Impact Factor
ISSN: 1837-9664
J Cancer 2012; 3:231-240. doi:10.7150/jca.4542 This volume Cite
Research Paper
A Pilot Study Assessing the Potential Role of non-CD133 Colorectal Cancer Stem Cells as Biomarkers
1. National Cancer Institute, National Institutes of Health, Bethesda, MD.
2. Biostatistics and Data Management Section, Office of the Clinical Director, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD.
3. Pathology/Histotechnology Laboratory, SAIC Frederick, Inc. Frederick National Laboratory for Cancer Research, Frederick, MD.
4. John Wayne Cancer Institute, Santa Monica, CA.
5. Department of Surgery, Division of Surgical Oncology, Walter Reed National Military Medical Center, Bethesda, MD.
6. Department of Surgery, Uniformed Services University of the Health Sciences, Bethesda, MD.
7. Bon Secours Cancer Institute, Richmond VA.
Received 2012-5-1; Accepted 2012-5-22; Published 2012-6-1
Abstract
Introduction: Over 50% of patients with colorectal cancer (CRC) will progress and/or develop metastases. Biomarkers capable of predicting progression, risk stratification and therapeutic benefit are needed. Cancer stem cells are thought to be responsible for tumor initiation, dissemination and treatment failure. Therefore, we hypothesized that CRC cancer stem cell markers (CRCSC) will identify a group of patients at high risk for progression.
Methods: Paraffin-embedded tissue cores of normal (n=8), and histopathologically well-defined primary (n= 30) and metastatic (n=10) CRC were arrayed in duplicate on tissue microarrays (TMAs). Expression profiles of non-CD133 CRCSC (CD29, CD44, ALDH1A1, ALDH1B1, EpCam, and CD166) were detected by immunohistochemistry and the association with clinicopathological data and patient outcomes was determined using standard statistical methodology. An independent pathologist, blinded to the clinical data scored the samples. Scoring included percent positive cells (0 to 4, 0 = <10%, 1 = 10 - 24%, 2 = 25 - 49%, 3 = 50 - 74%, 4 = 75 - 100%), and the intensity of positively stained cells (0 to 4; 0 = no staining, 1 = diminutive intensity, 2 = low intensity, 3 = intermediate intensity, 4 = high intensity). The pathologic score represents the sum of these two values, reported in this paper as a combined IHC staining score (CSS).
Results: Of 30 patients 7 were AJCC stage IIA, 10 stage IIIB, 7 stage IIIC and 6 stage IV. Median follow-up was 113 months. DFI was 17 months. Median overall survival (OS) was not reached. Stage-specific OS was: II - not reached; III - not reached; IV - 11 months. In a univariate analysis, poor OS was associated with loss of CD29 expression; median OS, 32 months vs. not reached for CSS 3-7 vs. >7.5, respectively; p=0.052 comparing entire curves, after adjustment. In a Cox model analysis, loss of CD29 exhibited a trend toward association with survival (p=0.098) after adjusting for the effect of stage (p=0.0076). Greater expression of ALDH1A1 was associated with increasing stage (p=0.042 over stages 2, 3b, 3c, and 4) while loss of CD29 expression exhibited a trend toward being associated with stages 3 and 4 (p=0.08). Compared to normal colon tissue, primary tumors were associated with increased expression of ALDH1B1 (p=0.008). ALD1H1B1 expression level differed according to whether the tumor was moderately or poorly differentiated, well differentiated, or mucinous; the highest expression levels were associated with moderately or poorly differentiated tumors (p=0.011). Lymph node metastases were associated with a trend toward decreased expression of EpCAM (p = 0.06) when comparing 0 vs. 1 vs. 2+ positive lymph nodes, as was CD29 (p = 0.08) when comparing 0 vs. any positive lymph nodes. Compared to normal colon tissue metastatic colon cancers from different patients were associated with increased ALDH1B1 expression (p=0.001) whereas CD29 expression was higher in normal colonic tissue (p=0.014).
Conclusion: CD29 may be associated with survival as well as clinical stage and number of lymph nodes. ALDH1B1 expression was associated with differentiation as well as type of tissue evaluated. ALDH1A1 was associated with clinical stage, and decreased EpCAM expression was found in patients with advanced lymph node stage. CRCSCs may be useful biomarkers to risk stratify, and estimate outcomes in CRC. Larger prospective studies are required to validate the current findings.
Keywords: colon cancer, staging, lymph node, cancer recurrence, overall survival, prognosis, biomarkers, cancer stem cells.
