3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(5):880-886. doi:10.7150/jca.18207 This issue Cite
Research Paper
CT120: A New Potential Target for c-Myc in Head and Neck Cancers
1. Istanbul University Cerrahpasa Medical Faculty, Department of Medical Biology;
2. Istanbul University Cerrahpasa Medical Faculty, Department of Otorhinolaryngology.
Received 2016-11-4; Accepted 2016-12-23; Published 2017-3-7
Abstract
Background: CT120 is a universally expressed protein with seven transmembrane domains. It functions in cell proliferation, survival and apoptosis by activating Raf/MEK/ERK and PI3K/Akt signaling pathways. Evidence suggests that CT120 plays important roles in lung carcinogenesis and oncogenic pathway activation. c-Myc is an important transcription factor modulating cell progression, apoptosis and cellular transformation. Previous studies have shown that MYC gene is amplified in many types of cancer including head and neck squamous cell carcinoma (HNSCC). Myc can regulate expression of many genes by binding to E-boxes. The aim of this study was to investigate the relationship between c-Myc protein and CT120 gene.
Methods: Tumor and normal tissue samples from 50 patients with HNSCC were investigated with chromatin immunoprecipitation assay (ChIP), Illumina MiSeq, bisulphite sequencing and qRT-PCR.
Results: c-Myc binds to all E-boxes except E-box 5 on CT120 promoter. The CpG dinucleotides were found to be partially methylated in all tumor and normal tissue samples. Bisulphite sequencing showed a 10% down-regulation in the methylation levels of the tumor tissues. CT120 gene was hypomethylated and up-regulated in 56% of the tumor tissue samples. Expression of c-Myc was significantly higher in tumor tissues than in non-cancerous tissue samples. MYC was overexpressed in 68% of the tumor tissue samples compared to normal tissues. The mean MYC levels were 2.42-fold higher in the tumor tissue samples. In 48% of the tumor tissues, MYC and CT120A mRNA were up- or down-regulated simultaneously (p<0.001).
Conclusion: We show that CT120 gene is a target of c-Myc and it contributes to cancer progression in HNSCC.
Keywords: CT120, FAM57A, c-Myc, HNSCC, bisulphite, sequencing, methylation.
Introduction
The novel human plasma membrane-associated gene CT120/FAM57A (GenBank accession No: AF 477201) has been isolated from chromosome 17p13.3 by positional cloning and rapid amplification of cDNA ends (RACE) by He et al [1]. The CT120 gene spans over 10.288 bp on chromosome 17p13.3 and is composed of 5 exons. Its main product is a 257 amino acid protein.
CT120 is universally expressed in different tissues except lung tissue and various human tumor cell lines [1]. In human two isoforms of CT120 have been identified [2]. Functional predictions based on the amino acid sequence of the CT120A gene using the SMART and TMHMM software revealed that the protein has seven transmembrane domains [1]. Homology comparison studies also revealed that the gene has been conserved during evolution which indicates that it exerts essential physiological functions. A variant of CT120, CT120B has been isolated from large cell lung carcinoma [2]. In this isoform, the fourth exon of CT120A is spliced. It consists of four exons and encodes a protein composed of 225 amino acids. By using a yeast two-hybrid system two partners of CT120 have been identified. These are the SLC3A2 (CD98) and GGTL3B proteins which function in the amino acid transport and glutathione metabolism, respectively [1].
In 2004, He et al. [3] have shown that two major signaling pathways are activated as a result of ectopic CT120 expression. These are the Raf/Mek/ERK and PI3K/Akt signal pathways, both of which are involved in cell proliferation, survival and apoptosis. CT120 may also contribute to the overexpression of metastasis-associated genes such as cathepsin B, cathepsin D, cathepsin L, and MMP-2/TIMP2 [3]. Silencing of CT120 enhances p53 and caspase 3 expression and reduces cyclin D1 and CDK4 expression which results in G1 arrest and apoptosis [4]. It has also been shown that knocking down CT120 in the SPCA1 human lung adenocarcinoma cell line results in reduced in vitro cell growth rates and in decreased tumorigenicity in nude mice [5]. These results indicate that CT120 may be a key modulator in lung carcinogenesis and might contribute to the activation of oncogenic pathways in human lung cancer cells. In a previous study we have observed that the CT120A isoform is overexpressed also in head and neck squamous cell carcinoma (HNSCC) tumors [6]. However, no detailed information is available on the structural features of the CT120 gene. Therefore, we first sequenced the promoter region of the CT120 gene to investigate the presence of any regulatory sequences.
