustafsson JA, Steffensen KR. The oxysterol receptor LXR inhibits proliferation of human breast cancer cells. Carcinogenesis. 2009;30:575-9
33. Chuu CP, Hiipakka RA, Kokontis JM, Fukuchi J, Chen RY, Liao S. Inhibition of tumor growth and progression of LNCaP prostate cancer cells in athymic mice by androgen and liver X receptor agonist. Cancer research. 2006;66:6482-6
34. Gabbi C, Kim HJ, Barros R, Korach-Andre M, Warner M, Gustafsson JA. Estrogen-dependent gallbladder carcinogenesis in LXRbeta-/- female mice. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14763-8
35. Rough JJ, Monroy MA, Yerrum S, Daly JM. Anti-proliferative effect of LXR agonist T0901317 in ovarian carcinoma cells. Journal of ovarian research. 2010;3:13
36. Bovenga F, Sabba C, Moschetta A. Uncoupling nuclear receptor LXR and cholesterol metabolism in cancer. Cell metabolism. 2015;21:517-26
37. Moschetta A. Nuclear receptor LXR as a novel therapeutic antitumoral target in glioblastoma. Cancer discovery. 2011;1:381-2
38. Guo D, Reinitz F, Youssef M, Hong C, Nathanson D, Akhavan D. et al. An LXR agonist promotes glioblastoma cell death through inhibition of an EGFR/AKT/SREBP-1/LDLR-dependent pathway. Cancer discovery. 2011;1:442-56
39. El Roz A, Bard JM, Valin S, Huvelin JM, Nazih H. Macrophage apolipoprotein E and proliferation of MCF-7 breast cancer cells: role of LXR. Anticancer research. 2013;33:3783-9
40. Uno S, Endo K, Jeong Y, Kawana K, Miyachi H, Hashimoto Y. et al. Suppression of beta-catenin signaling by liver X receptor ligands. Biochemical pharmacology. 2009;77:186-95
41. Zhang W, Jiang H, Zhang J, Zhang Y, Liu A, Zhao Y. et al. Liver X receptor activation induces apoptosis of melanoma cell through caspase pathway. Cancer cell international. 2014;14:16
42. Witjes JA, Comperat E, Cowan NC, De Santis M, Gakis G, Lebret T. et al. EAU guidelines on muscle-invasive and metastatic bladder cancer: summary of the 2013 guidelines. European urology. 2014;65:778-92
43. Clark PE, Spiess PE, Agarwal N, Bangs R, Boorjian SA, Buyyounouski MK. et al. NCCN Guidelines Insights: Bladder Cancer, Version 2.2016. Journal of the National Comprehensive Cancer Network: JNCCN. 2016;14:1213-24
44. Galsky MD, Stensland KD, Moshier E, Sfakianos JP, McBride RB, Tsao CK. et al. Effectiveness of Adjuvant Chemotherapy for Locally Advanced Bladder Cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2016;34:825-32
Author contact
![]() Corresponding authors: Prof. Ding-Wei Ye, M.D. (dwyelicom) and Dr. Yiping Zhu M.D. (qdzhuyipingcom), Department of Urology, Fudan University Shanghai Cancer Center, No. 270 Dong'an Road, Shanghai 200032, People's Republic of China. Tel: 86-21-64175590-2807; Fax: 86-21-64434556.
Corresponding authors: Prof. Ding-Wei Ye, M.D. (dwyelicom) and Dr. Yiping Zhu M.D. (qdzhuyipingcom), Department of Urology, Fudan University Shanghai Cancer Center, No. 270 Dong'an Road, Shanghai 200032, People's Republic of China. Tel: 86-21-64175590-2807; Fax: 86-21-64434556.

 Global reach, higher impact
Global reach, higher impact