Introduction
Colorectal cancer (CRC) is the third most common cause of cancer-related death in the US 1. One out of four patients presenting with conventionally staged node negative disease (AJCC Stage I and II), and over 50% of patients with Stage III disease will develop local recurrence and/ or metastases2,3. Across all stages, approximately 30% of patients will develop distant metastases2. Once metastases become clinically evident prognosis is often fatal. Moreover, in spite of the fact that modern systemic therapies for CRC have resulted in improved overall survival (OS), failure rate in the adjuvant setting is 30% for high-risk Stage II and Stage III patients, and overall response rate is only 60% for patients with Stage IV CRC 4,5,6. A significant advance in the care of patients will be realized by biomarkers that can accurately identify patients at-risk for disease recurrence and dissemination, and those that fail to respond to systemic therapy. These patients might benefit from early (preventative) treatment, alternative treatment strategies, and/or frequent surveillance for and early detection of disease recurrence.
Extensive CRC research over the last decade has suggested promising biomarkers7. Although many biomarkers exist, only a select few have provided prognostic data. This list includes markers such as tumor MSI-H expression (defects in DNA mismatch repair, MSI phenotype), 18q AI expression, p53 expression and KRAS mutation. MSI-H phenotype has been associated with improved clinical outcome (disease-free and overall survival) 8,9. Some published data supports the recommendation not to administer chemotherapy to Stage II patients with a MSI-H primary colon cancer 10. The overall predictive value of MSI-H phenotype is currently being tested in the Eastern Cooperative Oncology Group Trial 5202 (E5202) adjuvant CRC trial 7. This trial is specifically intended to identify patients with AJCC Stage II CRC most likely to respond to adjuvant systemic therapy.
Tumor expression of 18q AI has also been associated with decreased survival and is also being tested in the E5202 trial7. Furthermore, p53 mutation or overexpression has been associated with poor prognosis, including decreased disease free survival (DFS), recurrence free survival (RFS) and overall survival (OS)7. Although KRAS itself is not a prognostic factor in CRC, patients with wild-type KRAS tumors have been found to respond to and benefit from certain targeted adjuvant systemic therapies7,11,12. That being said, the overall poor survival of advanced CRC patients establishes the need for improved prognostic and predictive biomarkers.
The precise cell of origin of CRC remains unknown. Recently, compelling evidence has emerged in support of the cancer stem cell (CSC) hypothesis in several solid organ epithelial malignancies including CRC13,14,15-18. The CSC hypothesis posits that CSCs are responsible for tumor initiation, metastases and resistance to treatment leading to disease relapse following surgery and/or chemoradiotherapy19. The traditional, stochastic model of tumorigenesis suggests that all cells within a tumor are capable of tumor initiation and propagation20. The CSC hypothesis proposes a hierarchical model, in which only a small fraction of cells (CSC) are capable of tumor propagation20. The CSC hypothesis therefore raises questions regarding current diagnostic and therapeutic modalities, suggesting that the CSC is a rational target for the development of more efficacious screening, early detection, prevention, treatment and surveillance modalities and interventions20,21. Based on the tenets of the CSC hypothesis, identification, proper selection, characterization, testing, biological implications and validation of CRC derived CSC (CRCSC) are imperative for improving early detection, screening, risk stratification prognostication and individualized prediction of treatment response.
Properties that define potential CSCs are: [1] self-renewal; [2] the capacity for differentiation (allowing for recapitulation of all cell types of the original tumor); [3] tumor initiating capacity; and, [4] asymmetric cell division via non-random chromosomal co-segregation16,22. Investigators have used these properties and various membrane and cytoplasmic markers to isolate putative CRCSC: CD133, CD29, CD44, CD166 (ALCAM), EpCAM, ALDH1A1 and ALDH1B1 (Table 1). These markers represent all reported CRCSC. Despite the potential of CRCSC's to be utilized as clinically relevant biomarkers, little is known about the prognostic value of non-CD133 CRCSC markers23. Notwithstanding, the CSC hypothesis may herald a paradigm shift in screening and early detection in CRC once the precise role of CRCSC markers is further established. Therefore, we hypothesized that CRCSC markers can be used as biomarkers to predict disease progression, and identify patients at risk for recurrence.