The c-Myc protein belongs to the myc family of transcription factors and plays a fundamental role in cell cycle progression, apoptosis and cellular transformation [7, 8]. Amplification of the MYC gene is observed in many different types of cancer including HNSCC [9, 10]. In the human genome, the MYC gene is located on chromosome 8 and its protein product regulates expression of 15% of all genes through binding to the Enhancer box (E-box) sequences. An E-box is a specialized region that functions as a protein binding site and regulates gene expression [11-13]. The CANNTG consensus sequences of the E-box are known as the canonical E-boxes. There are several other non-canonical E-box motifs such as the GATGTG, CATGCG, CACGCG, CACGAG, CGCGAG and CAACGTG sequences [14, 15].
It is also well known that the gene expression rates are under the control of epigenetic modifications [16]. Methylation of the CpG islands which are found in the promoter regions is one of the most frequently studied epigenetic modifications. In this study, to investigate the possible epigenetic regulation of CT120 and the effect of the promoter methylation to the binding of c-Myc, we also investigated the methylation level of the CT120 promoter.
Materials and methods
Human tissue
Tumors and the corresponding normal tissue samples were obtained from 50 patients with HNSCC undergoing surgery at the Istanbul University Cerrahpasa Medical Faculty, Department of Otorhinolaryngology. The study was approved by the Ethics Committee of Istanbul University, Cerrahpasa Medical Faculty (No: 83045809/604.01/02-41972). Written informed consent was obtained from all patients prior to the study and the study was performed in accordance with the 1964 Declaration of Helsinki. The experiments have been performed twice to confirm results.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay was performed using the Tissue Acetyl-Histone H4 ChIP Kit (Abnova, Taipei City, Taiwan) according to the manufacturer's instructions. The strips were activated with the antibody buffer, then, Mouse monoclonal IgG (c-Myc antibody) (200 µg/0.1 ml ) (SC-40X, SantaCruz Biotechnology, Dallas TX, USA) and normal Mouse IgG (0.5 mg/ml) were added into the strip wells and incubated for 90 min. The tissue samples were cut into small pieces on ice and the histones and transcript-related proteins were cross-linked to DNA by 1% formaldehyde at room temperature on an orbital shaker for 20 min. After washing with ice-cold PBS, the tissue pieces were homogenized for 5 min using the Bullet Blender homogenizer (Next Advance, NY, USA). The samples were incubated on ice for 10 min after adding 1% of protease inhibitor cocktail. The DNA was sheared by 4-5 cycles of 20 s strokes using a Sonoplus HD2070 sonicator (Bandelin electronic GmbH&Co, Berlin, Germany), resting the samples on ice between each cycle. Unused samples were kept at -800C until used. The supernatant was diluted and 5 µl of the supernatant was labeled as input DNA. The antibody solutions were removed and the supernatant containing the protein was immunoprecipitated in the strip wells for overnight at 40C on a Janke Kunkel KS 250 rocking platform (IKA Works GmbH& Co. KG, Staufen, Germany). After washing the wells with 1X TE Buffer, reversal of the cross-linked DNA was performed by adding the DNA-release buffer containing Proteinase K and incubating the samples at 650C for 90 min. Purified and eluted DNA samples were used for PCR. PCR was performed using 10 ng/µl of DNA and appropriate primers which are given in Table 1. PCR reactions were as follows: 950C for 10 min, then 35 cycles of 940C for 30 s, 57-630C for 30 s (depending on the amplicon) and 720C for 30 s, with a final extension for 10 min at 720C (the annealing temperatures for E-box PCR are shown in Table 1).
Analysis of Promoter Methylation Levels by Targeted DNA Methylation Analysis
Promoter methylation levels of the CT120 gene were analyzed on the Illumina MiSeq system (Illumina Inc., CA, USA) and the methylation level of the 1st island was also confirmed by bisulphite sequencing. For this purpose genomic DNA was obtained from tumor and normal tissue samples using the Roche High Pure PCR Template Preparation Kit (Roche Diagnostics, Mannheim, Germany) and the samples were modified with sodium bisulphite using the EZ-DNA Methylation Gold Kit (Zymo Research, Irvine, CA, USA) according to the manufacturer's instructions.
