3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(12):2346-2355. doi:10.7150/jca.19465 This issue Cite
Research Paper
Development and Validation of a Serum Biomarker Panel for the Detection of Esophageal Squamous Cell Carcinoma through RNA Transcriptome Sequencing
1. State Key Laboratory of Oncology in Southern China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, China
2. Department of Clinical Laboratory, Sun Yat-sen University Cancer Center, Guangzhou, China
3. Department of Experimental Research, Sun Yat-sen University cancer center, Guangzhou, China
4. Department of Clinical Laboratory, Shaanxi Provincial People's Hospital, Xian, China
5. Guangdong Experimental High School, Guangzhou, China
6. Department of Laboratory Medicine, Sun Yat-sen University First Affiliated Hospital, Guangzhou, China
† Xing S, Zheng X and Wei LQ contributed equally to this work
Received 2017-2-3; Accepted 2017-4-30; Published 2017-7-21
Abstract
Serum tumor markers for the diagnosis of esophageal squamous cell carcinoma (ESCC) have low sensitivity. This study aims to identify new serum markers for ESCC diagnosis from RNA sequencing (RNA-seq) data.
RNA-seq was performed using six pairs of ESCC and matched normal tissues. The candidates for ESCC were screened from the differentially expressed genes. The candidates were analyzed by ELISA from the serum of a test group and a validation group. Real-time PCR, Western blotting and immunohistochemistry were used to detect the expression of the candidates in tumor cell lines and tumor tissues.
Ten genes were selected from the RNA-seq data. Serum levels of ADAM12, CHI3L1, MMP13 and SPP1 were significantly higher in the ESCC patients than in the healthy controls. A diagnostic model combining CHI3L1, MMP13, and SPP1 was established. The area under the curve (AUC) values for serum CHI3L1, MMP13, and SPP1 and the diagnostic model for discriminating ESCC patients from controls were 0.732, 0.881, 0.661 and 0.928, respectively. In the validation cohort, the AUC values were 0.753, 0.789, 0.696 and 0.843, respectively. Moreover, the AUC of the model for classifying patients with early ESCC was 0.918 in the test group and 0.857 in the validation group. Overexpression of CHI3L1, MMP13 and SPP1 was observed in the tumor cell lines and tissues.
The diagnostic model composed of CHI3L1, MMP13 and SPP1 discriminates ESCC patients with high sensitivity. Our data highlight the potential of this diagnostic model for the noninvasive diagnosis of ESCC.
Keywords: esophageal squamous cell carcinoma, RNA transcriptome sequencing, diagnostic biomarker, ESCC, early detection
Introduction
Esophageal cancer is one of the most aggressive cancers and represents the sixth leading cause of cancer death worldwide.[1] Approximately 70% of global esophageal cancer cases occur in China, with esophageal squamous cell carcinoma being the histopathological form found in the vast majority of cases.[2] The current 5-year survival rate of individuals diagnosed with ESCC is 10%,[2] reflecting that most cases are already at an advanced stage at the time of diagnosis. Like other cancers, the challenge in early detection lies in the relatively non-specific features of early symptoms, and detection requires that invasive physical procedures, such as gastrointestinal endoscopy, be performed on a regular basis, which may not be practical for general screening. The most ideal solution for early detection is to identify reliable markers that can detect the cancer through simple blood tests. Using epigenetics and miRNA microarray, a few new serum markers were recently found, including the aberrant methylation of genes such as FHIT[3] and the miRNAs miR-507 and miR-634, among others.[4, 5] However, despite their diagnostic power for ESCC, the associated complicated techniques limit their clinical application. In addition, some new serum markers for ESCC were identified using proteomic techniques.[6, 7] However, considering that the sample size in the study was rather small, more rigorous studies of these proposed markers are needed. The general challenge of the available proteomic studies of serum marker identification is that the potential markers likely have a substantially lower abundance in comparison to other proteins in the blood circulation. Consequently, their discovery is extremely difficult. Thus, we attempted to identify serum ESCC biomarkers from the transcriptome sequencing data of cancer tissues and matched non-cancerous tissues. RNA-seq is superior for the detection of low-abundance transcripts, differentiating biologically critical isoforms and allowing the identification of tumor markers.[8-10] To our knowledge, only a relatively small number of RNA-Seq analyses of the ESCC transcriptome have been reported,[11-13] and screening of serum biomarkers for ESCC diagnosis by ESCC transcriptome RNA-Seq analysis has not been investigated.
Here, we conducted a systematic study ultimately to identify serum markers for ESCC based on RNA-seq data using the following procedure. Transcriptome sequencing was performed using six pairs of ESCC tissues and the adjacent noncancerous tissues. Comparative analyses of the gene expression data were performed to identify the differentially expressed genes (DEGs) in cancer versus normal tissues. These genes were then subjected to computational analyses to determine whether their proteins are secreted into the blood circulation and thus could potentially serve as serum markers. To select markers that are reproducible in ESCC, the candidate genes were confirmed using published microarray gene-expression data for ESCC. An esophageal cancer signaling pathway analysis, an oncogenic protein function analysis, and a document retrieval retrospective analysis were used to verify the specificity of the predicted markers. We then established a double sandwich ELISA to detect the candidate proteins in the serum and set up a mathematical model to further evaluate the role of the candidate biomarkers for the diagnosis of ESCC.
Materials and Methods
Tissue and serum specimens
In this study, all selected ESCC patients met the following inclusion criteria: pathological examination confirmation of primary ESCC by the available biopsy samples; and no anticancer treatments given before surgery. The tumors were staged according to the 7th edition of the tumor-node-metastasis (TNM) classification for esophageal carcinoma (UICC, 2009).