Until more effective therapies can be developed, one strategy for improving outcomes while reducing the socio-economic burden of CRC is to develop novel strategies for CRC screening that will result in higher rates of early CRC detection. Another strategy to improve outcomes in this era of patient-centered, quality-driven, value-based purchasing in oncology is to identify those patients who would most benefit from adjuvant treatments and intensive post-treatment surveillance protocols. There is an acute need for highly sensitive and specific biomarkers capable of identifying patients with early CRC that are highly likely to recur and/ or metastasize, and patients who are likely to progress despite adjuvant therapy. Such risk stratification would: [1] spare low risk patients likely cured by surgery alone the toxicity of systemic therapy; [2] identify and treat with chemotherapy at-risk patients with early stage CRC; and, [3] limit treatments to patients with clinically latent (stable) residual disease and reserve additional therapy for treatment-responsive disease progression 24. The aim of this pilot study was to analyze the expression profile of non-CD133 putative CRCSC markers and determine a possible role for CRCSC as risk-stratifying, prognostic biomarkers.
Summary of putative CRC Stem Cell Markers.
| Marker | Gene | Function |
|---|---|---|
| CD29 | ITGB1 | - Integrin that mediates cell-ECM adhesion and is involved in homing to sites of inflammation -Involved in cell growth, differentiation, migration and death |
| CD44 | CD44 | -Cell surface glycoprotein involved in cell adhesion and migration -Associated with malignant progression (adenoma to carcinoma): involved in cell migration through the ECM -Enhanced expression in breast cancer epithelial cells facilitated tumor cell migration |
| CD166 | ALCAM | -Involved in neuronal extension, embryonic hemopoiesis, embryonic angiogenesis -Cell adhesion molecule -Associated with adenoma to carcinoma development |
| EpCAM | EPCAM | -Cell adhesion molecule -Linked to Cadherin-Cathenin pathway and Wnt pathway -Expression data linked to poorer survival times in several tumor types including breast cancer -Loss of expression associated with aggressive rectal cancer |
| ALDH1A1 | ALDH1A1 | -Detoxifying enzyme responsible for the oxidation of intracellular aldehydes -Early differentiation of stem cells -Involved in resistance to chemotherapy (alkylating agents) -Malignant prostate stem cells and predictor of prostate cancer patient outcome |
| ALDH1B1 | ALDH1B1 | -Detoxifying enzyme responsible for the oxidation of intracellular aldehydes -Early differentiation of stem cells -Higher expression in CRC |
Abbreviations: CD, cluster of differentiation; ALDH, aldehyde dehydrogenase-1; ECM, extra cellular matrix; CRC, colorectal cancer.
Methods
Patients
We used a colorectal Tissue Micro Array (TMA) with corresponding survival and clinicopathological data (catalog number C0951, US Biomax, Rockville, MD, USA). The TMA included 30 patients with double core tissue sites of primary colorectal cancers with matched normal colorectal tissue in eight patients and matched metastatic tissue in ten other patients. Patient characteristics are depicted in table 2.
Summary of patient characteristics (n = 30).
| Clinicopathological Feature | Outcome | Frequency N (%) |
|---|---|---|
| Age (years; n = 29) | Mean (range) | 56 (35 - 76) |
| Gender (n = 30) | Male | 17 (57) |
| Female | 13 (43) | |
| Location (n = 30) | Sigmoid colon | 11 (36.6) |
| Ascending colon | 8 (26.6) | |
| Transverse colon | 3 (10) | |
| Descending colon | 2 (6.6) | |
| Cecum | 2 (6.6) | |
| Rectum | 4 (13.3) | |
| Pathological Grade (n = 30) | Poorly differentiated | 1 (3.3) |
| Moderately differentiated | 18 (60) | |
| Well differentiated | 8 (26.6) | |
| Mucinous Adenocarcinoma | 3 (10) | |
| Stage (n = 30) | IIA | 7 (23.3) |
| IIIB | 10 (33.3) | |
| IIIC | 7 (23.3) | |
| IV | 6 (20) | |
| T Classification (n = 30) | pT3 | 25 (83.3) |
| pT4 | 5 (16.6) | |
| N Classification (n = 30) | N0 | 7 (23.3) |
| N1 | 13 (43.3) | |
| N2 | 10 (33.3) | |
| Sites of Metastatic Tissue (n = 8) | Lung | 2 |
| Lymph node | 5 | |
| Ovary | 3 | |
| Follow Up (months) | Median (Range) | 113 (0 - 130) |
Immunohistochemistry
Immunohistochemistry (IHC) was performed using previously described techniques25. We performed immunohistochemistry for CD29, CD44, CD166, EpCam, ALDH1A1 and ALDH1B1 (Table 1). Staining was performed on Leica Biosystems (IL, USA) Bond Autostainer. The primary antibodies used in this study included anti-CD29 (1:200, Abcam, Cambridge, MA, USA), anti-CD44 (1:400, Abcam, Cambridge, MA, USA), anti-CD166 (1:40, Abcam, Cambridge, MA, USA), anti-EpCam (1:100, Cell Signaling, Danvers, MA, USA), anti-ALDH1A1 (1:250, Epitomics, Burlingame, CA, USA), anti-ALDH1B1 (1:400, Sigma, St. Louis, MO, USA). TMAs were single stained for each biomarker with matched positive and negative controls25. A pathologist who was blinded to the clinical outcomes evaluated individual samples within the TMAs. Each individual sample was scored quantitatively evaluating the proportion of positive cells, and the intensity of positively stained cells. The percent of positive cells was graded using the following rubric: 0 = <10%, 1 = 10 - 24%, 2 = 25 - 49%, 3 = 50 - 74%, 4 = 75 - 100%. The intensity of positive stained cells was graded using the following rubric: 0 = no staining, 1 = diminutive intensity, 2 = low intensity, 3 = intermediate intensity, 4 = high intensity. The pathologic score for each sample is represented by the sum of these two values: this sum is referred to as the combined IHC staining score (CSS).