The PCR products were purified and eluted for Nextera XT library preparation with dual indexing. The libraries were multiplexed and sequenced on the Illumina MiSeq (Illumina, San Diego, CA, USA) following the manufacturer's protocol. The sequence data was demultiplexed and assessed for sample read coverage, mapping efficiency, unique CpG coverage and bisulfite conversion rate.
The coordinates, methylated CpG counts and total CpG counts were used to investigate the methylation status on the various positions of the promoter region. The methylated and unmethylated regions were investigated using USCS Genome Browser on Human Feb. 2009 (GRCh37/hg19). Assembly and the reports were generated on Excel files. The methylation ratio of the promoter region and the methylation status of each CpG island on each position on Chr 17:633812-635936 were determined.
Bisulphite Sequencing
To confirm the results of the 1st CpG island in the CT120 promoter region targeted bisulphite sequencing was performed. For cycle sequencing modified DNA samples were amplified by PCR specific to the methylated promoter regions of CT120 (primers are given in Table 1) and the products were analyzed on 2% agarose gels. After purification of the PCR samples with Ethanol/EDTA/Sodium Acetate, cycle sequencing was performed using the ABI 3100 Genetic Analyzer capillary sequencing system (Applied Biosystems, Waltham, MA, USA). The PCR conditions for cycle sequencing were as follows: Denaturation at 96oC for 30 sec, hybridization at 55oC for 30 sec and an extension step at 60oC for 2 min for 44 cycles.
RT-PCR and Real-Time Quantitative RT-PCR
Total RNA from the tissues was isolated by using the PureLink RNA Mini Kit (Ambion, USA) according to the manufacturer's instructions. cDNAs were prepared from 200 ng of total RNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche Diagnostics, Mannheim, Germany). Expression levels of the MYC gene in the tumors and non-cancerous tissue samples were analyzed by quantitative Real Time PCR (qRT-PCR) using the LightCycler 480 system (Roche Diagnostic, Mannheim, Germany). PCR reactions were performed in a final volume of 20 µl containing 1x Master Mix, 200 nM gene specific primers (forward: 5'- GCTGCTTAGACGCTGGATTT-3' and reverse: 5'- TAACGTTGAGGGGCATCG-3') and 200 nM hydrolysis probe (UPL Probe No.66) for MYC that was 5'-labeled with fluorescein (FAM) and 3'- labeled with a dark quencher dye. The Succinate Dehyrogenase (SDHA) gene was used as the reference to normalize the quantification of mRNA levels. Primers (forward: 5'- GGACCTGGTTGTCTTTGGTC-3' and reverse: 5'- CCAGCGTTTGGTTTAATTGG-3') and 200 nM hydrolysis probe (UPL Probe No: 80) that is 5'-labeled with fluorescein (FAM) and 3'-labeled with dark quencher dye. Relative mRNA levels were calculated using the 2-∆∆Ct method [17].
Sequences of the primers for ChIP analysis and bisulphite sequencing.
| Primer | Sequence | Annealing Temperature | Amplicon length | Location |
|---|---|---|---|---|
| E-box-1 | F-5'- AGGAAGATACAGACCAAAGCAAA -3' R- 5'- GGACAGGCGTCTAAACTTGAAT -3' | 61 °C | 146 bp | -1405/-1259 |
| E-box-2/3 | F-5'- TAGACGCCTGTCCCGATAAC -3' R-5'- AGACACCATCCAGAGGCGTA -3' | 62 °C | 389 bp | -1272/ -883 |
| E-box-4 | F-5'- CAGAAGCCGGGAGCCGCG -3' R-5'- ATAGGGCGGGGCGGGGTC -3' | 64 °C | 207 bp | -488/-281 |
| E-box-5 | F-5'- GACCCCGCCCCGCCCTAT -3' R-5'- GCCGCGCGATTTCAACCCT -3' | 58 °C | 123 bp | -299/-176 |
| E-box-6 | F-5- AGGGTTGAAATCGCGCGG -3' R-5'- AGTCGGTGCGGCTCCAT -3' | 57 °C | 190 bp | -195/-5 |
| 1st CgP Island | F-5'- AGGTATTATTATATTTGGTTAATTTTGTAT-3' R-5'-AAAAACTACAAACTCTTACTACCTCTC -3' | 58 °C | 255 bp | -1093/-838 |
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 6 for Windows, (GraphPad Software, La Jolla California USA). We used One-way ANOVA test to analyze the association between gene expression levels and promoter methylation. p<0.05 was considered statistically significant.