Six pairs of ESCC and adjacent non-cancerous tissue samples were obtained from patients with ESCC who had undergone surgery at the Cancer Center of Sun Yat-Sen University between 2005 and 2011. The samples were used for RNA-seq, real-time RT-PCR and Western blotting. A total of 20 formalin-fixed and paraffin-embedded ESCC tumor specimens for immunochemistry were obtained from the Sun Yat-sen University Cancer Center (SYSUCC). The corresponding normal esophageal tissue specimens (n = 20) were collected from areas at a standard distance (8 cm) from the corresponding resected tumors. All ESCC and adjacent non-cancerous tissue samples were collected immediately after surgical resection and confirmed by pathological review.
Serum samples were classified into two groups: the test cohort and the validation cohort. The 80 preliminary screening samples were selected randomly from the test cohort.
None of the selected healthy controls had esophageal diseases, inflammatory diseases or other diseases, based on a physical examination. All selected patients with benign esophageal disease were confirmed by endoscopy.
The test cohort consisted of 150 ESCC patients, 96 healthy controls and 44 patients with benign esophageal disease selected from SYSUCC between 2013 and 2015. The validation cohort was enrolled between 2014 and 2015 and comprised 169 ESCC patients, 80 healthy controls and 74 patients with benign esophageal disease from Shantou central hospital (STCH), Shaanxi provincial people's hospital (SPPH), and SYSUCC. More details about the patients are provided in the supplementary material.
Venous blood (3-5 ml) was obtained at the time of diagnosis before treatment, clotted at room temperature, centrifuged at 3000 r/min for 10 min, processed into serum aliquots within 3 hr and stored at -80°C until use.
Prior to the use of these serum and tissues, informed consent was obtained from each participant. This experiment was approved by the Institute Research Ethics Committee of the Cancer Center of SYSUCC, Guangzhou, China, STCH, Shantou, China and the SPPH, Shanxi, China.
Cell lines
The immortalized esophageal epithelial cell line, NE-3, induced by the human papillomavirus type 16 E6/E7, was obtained from Dr. Jin (the University of Hong Kong, P.R. China) and cultured in Keratinocyte-SFM (Invitrogen, Carlsbad, CA) medium. The ESCC cell lines, Eca-109, Kyse30, Kyse140, Kyse180, Kyse510 and Kyse520 (Chinese Academy of Sciences, Shanghai, China), were grown in RPMI 1640 (Invitrogen, USA) supplemented with 10% fetal bovine serum. All cell lines were obtained between 2012 and 2014, and they were authenticated by qRT-PCR analysis for the expression of signature genes.
RNA isolation, cDNA library preparation, and RNA-seq analysis
RNA-Seq analysis of the transcriptome of ESCC tissues and matched non-cancerous tissues was performed by BGI (formerly Beijing Genomics Institute). Additional details are provided in the supplementary material.
Real-time RT-PCR
Reverse transcription of total RNA (2 μg) was performed using SuperScript II reverse transcriptase. The target and reference (GAPDH) genes were quantified in triplicate on a LightCycler® 480 II (Roche, Applied Science) using a SYBR green-based assay (Bio-Rad, USA). The primers used in the real-time RT-PCR reaction are shown in Table S1.
Western blot analysis
The Western blot analysis was performed using standard protocols with antibodies against CHI3L1 (1:1000; Abcam, UK), MMP13 (1:1000; R&D systems, USA), SPP1 (1:500; Abcam, UK) and α-tubulin (1:3000, Abcam, UK).
Immunohistochemistry
Immunohistochemistry was performed using standard protocols with antibodies against CHI3L1 (1:50; Abcam, UK), MMP13 (1:100; R&D systems, USA), and SPP1 (1:100; Abcam, UK). Further details are provided in the supplementary material.
ELISA
Serum biomarker levels were determined using a double-antibody sandwich ELISA according to the manufacturer's instructions (CHI3L1, R&D systems, USA; MMP13, CUSABIO, China; SPP1, ebioscience, USA). They were analyzed blinded to clinical parameters and study endpoints at the end of the study.
Briefly, 100 μl of the test samples (1:100 diluted for CHI3L1, original for MMP13, 1:5 diluted for SPP1) were added and incubated for 2 h at room temperature. Subsequently, 100 μl/well of the detection antibody was added and incubated for 2 h at room temperature. Next, 100 μl/well of Streptavidin-HRP was added and incubated for 20 min at room temperature. Finally, the substrate (tetramethylbenzidine) solution was added, and the reaction was stopped with 2 N H2SO4 and read at an OD of 450 nm. For Intra-assay Precision: CHI3L1, CV: 5.2% (0.25 ng/ml) and CV: 3.6% (2 ng/ml); MMP13, 4.9% (2 ng/ml) and 2.3% (10 ng/ml); SPP1, 4.1% (2 ng/ml) and 2.9% (10 ng/ml). For Inter-assay Precision: CHI3L1, 10.1% and 7.5%; MMP13, 8.7% and 6.1%; SPP1, 6.6% and 5.6%, respectively.
CEA assay
The concentrations of CEA in the serum were assessed using electrochemiluminescence immunoassay (ECLIA) kits (Roche, German) on a Roche E170 analyzer (Roche, German). The test included a standard control (CV < 5%).
Statistical analysis
The statistical analyses were performed using SPSS 16.0 (SPSS Inc.). The comparisons of each protein concentration among the different groups were assessed using the Mann-Whitney test. The efficacy of each protein was evaluated by the AUC. To assess whether the combined use of biomarkers was better than either of these biomarkers alone, a new variable was used to predict the probability (p) for ESCC based on an equation obtained by binary logistic regression (all ESCC versus all control groups in the test cohort). The cutoff value for each protein was defined when the sensitivity reached 90% in the test cohort. Furthermore, the sensitivity (Sen), specificity (Spe), positive predictive value (PPV) and negative predictive value (NPV) were used to compare the diagnostic efficiency. All statistical tests were two-sided, and P < 0.05 was considered statistically significant.