Statistical Analyses
CSS was computed for each sample and each marker. In cases in which there were two samples obtained for a given patient/site, the average of the sum of the two scores was used. The association between the pathologic score and overall survival (OS) of patients was determined based on data from the 30 primary sites. The probability of survival as a function of time was determined by the Kaplan-Meier method, and the statistical significance was determined by the log-rank method. In cases for which the initial grouping of patients' scores or clinical data was reduced to two groups in order to identify a better grouping of subjects based on prognostic ability, the p-values for the revised analysis of dichotomized groups were adjusted for multiple testing, which would be performed to arrive at the identified final grouping. A Cox proportional hazards model analysis was performed to determine the joint association of CSSs found to be significantly associated with OS in the initial univariate analyses after adjusting for important clinical factors.
Within the 30 primary specimens, the association between the CSS and tumor grade was determined with an exact Kruskal-Wallis test. The association between the CSS and stage was determined by a Jonckheere-Terpstra test for trend. The association between the CSS and sex was determined by an exact Wilcoxon rank sum test, while Spearman rank correlation was used to determine the correlation between the CSS and age. Analyses to compare the CSS between the matched primary sites and the metastatic sites, or the primary sites and the normal sites, were performed on paired samples using a Wilcoxon signed rank test. Comparisons of CSS between metastatic and normal samples were made with a Wilcoxon rank sum test, since these data are only from different patients. All p-values are two-tailed and presented without adjustment for multiple comparisons.
Results
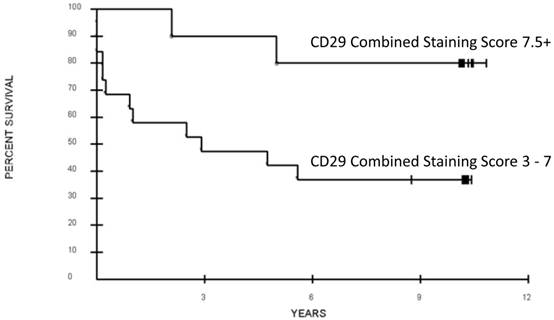
All six CRCSC markers were assessed for association with overall survival. Out of the six, only loss of CD29 expression was potentially associated with a significant difference in OS (CSS 3-7 vs. CSS > 7.5, median 32 months vs. not reached, respectively; p = 0.052 after adjustment for the grouping identified, Figure 1, 2). Increased ALDH1A1showed a trend towards decreased OS (p=0.16, Table 3). Age, sex, and pathologic grade (poorly, moderately, well differentiated) were not significantly associated with OS. Stage of disease was associated with OS; patients with AJCC Stage 2 or 3 disease had a significantly better prognosis than those with Stage 4 disease (Median OS: not reached vs. 11 months; p = 0.0009, adjusted for the grouping of stages). The joint association between CD29 and Stage (2-3 vs. 4) was evaluated in a Cox proportional hazards model. After adjusting for stage (p=0.0076, Hazard ratio 5.06; 95% CI for HR: 1.54 to 16.65), loss of CD29 expression was marginally associated with poor OS (p = 0.098; Hazard ratio=0.28; 95% CI for HR: 0.06 to 1.27; Figure 2).