Results
Characterization of the Promoter Region of the CT120 Gene
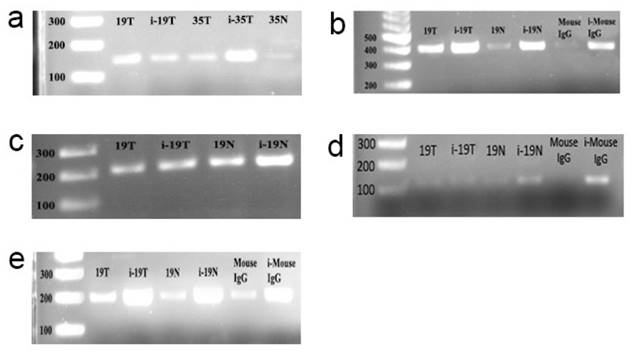
Using sequence analysis we identified 6 E-boxes in the promoter region of the CT120 gene (Fig. 1). To investigate binding of c-Myc to the CT120 gene promoter we performed ChIP assays. Our data have shown that c-Myc binds to all E-boxes which are located in the CT120 promoter except the E-box 5 (Fig. 2).
Nucleotide sequence of the promoter region (-1334 to +14) of the CT120 gene. The putative binding sites for c-Myc are highlighted in bold. The start codon (ATG) is italicized and underlined

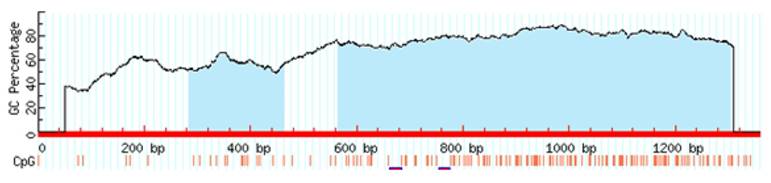
Target sequences of the transcription factors contain CpG islands and methylation of the CpG dinucleotides which are located in these regions may affect the binding of the factors to DNA. Therefore, to predict CpG islands in the promoter region of CT120 we analyzed the target sequences using the MethPrimer 3 software. We identified two putative CpG-rich regions in the CT120 gene promoter (Fig. 3).
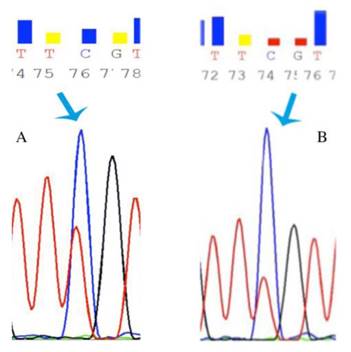
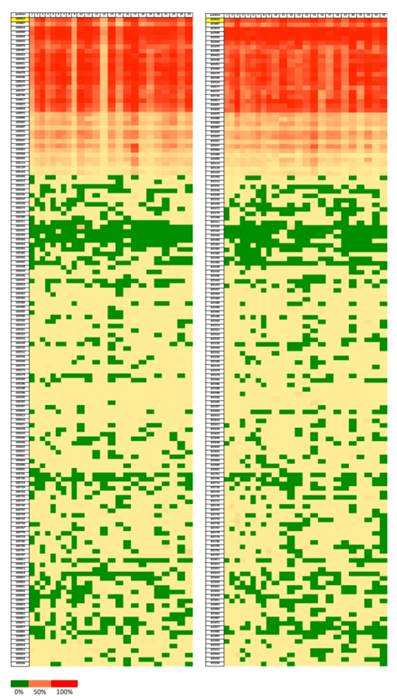
In order to reveal whether CT120 expression or binding of c-Myc is attributable to epigenetic mechanisms, the methylation status of the CT120 promoter was analyzed. In all tumors and non-cancerous tissue samples the CpG dinucleotides were partially methylated (Fig 4). However, there were no differences in the methylation levels of the CpG dinucleotides which are located in the E-boxes 4, 5 and 6. The CpG dinucleotides of the E-boxes 1, 2 and 3 were hypomethylated in tumor tissues compared to non-cancerous tissues (Fig. 5). Bisulphite sequencing also showed that there was a 10% down-regulation in the methylation levels of the tumor tissues with respect to their normal counterparts. In all of our study samples the CT120 expression levels had been analyzed previously. Therefore, we also analyzed the methylation levels in association with CT120 expression. Our results showed that 28 (56%) of the tumor tissue samples which displayed CT120 up-regulation were hypomethylated (Table 2 and Fig. 6).