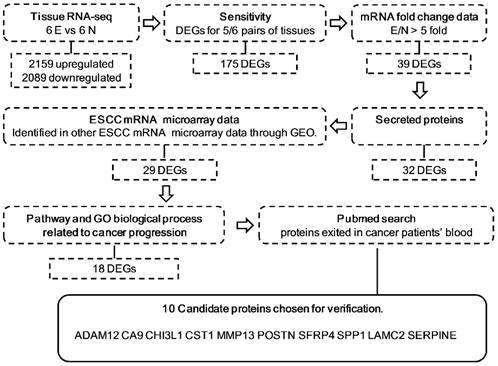
Schematic representation of the approach used for candidate biomarker selection in this study. Secreted proteins were selected using SignalP 4.1 and Secretome 2.0 software. The expression levels of the candidates were tested by examining 3 mRNA expression microarrays: GSE23400, GSE20347, and GSE33810. Pathway and GO biological processes related to cancer progression were determined with Genecards and DAVID. E, ESCC tissues; N, ESCC adjacent normal tissues; DEG, differentially expressed genes; GEO, Gene Expression Omnibus.
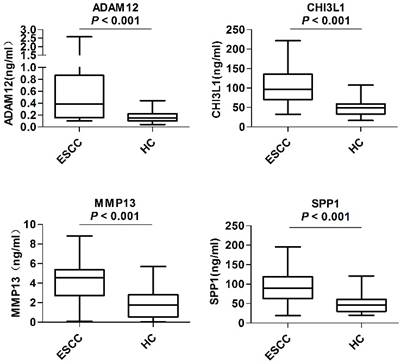
Serum levels of candidate biomarkers in the preliminary screening phase. Levels of serum ADAM12, CHI3L1, MMP13 and SPP1 were compared between 40 ESCC patients (ESCC) and 40 healthy controls (HC). The Mann-Whitney U test was performed for comparisons between groups. P < 0.05 was considered statistically significant.
Results
Candidate selection
The procedure and criterion for selecting the candidate biomarkers from the RNA-seq database are described in Figure 1. There were 2159 upregulated and 2089 downregulated DEGs, on average, based on an analysis of the RNA transcriptomes of six pairs of ESCC tissues. To guarantee the sensitivity and reliability of the selected markers for the diagnosis of ESCC, we first selected 175 genes, which were differentially expressed in at least 5 out of 6 tissue pairs (Figure S1). Because most of the traditional tumor markers used in the clinical laboratory, such as CEA, AFP and PSA, were upregulated in cancer patient serum and upregulated makers are easier to detect in serum, we selected 39 DEGs that were upregulated at least 5-fold in tumor tissues compared with non-cancerous tissues (Figure S2). Among these 39 DEGs, we selected 32 secretory proteins using SignalP4.1 and SecretomeP 2.0 (Figure S3). To confirm that these markers were indeed upregulated in ESCC, the expression levels of the candidates were tested by examining 3 mRNA expression microarrays of ESCC tissues and normal tissues in the PubMed GEO database (Figure S4). Twenty-nine candidates were used for further verification, including 28 that were upregulated in most data and AMTN, which was not detected in any of the three data sets because the microarrays may not encompass this gene. To further refine our candidate list, we selected genes related to ESCC pathways. Table S2 shows that 18 of the 29 candidates were related to ESCC pathways. Table S3 shows that these 18 candidates were associated with tumor biological processes such as cell adhesion, angiogenesis and cell growth. Furthermore, 10 of the 18 candidates, ADAM12, CA9, CHI3L1, CST1, LAMC2, POSTN, SFRP4, SPP1, MMP13, WISP1 and SERPINE1, showed evidence that they were present in the serum of ESCC or other cancer patients (Table S4). These ten candidates were subjected to an ELISA analysis using serum from patients with ESCC.
The preliminary screening phase
In the preliminary screening phase, the levels of the above-mentioned 10 candidates in the serum samples from 40 ESCC patients and 40 healthy controls were examined. As shown in Figure 2, the serum levels of ADAM12 (P < 0.001), CHI3L1 (P < 0.001), MMP13 (P < 0.001) and SPP1 (P < 0.001) were significantly elevated in the ESCC patients compared with the healthy controls. The remaining six proteins, CA9, CST1, POSTN, SFRP4, LAMC2 and SERPINE1, provided poor discriminatory performance and thus were excluded from subsequent analyses (Figure S5).