Univariate analyses of the CSS were performed for the 30 primary tissue specimens in order to assess the interaction between the CRCSC biomarkers and the level of pathologic differentiation (poorly, moderately, well differentiated). Out of the six CRCSC, only ALDH1B1 showed a statistically significant association with tumor differentiation. ALDH1B1 expression was significantly increased in poorly and/or moderately differentiated tumors as compared to well-differentiated or mucinous tumors (p = 0.011). Analysis of the association between CSS and stage was performed. Only increased ALDH1A1 showed an association between increasing expression level and increasing stage (p = 0.04). The combination of CD29 and ALDH1B1 was not associated with a difference in OS.
When comparing normal colon tissue to metastatic tissue sites from different patients, CD29 expression was higher in normal tissue (p = 0.014), whereas ALDH1B1 expression was significantly higher in metastatic tissue (p = 0.001). In comparing primary tumors to their matched patient metastatic sites, none of the CSS differed significantly between two samples from the same patient. However, comparison of normal colon tissue to primary tumor showed that increased ALDH1B1 expression was associated with the primary tumors (p = 0.008).
Association of CRCSC with survival and clinical characteristics. (Overall Survival Statistics)
| Marker | Expression | p-value | |
|---|---|---|---|
| Overall Survival Statistics | |||
| CD29 | Decreased | 0.052 (adjusted for grouping) | |
| CD44 | - | 0.69 | |
| CD166 | - | 0.33 | |
| EpCAM | - | 0.77 | |
| ALDH1A1 | - | 0.16 | |
| ALDH1B1 | - | 0.27 | |
| Pathologic Differentiation (poorly/moderately differentiated vs. well differentiated vs. mucinous) | |||
| CD29 | - | 0.55 | |
| CD44 | - | 0.94 | |
| CD166 | - | 0.75 | |
| EpCAM | - | 0.09 | |
| ALDH1A1 | - | 0.30 | |
| ALDH1B1 | Increased | 0.011 | |
| Increasing Stage of Disease | |||
| CD29 | - | 0.19 | |
| CD44 | - | 0.38 | |
| CD166 | - | 0.77 | |
| EpCAM | - | 0.34 | |
| ALDH1A1 | Increased | 0.04 | |
| ALDH1B1 | - | 0.97 | |
| Normal vs. Metastatic Tissue | |||
| CD29 | Decreased | 0.014 | |
| CD44 | - | 0.26 | |
| CD166 | - | 0.23 | |
| EpCAM | - | 0.10 | |
| ALDH1A1 | - | 0.24 | |
| ALDH1B1 | Increased | 0.001 | |
| Normal vs. Primary Tumor Tissue | |||
| CD29 | - | 0.44 | |
| CD44 | - | 0.84 | |
| CD166 | - | 0.13 | |
| EpCAM | - | 0.50 | |
| ALDH1A1 | Increased | 0.06 | |
| ALDH1B1 | Increased | 0.008 | |
| Lymph Node Stage | |||
| CD29 | Decreased | 0.08 (0 vs. 1+2; p=0.11 for 0 vs. 1 vs. 2) | |
| CD44 | - | 0.57 | |
| CD166 | - | 0.52 | |
| EpCAM | Decreased | 0.06 | |
| ALDH1A1 | - | 0.47 | |
| ALDH1B1 | - | 0.65 | |
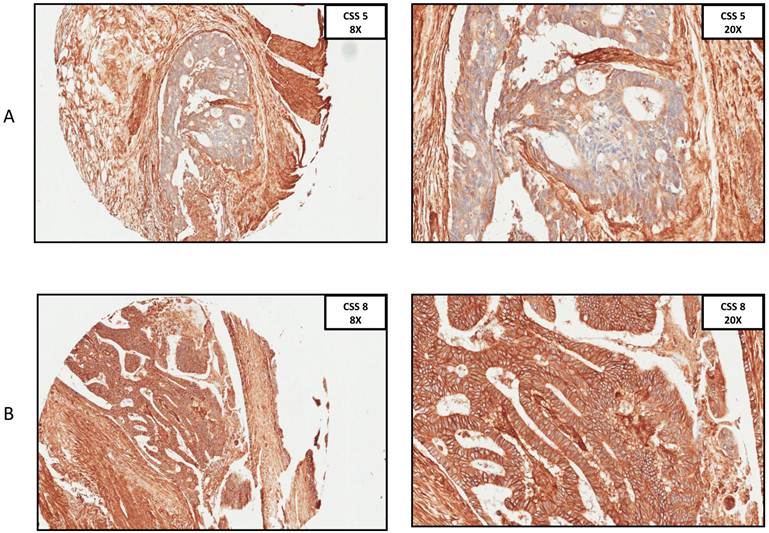
A. Immunohistochemistry anti-CD29 staining, CSS 5 at 8x and 20x ocular. B. Immunohistochemistry anti-29 staining, CSS 8 at 8x and 20x ocular. (CSS: combined staining score).