MYC Expression
In order to determine the level of MYC expression qRT-PCR was performed using the tumor and adjacent non-cancerous tissue samples. Expression of MYC was significantly higher in tumor tissues than in non-cancerous tissue samples (Table 3). We observed MYC overexpression in 68% (34/50) of the tumor tissue samples when compared to non-cancerous tissues. The mean MYC expression levels were 2.42-fold higher in the tumor tissue samples (Table 3).
To understand the effect of c-Myc on the CT120 expression levels we evaluated the correlation between these two genes. As a result of this analysis we observed a concordant change in the expression levels of both genes (Table 4). In 48% of the tumor tissues, MYC and CT120A mRNA were up- or down-regulated concurrently. This correlation was statistically significant (p<0.001). In 40% (20/50) and 8% (4/50) of the samples an increase or decrease in the c-Myc level resulted in a concurrent increase or decrease in the CT120A level, respectively.
Correlation between CT120 gene expression and promoter methylation
| CT120 Expression | |||||
|---|---|---|---|---|---|
| Increased | Decreased | No change | |||
| CT120 Methylation | Increased | 13 (26%) | 6 (12%) | 3 (6%) | p<0.001 |
| Decreased | 16 (32%) | 11 (22%) | 1 (2%) | ||
MYC expression levels in tumors and non-cancerous tissue samples from HNSCC patients
| Ct Reference | Ct Target | ∆Ct | ∆∆Ct | 2-∆∆Ct | p | |
|---|---|---|---|---|---|---|
| Tumor | 27.91 | 26.46 | -1.451 | -1.27 | 2.42 | 0.001 |
| Normal | 28.22 | 28.04 | -0.174 | 0 | 1 |
Correlation between CT120 and MYC expression in HNSCC tumors
| CT120 Expression | |||||
|---|---|---|---|---|---|
| Increased | Decreased | No change | |||
| MYC Expression | Increased | 20 (40%) | 11 (22%) | 3 (6%) | p<0.001 |
| Decreased | 7 (14%) | 4 (8%) | 1 (2%) | ||
| No change | 2 (4%) | 2 (4%) | - | ||
ChIP assay electrophoregrams to demonstrate that c-myc binds to the E-boxes which are found in the promoter of the CT120 gene. i-19T: input DNA, T. Tumor, N: Normal, Mouse IgG: PCR product derived from DNA template immunoprecipitated by normal IgG. First lanes are 100 bp markers. a: PCR products which were amplified using E-box 1-specific primers; b: PCR products which were amplified using E-box 1-2-specific primers; c: PCR products which were amplified using E-box 4-specific primers; d: PCR products which were amplified using E-box 5-specific primers; e: PCR products which were amplified using E-box 6-specific primers.
CpG islands of the CT120 promoter
Representative chromatograms of bisulphide sequencing of the promoter region. a: 21 T partial methylation; b: 21 N partial methylation. Blue arrow shows hypomethylated region.
Discussion
The CT120 gene is located on chromosome 17p13.3 region and codes for a human plasma membrane-associated protein [1]. Although it has been characterized and associated with tumor progression more than a decade ago its exact mechanism of action has not been investigated in detail. Depending on its partners SLC3A2 and GGTL3B a function in the amino acid transport and glutathione metabolism has been suggested [1]. On the other hand, some investigators reported that CT120 is involved in major signaling pathways such as PI3K/Akt and Ref/Mek/ERK which are de-regulated in carcinogenesis [3]. Until recently CT120 had been associated only with lung cancer [3-5]. In our previous report we have shown that CT120 acts as an oncogene in HNSCC [6]. Depending on our previous data we hypothesized that CT120 may have important functions in the progression of HNSCC needing further investigation.
Heatmap showing the ratios of methylation on each position of the CpG dinucleotides (Chr 17:633812-635936). Green color represents lower methylation ratio while red color shows increased methylation up to 100%. Left for tumor samples and right for normal tissue samples.
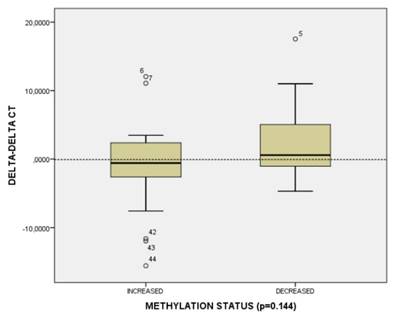
CT120 methylation and gene expression distribution in HNSCC tumors.