Clinical characteristics of the ESCC patients, healthy controls and patients with esophageal benign disease in the test and validation cohorts
| Characteristics | Test cohort (n=290) | Validation cohort (n=323) | ||||||
|---|---|---|---|---|---|---|---|---|
| n | % | n | % | P | ||||
| Healthy controls | 96 | 80 | ||||||
| Age, years | 0.0841 | |||||||
| Median (Range) | 53(25-73) | 56(34-79) | ||||||
| Gender | 0.3463 | |||||||
| Male/ Female | 52/44 | 54/46 | 49/31 | 61/39 | ||||
| ESCC | 150 | 169 | ||||||
| Age, years | 0.9840 | |||||||
| Median (Range) | 60.5(39-83) | 60(39-82) | ||||||
| Gender | 0.1546 | |||||||
| Male/ Female | 117/33 | 78/22 | 120/49 | 71/29 | ||||
| pT classification | 0.0569 | |||||||
| pT1 | 21 | 14 | 18 | 11 | ||||
| pT2 | 23 | 15 | 27 | 16 | ||||
| pT3 | 99 | 66 | 98 | 58 | ||||
| pT4 | 7 | 5 | 26 | 15 | ||||
| pN classification | 0.445 | |||||||
| pN0 | 65 | 43 | 78 | 46 | ||||
| pN1 | 56 | 37 | 69 | 41 | ||||
| pN2 | 25 | 17 | 13 | 8 | ||||
| pN3 | 3 | 2 | 9 | 5 | ||||
| Metastasis | 0.1634 | |||||||
| pM0 | 142 | 95 | 153 | 91 | ||||
| pM1 | 8 | 5 | 16 | 9 | ||||
| Clinical stage | 0.6276 | |||||||
| Stage I | 20 | 13 | 13 | 8 | ||||
| Stage II | 51 | 34 | 71 | 42 | ||||
| Stage III | 71 | 47 | 69 | 41 | ||||
| Stage IV | 8 | 5 | 16 | 9 | ||||
| Benign disease patients 44 | 74 | |||||||
| Age, years | 0.5909 | |||||||
| Median (Range) | 52(28-78) | 53(21-78) | ||||||
| Gender | 0.4422 | |||||||
| Male/ Female | 27/17 | 61/39 | 40/34 | 54/46 | ||||
Abbreviations: ESCC:esophageal squamous cell carcinoma
Performance of 4 markers in the test and validation cohorts
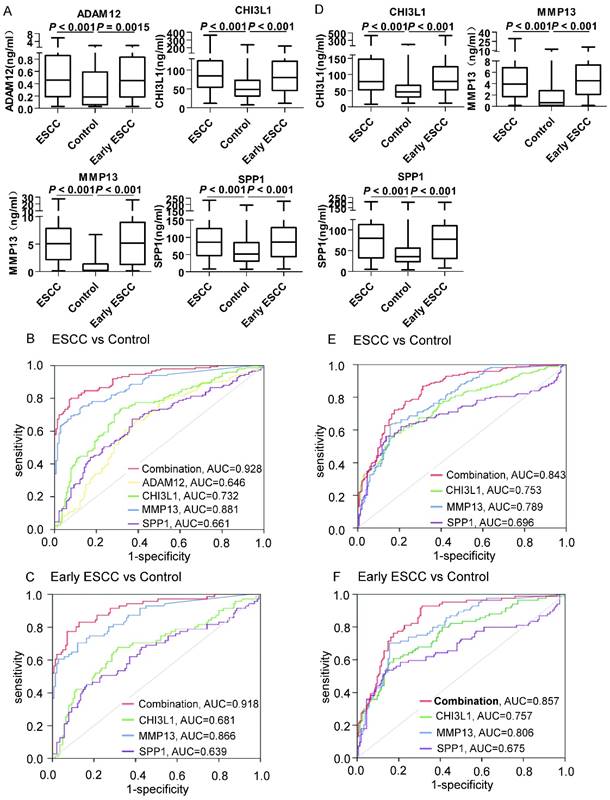
We utilized serum samples from 150 ESCC patients and 140 controls (44 patients with benign esophageal disease and 96 healthy controls, Table 1) to validate the discriminatory capability of ADAM12, CHI3L1, MMP13 and SPP1. As shown in Figure 3A, the serum levels of these four markers were significantly higher in the ESCC patients than in the controls (P < 0.001). Moreover, the levels of the 4 proteins were significantly increased even in comparisons of only early-stage ESCC patients and controls. Next, we generated ROC curves to evaluate the performance of the four proteins for the detection of ESCC. The AUC values for serum ADAM12, CHI3L1, MMP13 and SPP1 in discriminating the ESCC patients from the controls (Figure 3B) were 0.646 (95% CI, 0.582 to 0.710), 0.732 (0.673 to 0.790), 0.881 (0.842 to 0.919) and 0.661 (0.598 to 0.724), respectively. Because we aimed to screen ESCC, the appropriate cutoffs for each marker were obtained when the sensitivity achieved 90%, as shown in Table 2. The details of cut-off values for CHI3L1, MMP13, and SPP1 for each stage are shown in the Table S5 and supplementary materials. Based on the ROC analysis in the test cohort comparing all ESCC (n=150) versus all controls (n=140), the highest specificity was 57.86%, stemming from MMP13, and the details of the specificity, PPV and NPV are shown in Table 2.
The diagnostic performance of ADAM12, CHI3L1, MMP13, SPP1 and their combination (Logit(p=ESCC)=-4.583+0.017×CHI3L1+0.018×SPP1+0.821×MMP13) in discriminating ESCC, early stage ESCC and controls (healthy controls and patients with esophageal benign disease) in the test cohort and validation cohort.
| Markers(ng/ml) | AUC | Cut-off | Sen (%) | Spe (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|
| ESCC in the test cohort | ||||||
| ADAM12 | 0.646 | 0.068 | 90 | 29.29 | 57.69 | 73.21 |
| CHI3L1 | 0.732 | 33.100 | 90 | 25.71 | 56.49 | 70.59 |
| MMP13 | 0.881 | 0.438 | 90 | 57.86 | 69.59 | 84.38 |
| SPP1 | 0.661 | 23.920 | 90 | 15.00 | 53.15 | 58.33 |
| Combination | 0.928 | 0.282 | 90 | 72.14 | 77.59 | 87.07 |
| Early Stage ESCC in the test cohort | ||||||
| CHI3L1 | 0.681 | 33.100 | 83.10 | 25.71 | 36.20 | 75.00 |
| MMP13 | 0.866 | 0.438 | 88.73 | 57.86 | 51.64 | 91.01 |
| SPP1 | 0.639 | 23.920 | 84.51 | 15.00 | 33.52 | 65.63 |
| Combination | 0.918 | 0.282 | 90.14 | 72.14 | 62.14 | 93.52 |
| ESCC in the validation cohort | ||||||
| CHI3L1 | 0.753 | 33.100 | 89.35 | 29.87 | 58.30 | 71.88 |
| MMP13 | 0.789 | 0.438 | 91.12 | 44.81 | 64.43 | 82.14 |
| SPP1 | 0.696 | 23.920 | 80.47 | 25.97 | 54.40 | 54.79 |
| Combination | 0.843 | 0.282 | 84.02 | 69.48 | 75.13 | 79.85 |
| Early Stage ESCC in the validation cohort | ||||||
| CHI3L1 | 0.757 | 33.100 | 89.29 | 29.87 | 40.98 | 83.63 |
| MMP13 | 0.806 | 0.438 | 92.86 | 44.81 | 47.85 | 92.00 |
| SPP1 | 0.675 | 23.920 | 79.76 | 25.97 | 37.02 | 70.18 |
| Combination | 0.857 | 0.282 | 90.48 | 69.48 | 61.79 | 93.04 |
Abbreviations: ESCC:esophageal squamous cell carcinoma; Sen, sensitivity; Spe, specificity; PPV, positive predictive value; NPV, negative predictive value; HC,healthy control.
