3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(7):1916-1928. doi:10.7150/jca.91546 This issue Cite
Research Paper
Identification of a lncRNA/circRNA-miRNA-mRNA network in Nasopharyngeal Carcinoma by deep sequencing and bioinformatics analysis
1. NHC Key Laboratory of Cancer Proteomics, Department of Oncology, Xiangya Hospital, Central South University, Changsha, Hunan, P.R. China, 410008.
2. National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan, P.R. China, 410008.
3. Department of Pathology, Changsha Medical College, Changsha, Hunan, P.R. China, 410219.
4. Department of Otolaryngology Head and Neck Surgery, The Second Xiangya Hospital, Central South University, Changsha, Hunan, P.R. China, 410011.
# These authors contributed equally to this work.
Abstract
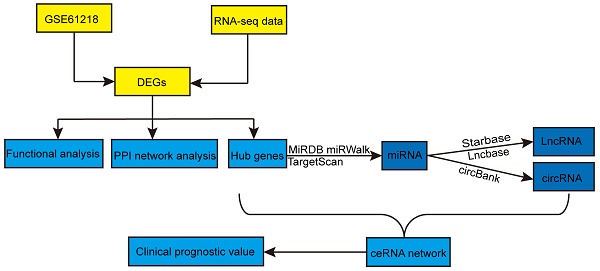
Background: Accumulating evidence indicates that non-coding RNAs (ncRNA), including long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs), can function as competitive endogenous RNAs (ceRNAs) by binding to microRNAs (miRNAs) and regulating host gene expression at the transcriptional or post-transcriptional level. Dysregulation in ceRNA network regulation has been implicated in the occurrence and development of cancer. However, the lncRNA/circRNA-miRNA-mRNA regulatory network is still lacking in nasopharyngeal carcinoma (NPC).
Methods: Differentially expressed genes (DEGs) were obtained from our previous sequencing data and Gene Expression Omnibus (GEO). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes pathway (KEGG) were used to explore the biological functions of these common DEGs. Through a series of bioinformatic analyses, the lncRNA/circRNA-miRNA-mRNA network was established. In additional, the external data GSE102349 was used to test the prognostic value of the hub mRNAs through the Kaplan-Meier method.
Results: We successfully constructed a lncRNA/circRNA-miRNA-mRNA network in NPC, consisting of 16 lncRNAs, 6 miRNAs, 3 circRNAs and 10 mRNAs and found that three genes (TOP2A, ZWINT, TTK) were significantly associated with overall survival time (OS) in patients.
Conclusion: The regulatory network revealed in this study may help comprehensively elucidate the ceRNA mechanisms driving NPC, and provide novel candidate biomarkers for evaluating the prognosis of NPC.
Keywords: bioinformatics, competitive endogenous RNA, deep sequencing, nasopharyngeal carcinoma, non-coding RNAs

 Global reach, higher impact
Global reach, higher impact