The sequence analysis showed that 6 E-boxes were present in the promoter region of the CT120 gene. To test the binding of c-Myc to the CT120's promoter we performed ChIP assays. The result of this analysis confirmed binding of c-Myc to the E-boxes 1,2,3,4, and 6 which are located on CT120's promoter. Our results show for the first time that CT120 is a new target of c-Myc.
On the other hand, Myc-target sites are rich in CpG islands [18]. When we investigated the promoter region of the CT120 gene we identified two CpG-rich regions. c-Myc was found to strongly bind to the E-boxes 2 and 3 which are in the first CpG-rich region. Aberrant DNA methylation within the CpG islands is among the earliest and most common alterations in human cancers, leading to abnormal expression of a broad spectrum of genes. Methylation of the CpG dinucleotides prevents binding of transcription factors. It is also known that CpG-dinucleotides within the E-boxes prevent the binding of c-Myc [18-20]. Therefore, a decrease in the methylation level of the promoter region in the tumor samples leads to an increase in the expression level by allowing binding of the c-Myc protein. Parallel changes in the expression of the MYC and CT120 genes supports this correlation.
Dysregulated expression or function of c-Myc is one of the most common abnormalities in human malignancies including HNSCC. Several studies have reported that MYC amplification is a frequent event in HNSCC [9, 10, 21, 22]. However, reported amplification frequencies vary from 6 to 38% [23-28]. In our previous study we detected MYC amplification in 10% of the HNSCC tumor samples by MLPA analysis [29]. Therefore, in this study we investigated the expression of c-Myc to identify whether over-expression of c-Myc occurs as a result of amplification in HNSCC. However, in our study cohort overexpression of c-Myc was more frequent than amplification. Overexpression of MYC gene at the mRNA and protein level has been reported in HNSCC with different frequencies (21-68%) [21-29].
In our study overexpression of the MYC gene was higher than the previous reports [21-28]. This may be due to the low sensitivity of the methods which have been used in previous studies. In most of these studies the c-Myc protein levels were investigated using immunohistochemistry. As to our knowledge this is the first report in the literature investigating the MYC mRNA using a PCR-based method.
As a result, this is the first report indicating the CT120 gene as the target of the c-Myc protein. Our data suggest that CT120 participates in HNSCC progression under the control of c-Myc and epigenetic regulations.
Abbreviations
HNSCC: Head and neck squamous cell carcinoma; E-box: Enhancer box; ChIP: Chromatin Immunoprecipitation; qRT-PCR: quantitative Real Time PCR.
Acknowledgements
This work was supported by the Scientific and Technological Research Council of Turkey (TUBITAK) with the project number: 115Z807
Competing Interests
The authors have declared that no competing interest exists.
References
1. He X, Di Y, Li J, Xie Y, Tang Y, Zhang F. et al. Molecular cloning and characterization of CT120, a novel membrane-associated gene involved in amino acid transport and glutathione metabolism. Biochem Biophys Res Commun. 2002;297:528-36
2. Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS. et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA. 2002;99:16899-903
3. He XH, Li JJ, Xie YH, Tang YT, Yao GF, Qin WX. et al. Altered gene expression profiles of NIH3T3 cells regulated by human lung cancer associated gene CT120. Cell Res. 2004;14:487-96
4. Li Z, Shao S, Xie S, Jiao F, Ma Y, Shi S. Silencing of CT120 by antisense oligonucleotides could inhibit the lung cancer cells growth. Ir J Med Sci. 2010;179:217-23
5. Pan DN, Li JJ, Wei L, Yao M, Wan DF, Gu JR. Inhibitory effect of CT120B, an alternative splice variant of CT120A, on lung cancer cell growth. Acta Biochim Biophys Sin (Shanghai). 2005;37:588-92
6. Baltaci E, Ekizoglu S, Sari E, Karaman E, Ulutin T, Buyru N. CT120A Acts as an oncogene in head and neck squamous cell carcinoma. J Cancer. 2015;6:1255-59