Moreover, a binary logistic regression analysis was applied to establish a diagnostic model: Logit(p=ESCC)=-4.583+0.017×CHI3L1+0.018×SPP1+0.821×MMP13 (ADAM12 was ruled out because it provided a P > 0.05). The model combined CHI3L1, MMP13 and SPP1, improving the performance of an individual marker, with an AUC of 0.928 (95% CI, 0.900 to 0.956), a sensitivity of 90%, and a specificity of 72.14%. To discriminate early-stage ESCC patients from controls (Figure 3C), the AUC value for the combination was 0.918 (95% CI, 0.876 to 0.959) and the specificity was 72.14%; the diagnostic details of the individual markers for discriminating early-stage ESCC are shown in Table 2. Similarly, when applied to the blinded validation cohort (169 patients with ESCC, 74 patients with benign esophageal disease and 80 healthy controls, Table 1), the three markers and their combination were comparable for distinguishing ESCC from controls (Figure 3D-E, AUC combination = 0.843; Table 2, Specificity combination= 69.48%;) and early-stage ESCC from controls (Figure 3F, AUC combination = 0.857; Table 2, Specificity combination= 69.48%). The combination panel performed better than the marker CEA. The details were seen in the supplementary materials and Figure S6.
The associations between the median serum CHI3L1, MMP13, SPP1 levels and the clinicopathological parameters are presented in Table S6. Serum CHI3L1 and MMP13 were only significantly associated with age (P = 0.0002 and P = 0.0267, respectively). Serum SPP1 was only correlated with gender and T classification (P = 0.0037 and P =0.0165, respectively). More details are provided in the supplementary materials.
Serum ADAM12, CHI3L1, MMP13 and SPP1 in the test cohort and the validation cohort. A: Levels of biomarkers were compared between 150 ESCC patients (ESCC), 140 controls (96 healthy controls and 44 patients with benign esophageal disease) and 71 early-stage ESCC patients. Statistical analyses were performed using the Mann-Whitney U test. B: ROC curves for biomarkers and their combination (Logit(p=ESCC)=-4.583+0.017×CHI3L1+0.018×SPP1+0.821×MMP13) for the discrimination of 150 patients with ESCC and 140 controls. C: ROC curves for biomarkers and their combination for the discrimination of 71 patients with early-stage ESCC and 140 controls. D: Levels of biomarkers were compared in 169 ESCC patients (ESCC), 154 controls (80 healthy controls and 74 patients with benign esophageal disease) and 84 early-stage ESCC patients. Statistical analyses were performed using the Mann-Whitney U test. E: ROC curves for biomarkers and their combination for the discrimination of 169 patients with ESCC and 154 controls. F: ROC curves for biomarkers and their combination (Logit(p=ESCC)=-4.583+0.017×CHI3L1+0.018×SPP1+0.821×MMP13) for the discrimination of 84 patients with early-stage ESCC and 154 controls. P < 0.05 was considered statistically significant.
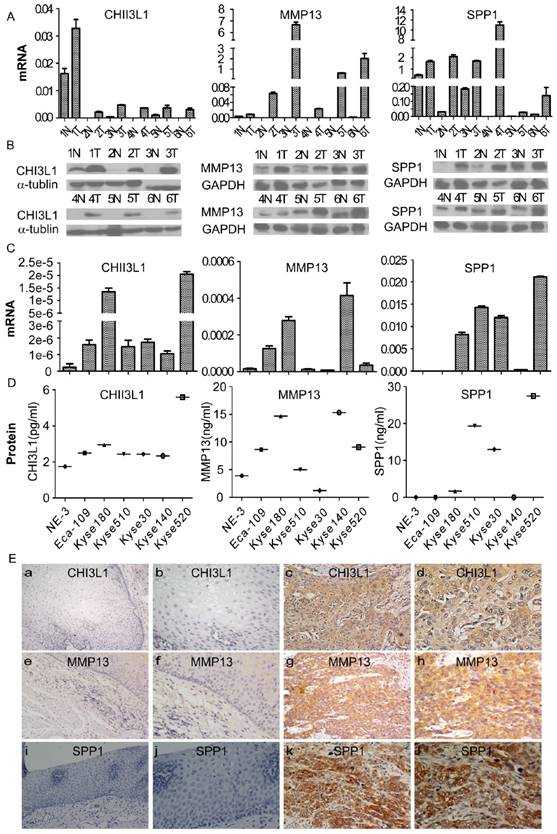
Expression of CHI3L1, MMP13 and SPP1 mRNA or protein in ESCC cell lines and tissues and their locations in tissue. The mRNA and protein expression levels in six pairs of matched ESCC and noncancerous tissues was analyzed by real-time PCR and Western blotting, respectively (A, B), and in an immortalized esophageal epithelial cell line (NE-3) and esophageal carcinoma cell lines by real-time PCR and ELISA, respectively (C, D). The expression level was normalized to the expression of GAPDH and α-tubulin, respectively. Error bars represent standard deviations (SD) calculated from three parallel experiments. The locations of CHI3L1, MMP13 and SPP1 were determined by immunohistochemistry (E). The normal esophageal epithelial tissue showed no expression of CHI3L1, MMP13 or SPP1, respectively (E a, e, i, 200 × and b, f, j, 400×). The ESCC tissues exhibited high expression levels of these three biomarkers (E c, f, k, 200 × and d, h, l, 400×).
Expression of MMP13, SPP1 and CHI3L1 in esophageal carcinoma cell lines and tumor tissues
To investigate the expression of MMP13, SPP1 and CHI3L1 in ESCC, the mRNA and protein levels of the 3 markers in six pairs of matched ESCC tissues and adjacent noncancerous tissues were detected by real-time RT-PCR and Western blot analysis. As shown in Figure 4A and 4B, the expression levels of all three markers in the six ESCC samples were much higher compared with the paired adjacent noncancerous tissues both at the mRNA and protein level.
Furthermore, a comparative analysis was performed to evaluate the levels of CHI3L1, MMP13 and SPP1 in cells; the mRNA and protein levels of the 3 markers in 6 esophageal carcinoma cell lines and the immortalized esophageal epithelial cell line NE-3 were detected by real-time RT-PCR and Western blotting, as shown in Figure 4C and 4D. The CHI3L1 mRNA and protein levels were elevated in all tumor cell lines compared with the immortalized cell line NE-3. By comparison, the mRNA and protein levels of MMP13 were significantly increased in Eca-109, Kyse140, and Kyse180. Similarly, SPP1 mRNA and protein levels were highly expressed in Kyse30, Kyse180, Kyse510 and Kyse520.
To further investigate the precise expression state of CHI3L1, MMP13 and SPP1 in vivo, the protein expression of the three markers was determined by immunohistochemistry (Figure 4E). CHI3L1 was mainly located in the cytoplasm of tumor cells, as well as in tumor stromal cells. MMP1 and SPP1 were located in the cytoplasm of tumor cells. The expression levels of the three markers in the tumor cells were observed at various levels. Protein expression of CHI3L1, MMP13 and SPP1 was detected in 17 of 20 (85%), 16 of 20 (80%) and 18 of 20 (90%) ESCC samples, respectively, whereas the detection levels were 10%, 15% and 10% in the noncancerous tissues. In conclusion, the expression levels of CHI3L1, MMP13 and SPP1 were elevated in ESCC tissues compared with the noncancerous tissues.
Discussion
In this study, we screened out three markers, CHI3L1, MMP13, and SPP1, from more than 4000 differentially expressed genes in the ESCC transcriptome database. These markers were not only significantly elevated in ESCC patient serum, but they also displayed a relatively high expression level in esophageal cancer cells in comparison to normal esophageal cells and were shown to be useful for the diagnosis of ESCC.
For tumor detection, biomarkers in the serum are most applicable for clinical routine assessments because, in general, the tests are non-invasive and low cost, have a low dependence on operator expertise, and are highly reproducible.[14-16] CEA, CYFRA21-1 and SCCA are the most commonly used protein tumor markers for ESCC diagnosis.[17] However, these markers exhibit a high specificity but a low sensitivity for ESCC detection.[18-20] Our group previously found that CEA, CYFRA21-1, and SCCA, applied for the diagnosis of ESCC, only had a sensitivity of 10.64%, 40.43%, and 32.67%, respectively.[21] We also confirmed the low sensitivity of CEA in all test and validation samples. To increase the sensitivity, some studies have used proteomics technology to screen tumor markers from serum. However, proteins with a low abundance may be missed due to interference from highly abundant proteins in serum. Because the variation in mRNA expression could reflect the protein level,[22] it is reasonable to speculate that an increase in mRNA expression could lead to overexpression of the protein, and screening tumor markers based on their cancer-specific mRNA expression is feasible. In fact, recent studies have discovered multiple protein tumor markers by using mRNA microarray analysis of tumor tissues and non-cancerous tissues.[23, 24] We attempted to identify serum tumor markers for the diagnosis of ESCC using an RNA transcriptome sequencing database of tumor tissues and non-cancerous tissues. Our hypothesis is based on the following observations. 1) Tumor markers are produced directly by the tumor or by non-tumor cells as a response to the presence of a tumor.[25] The RNA transcriptome sequencing database of ESCC tissues contains almost all of the tumor-related markers, ensuring that the candidates with a low abundance are not missed. Tumor markers could be screened out by analyzing the DEGs in the tumor tissues versus the adjacent normal tissues. 2) Secretory proteins are derived from DEGs, which are highly expressed in ESCC tissues and may be detected at higher levels in blood. 3) Appropriate filtering rules, including an expression frequency of 80% and upregulation by at least 5-fold in ESCC tissues, were used to select biomarkers with a high sensitivity and reliability for the detection of ESCC. 4) Because reproducibility is one of the most important components in the discovery of clinically relevant biomarkers, third-party validation was applied to ensure the specificity of the candidates. Other mRNA expression microarrays of ESCC tissues and non-cancerous tissues were used to test the repeatability of the candidate markers. This procedure enabled us to choose markers that have been identified by independent researchers using different patient cohorts. In addition, we used an esophageal cancer-associated signaling pathway analysis, a tumor-associated functional analysis and a retrospective analysis of the literature to identify useful tumor markers for the diagnosis of ESCC. This strategy ensured the selection of a few potential tumor markers from a large number of DEGs.