7. Henriksson M, Lüscher B. Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv Cancer Res. 1996;68:109-82
8. Levens DL. Reconstructing MYC. Genes Dev. 2003;17:1071-77
9. Bhattacharya N, Roy A, Roy B, Roychoudhury S, Panda CK. MYC gene amplification reveals clinical association with head and neck squamous cell carcinoma in Indian patients. J Oral Pathol Med. 2009;38:759-63
10. Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004-16
11. Walhout AJ, Gubbels JM, Bernards R, van der Vliet PC, Timmers HT. c-Myc/Max heterodimers bind cooperatively to the E-box sequences located in the first intron of the rat ornithine decarboxylase (ODC) gene. Nucleic Acids Res. 1997;25:1493-501
12. Amati B, Dalton S, Brooks MW, Littlewood TD, Evan GI, Land H. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature. 1992;359:423-6
13. Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211-7
14. Blackwell TK, Huang J, Ma A, Kretzner L, Alt FW, Eisenman RN, Weintraub H. Binding of myc proteins to canonical and noncanonical DNA sequences. Mol Cell Biol. 1993;13:5216-24
15. Haggerty TJ, Zeller KI, Osthus RC, Wonsey DR, Dang CV. A strategy for identifying transcription factor binding sites reveals two classes of genomic c-Myc target sites. Proc Natl Acad Sci U S A. 2003;100:5313-8
16. Medvedeva YA, Khamis AM, Kulakovskiy IV, Ba-Alawi W, Bhuyan MS, Kawaji H. et al. Effects of cytosine methylation on transcription factor binding sites. BMC Genomics. 2014;15:119
17. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101-8
18. Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J. et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115-29
19. Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB. Analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer. 2004;4:562-8
20. Prendergast GC, Ziff EB. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991;251:186-9
21. Field JK, Spandidos DA, Stell PM, Vaughan ED, Evan GI, Moore JP. Elevated expression of the c-myc oncoprotein correlates with poor prognosis in head and neck squamous cell carcinoma. Oncogene. 1989;4:1463-8
22. Riva C, Lavieille JP, Reyt E, Brambilla E, Lunardi J, Brambilla C. Differential c-myc, c-jun, c-raf and p53 expression in squamous cell carcinoma of the head and neck: implication in drug and radioresistance. Eur J Cancer B Oral Oncol. 1995;31:384-91
23. Gapany M, Pavelic ZP, Kelley DJ, Pavelic L, Gapany SR, Craven JM. et al. Immunohistochemical detection of c-myc protein in head and neck tumors. Arch Otolaryngol Head Neck Surg. 1994;120:255-9
24. Merritt WD, Weissler MC, Turk BF, Gilmer TM. Oncogene amplification in squamous cell carcinoma of the head and neck. Arch Otolaryngol Head Neck Surg. 1990;116:1394-8
25. Leonard JH, Kearsley JH, Chenevix-Trench G, Hayward NK. Analysis of gene amplification in head-and-neck squamous-cell carcinoma. Int J Cancer. 1991;48:511-5
26. Haughey BH, von Hoff DD, Windle BE, Wahl GM, Mock PM. c-myc oncogene copy number in squamous carcinoma of the head and neck. Am J Otolaryngol. 1992;13:168-71
27. Rodrigo JP, Lazo PS, Ramos S, Alvarez I, Suárez C. MYC amplification in squamous cell carcinomas of the head and neck. Arch Otolaryngol Head Neck Surg. 1996;122:504-7
28. Waitzberg AF, Nonogaki S, Nishimoto IN, Kowalski LP, Miguel RE, Brentani RR. et al. Clinical significance of c-myc and p53 expression in head and neck squamous cell carcinomas. Cancer Detect Prev. 2004;28:178-86
29. Baltaci E, Karaman E, Dalay N, Buyru N. Analysis of gene copy number changes in head and neck cancer. Clin Otolaryngol. 2016 doi: 10.1111/coa.12686
Author contact
![]() Corresponding author: Prof. Dr. Nur Buyru, Istanbul Universitesi Cerrahpasa Tip Fakultesi Yemel Bilimler Binasi Kat: 6 Kocamustafapasa, 34098, Istanbul, Turkey E-mail: nbuyrucom GSM: +90 505 5249471
Corresponding author: Prof. Dr. Nur Buyru, Istanbul Universitesi Cerrahpasa Tip Fakultesi Yemel Bilimler Binasi Kat: 6 Kocamustafapasa, 34098, Istanbul, Turkey E-mail: nbuyrucom GSM: +90 505 5249471

 Global reach, higher impact
Global reach, higher impact