Using this strategy, ten candidates were screened from 4248 DEGs. Subsequently, the preliminary screening phase analysis indicated that 4 of the 10 markers, including ADAM12, CHI3L1, MMP13 and SPP1, were present at high levels in serum from ESCC patients and could significantly classify patients with cancer versus healthy controls. Next, we analyzed a cohort of 613 serum samples in the test and validation cohorts. There were no significant differences in serum CHI3L1, MMP13 and SPP1 levels between patients with early-stage tumors (I-II) and those with advanced-stage tumors (III-IV). These results indicate that the three markers can be used for the detection of early ESCC as well as advanced ESCC. All four markers independently possess a certain diagnostic power for discriminating patients with ESCC from controls. Overexpression of CHI3L1, MMP13 and SPP1, both in ESCC tumor cell lines and in tumor tissues, might contribute to their high serum levels. In agreement with our data, an elevated level of serum MMP-13 was detected in the patients with ESCC, and a high level of serum MMP-13 is associated with tumor progression and poor survival.[26] It is noteworthy that MMP13 plays a role in destroying the extracellular matrix and basement membrane, which leads to tumor cell invasion and distant metastasis, mediating tumor angiogenesis, and adjusts the adhesion of tumor cells and the matrix.[27, 28] Previous studies have shown that SPP1 is widely upregulated and considered to be a carcinogenic gene in tumors, such as liver and lung cancer, colorectal cancer, and esophageal tumors, participating in cell adhesion, apoptosis, angiogenesis, tumor-associated inflammation, and tumor metastasis.[29] Our prior report showed that CHI3L1 is expressed at high levels in ESCC, and serum CHI3L1 achieved a higher diagnostic performance than any of the traditional tumor markers, including CEA, CYFRA21-1, and SCCA.[21] ADAM12 is also overexpressed in many tumors, such as lung cancer, gastric cancer and colorectal cancer.[30] In the present study, ADAM12 demonstrated an AUC of 0.7 for the diagnosis of ESCC, whereas when we used logistic regression to develop a diagnostic model for binary outcomes, it was eliminated because it did not improve the diagnostic effect when used in combination with the other three biomarkers. The diagnostic capabilities of these markers were generally similar in the test and validation cohorts. Among them, MMP13 displayed the best performance; however, the false-positive rate still needs improvement. By performing the logistic regression, we were able to combine an appropriate panel using CHI3L1, MMP13 and SPP1, which appeared to have greater diagnostic power than any individual marker, reducing the false-positive rate with the same missing positive rate. The effect of the combination was evaluated and validated in a multicenter trial of independent samples. Importantly, this diagnostic model identified approximately 90% of the patients with early-stage ESCC, suggesting that this model may be useful for ESCC screening. The limitation of this study is that the downregulated markers were not taken into consideration, and thus some potential tumor markers might have been missed. Additionally, the prognostic value of CHI3L1, MMP13 and SPP1 requires further exploration in a follow-up study. In addition, further investigations with larger patient numbers are required to refine the diagnostic algorithms with the expectation that an efficient, optimal diagnostic strategy will improve patient outcomes.
In summary, we have screened out three serum biomarkers, CHI3L1, MMP13, and SPP1, for the diagnosis of ESCC by analyzing the RNA transcriptome database of tumor tissues and non-cancerous tissues. A combination of CHI3L1, MMP13 and SPP1 may be a robust predictor of the occurrence of ESCC.
Abbreviations
ESCC: esophageal squamous cell carcinoma;
RNA-seq: RNA sequencing;
AUC: area under the curve;
DEGs: differentially expressed genes;
SYSUCC: Sun Yat-sen University Cancer Center;
STCH: Shantou central hospital;
SPPH: Shaanxi provincial people's hospital;
Sen: sensitivity;
Spe: specificity;
PPV: positive predictive value;
NPV: negative predictive value;
HC: healthy control;
ADAM12: ADAM metallopeptidase domain 12;
CHI3L1: chitinase 3 like 1;
MMP13: matrix metallopeptidase 13;
SPP1: secreted phosphoprotein 1;
CA9: carbonic anhydrase IX;
CST1: cystatin SN;
LAMC2: laminin subunit gamma 2;
POSTN: periostin, osteoblast specific factor;
SFRP4: secreted frizzled-related protein 4;
WISP1: WNT1 inducible signaling pathway protein 1;
SERPINE1: serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
We would like to thank all of the study participants for agreeing to participate in medical research.
Financial support
This work was supported by the National Natural Science Foundation of China (Wan-Li Liu, No.81271902 and No.81472008). The funding organizations played no role in the design of the study, choice of enrolled patients, review and interpretation of data, or preparation or approval of manuscript.
Ethics approval and consent to participate
Prior to use of these serum and tissues, informed consent was obtained from each of the participants. This experiment was approved by the Institute Research Ethics Committee of the Cancer Center of SYSUCC, Guangzhou, China, STCH, Shantou, China and the SPPH, Shanxi, China.
Authors' contributions
SX, XZ participated in the experiment designs and carried out the experiment.
SS, DL, NX and LW collected the samples.
LW provided the samples from the validation cohort.
SX and WL drafted the manuscript and interpreted data.
MW and CH performed the statistical analysis.
QZ and XL helped to collect the clinical data of patients.
WL designed the experiment and revised the manuscript.
MZ provided the equipment, materials.
All authors read and approved the final manuscript.
Competing Interests
The authors declare no potential conflicts of interest.
References
1. Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: Defining priorities to reduce cancer disparities in different geographic regions of the world[J]. J Clin Oncol. 2006;24(14):2137-2150
2. Song Y, Li L, Ou Y. et al. Identification of genomic alterations in oesophageal squamous cell cancer[J]. Nature. 2014;509(7498):91-95
3. Kuroki T, Trapasso F, Yendamuri S. et al. Allele loss and promoter hypermethylation of VHL, RAR-beta, RASSF1A, and FHIT tumor suppressor genes on chromosome 3p in esophageal squamous cell carcinoma[J]. Cancer Res. 2003;63(13):3724-3728
4. Yamamoto S, Inoue J, Kawano T. et al. The impact of miRNA-based molecular diagnostics and treatment of NRF2-stabilized tumors[J]. Mol Cancer Res. 2014;12(1):58-68
5. Zhang C, Wang C, Chen X. et al. Expression profile of microRNAs in serum: A fingerprint for esophageal squamous cell carcinoma[J]. Clin Chem. 2010;56(12):1871-1879
6. Fan NJ, Gao CF, Zhao G. et al. Serum peptidome patterns for early screening of esophageal squamous cell carcinoma[J]. Biotechnol Appl Biochem. 2012;59(4):276-282
7. Liu LH, Shan BE, Tian ZQ. et al. Potential biomarkers for esophageal carcinoma detected by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry[J]. Clin Chem Lab Med. 2010;48(6):855-861
8. Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics[J]. Genomics. 2008;92(5):255-264
9. Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing[J]. Nat Rev Genet. 2010;11(10):685-696
10. Trapnell C, Williams BA, Pertea G. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation[J]. Nat Biotechnol. 2010;28(5):511-515
11. Ma S, Bao JY, Kwan PS. et al. Identification of PTK6, via RNA sequencing analysis, as a suppressor of esophageal squamous cell carcinoma[J]. Gastroenterology. 2012;143(3):675-686
12. Tong M, Chan KW, Bao JY. et al. Rab25 is a tumor suppressor gene with antiangiogenic and anti-invasive activities in esophageal squamous cell carcinoma[J]. Cancer Res. 2012;72(22):6024-6035
13. Yang LX, Li BL, Liu XH. et al. RNA-seq reveals determinants of sensitivity to chemotherapy drugs in esophageal carcinoma cells[J]. Int J Clin Exp Pathol. 2014;7(4):1524-1533
14. Malaguarnera G, Giordano M, Paladina I. et al. Serum markers of hepatocellular carcinoma[J]. Dig Dis Sci. 2010;55(10):2744-2755
15. Brawley OW, Ankerst DP, Thompson IM. Screening for prostate cancer[J]. CA Cancer J Clin. 2009;59(4):264-273
16. Locker GY, Hamilton S, Harris J. et al. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer[J]. J Clin Oncol. 2006;24(33):5313-5327
17. Mealy K, Feely J, Reid I. et al. Tumour marker detection in oesophageal carcinoma[J]. Eur J Surg Oncol. 1996;22(5):505-507
18. Dong J, Zeng BH, Xu LH. et al. Anti-CDC25B autoantibody predicts poor prognosis in patients with advanced esophageal squamous cell carcinoma[J]. J Transl Med. 2010;8:81
19. Kawaguchi H, Ohno S, Miyazaki M. et al. CYFRA 21-1 determination in patients with esophageal squamous cell carcinoma: Clinical utility for detection of recurrences[J]. Cancer. 2000;89(7):1413-1417
20. Yamamoto K, Oka M, Hayashi H. et al. CYFRA 21-1 is a useful marker for esophageal squamous cell carcinoma[J]. Cancer. 1997;79(9):1647-1655
21. Zheng X, Xing S, Liu XM. et al. Establishment of using serum YKL-40 and SCCA in combination for the diagnosis of patients with esophageal squamous cell carcinoma[J]. Bmc Cancer. 2014;14:490
22. Schwanhausser B, Busse D, Li N. et al. Global quantification of mammalian gene expression control[J]. Nature. 2011;473(7347):337-342
23. Zen K, Yasui K, Gen Y. et al. Defective expression of polarity protein PAR-3 gene (PARD3) in esophageal squamous cell carcinoma[J]. Oncogene. 2009;28(32):2910-2918
24. Yamabuki T, Takano A, Hayama S. et al. Dikkopf-1 as a novel serologic and prognostic biomarker for lung and esophageal carcinomas[J]. Cancer Res. 2007;67(6):2517-2525
25. Patz EJ, Campa MJ, Gottlin EB. et al. Panel of serum biomarkers for the diagnosis of lung cancer[J]. J Clin Oncol. 2007;25(35):5578-5583
26. Etoh T, Inoue H, Yoshikawa Y. et al. Increased expression of collagenase-3 (MMP-13) and MT1-MMP in oesophageal cancer is related to cancer aggressiveness[J]. Gut. 2000;47(1):50-56
27. Meierjohann S, Hufnagel A, Wende E. et al. MMP13 mediates cell cycle progression in melanocytes and melanoma cells: In vitro studies of migration and proliferation[J]. Mol Cancer. 2010;9:201
28. Ala-aho R, Ahonen M, George SJ. et al. Targeted inhibition of human collagenase-3 (MMP-13) expression inhibits squamous cell carcinoma growth in vivo[J]. Oncogene. 2004;23(30):5111-5123
29. Rangaswami H, Bulbule A, Kundu GC. Osteopontin: Role in cell signaling and cancer progression[J]. Trends Cell Biol. 2006;16(2):79-87
30. Mochizuki S, Okada Y. ADAMs in cancer cell proliferation and progression[J]. Cancer Sci. 2007;98(5):621-628
Author contact
![]() Corresponding authors: Wan-Li Liu. Email: liuwlorg.cn; Phone: 86-020-87343196. Department of Clinical Laboratory, Sun Yat-sen University Cancer Center, 651 Dongfeng Road East,Guangzhou 510060, P.R. China. Mu-Sheng Zeng; Email: ngmshorg.cn Phone: +86-020-87343192. Department of Experimental Research, Sun Yat-sen University cancer center, 651 Dongfeng Road East,Guangzhou 510060, P.R.China
Corresponding authors: Wan-Li Liu. Email: liuwlorg.cn; Phone: 86-020-87343196. Department of Clinical Laboratory, Sun Yat-sen University Cancer Center, 651 Dongfeng Road East,Guangzhou 510060, P.R. China. Mu-Sheng Zeng; Email: ngmshorg.cn Phone: +86-020-87343192. Department of Experimental Research, Sun Yat-sen University cancer center, 651 Dongfeng Road East,Guangzhou 510060, P.R.China

 Global reach, higher impact
Global reach, higher impact