Association of overall survival and CD29 expression of the primary colorectal cancer tumor. Combined staining score was calculated as the sum of antibody intensity (0 - 4) and percent cells positive (0 - 4).
Assessment of the six CRCSC markers and their relation to lymph node status was performed. Advanced AJCC lymph node stage (N0 vs. N1-2) was associated with decreased CD29 (p = 0.08) expression, while EpCAM tended to decrease with increased nodal stage, from N0 to N1 to N2; p=0.06.
Localization of expression was different between the various CRCSC markers, and consistent with previous reports and their associated functions. ALDH1A1 and ALDH1B1 expression was intracellular, while EpCAM expression was at the cell surface. CD29, CD44 and CD166 had a combination of intracellular and cell surface expression.
Discussion
Two fundamental issues regarding colorectal carcinogenesis remain unanswered. First, the level of differentiation in the initiating neoplastic cell has not been well described i.e. colonic stem cell vs. differentiated mature colonic mucosal epithelial cell. Second, since tumors are well known to be composed of a heterogeneous group of cells, the specific identity of tumor cells that lead to lymph-node involvement, and metastatic disease is not well characterized.20 Recently, attempts to address both of these issues pragmatically, which are critical to our understanding of tumor biology, have resulted in the description of cancer stem cells. While the stochastic model of tumorigenesis holds that every cell within the tumor population is capable of tumor initiation and propagation, the cancer stem cell model proposes that only a small fraction of cells possesses the ability to initiate cancer growth and promote metastatic dissemination20.
There are various methods of CSC identification in vitro; however, there is currently no consensus on a universally acceptable method15-18,20,23. For technical reasons, the most commonly utilized method of identification involves separating cells based on proteins thought to be associated with the cancer stem cell phenotype26. Detection of sub-populations of cells such as CRCSC in regional nodes negative for tumor cells by conventional histopathology, or as circulating cells in the blood or bone marrow, is likely to increase accuracy of both cancer staging and prognosis. While some studies have linked CSC markers to prognosis, there remains no definitive association between putative CSC markers and disease behavior, disease progression, or survival. An association between putative CRCSC markers and survival could not only improve cancer screening and early detection, but also help define optimal post-treatment follow up. Equally as important, CRCSC markers could potentially tailor more efficacious treatment modalities to those patients who stand to benefit most, such as at-risk node-negative (early stage) CRC patients who are prone to recur.
Molecular biomarkers provide potential benefits because they enable identification of specific cell types and cell populations that are associated with disease behavior and clinical outcomes 26. Methods of cancer stem cell identification are based on markers specific for normal progenitor or stem cells in the same organ27. The adult stem cells of the colon are of particular interest because they sustain the perpetual self-renewal of healthy colonic epithelium and are therefore able to acquire the number of mutations required for carcinogenesis26. Although initial reports identified CD133 as a reliable CSC marker in colorectal cancer, subsequent studies have shown that CD133 expression is not restricted to rare cell subsets and it is detectable in a large majority of tumor cells, irrespective of their tumorigenicity23. Lugli et al also stated that neither over-expression nor loss of CD133 was significantly associated with tumor progression or survival23. Alternatively, CD29, C44, EpCAM, CD166, ALDH1A1 and ALDH1B1 were reported to identify another CSC pool23,28. Presence of these proteins has been associated with characteristics of stemness both in vitro and in vivo 20. Cells positive for these markers have been shown to generate tumors recapitulating the primary tumor with increased clonogenic ability and multi-lineage potential 20. This subset of cells has also been associated with tumor stage, differentiation, invasiveness, metastasis formation, and prognosis20. However, there is a paucity of longitudinal data in order to more fully understand the related disease biology, as is presented in our study.
Uncertainty remains as to whether CSC markers exclusively delineate the stem cell population and whether they can predict disease behavior 26, 4. In normal colon, CD29 has been observed in the lower parts of the crypts and therefore has been hypothesized to be a mucosal progenitor cell marker29. The combination of CD24 and CD29 has been suggested to identify the tumor initiating fraction in mouse colon carcinomas29 and it is proposed that CD29 may promote cancer progression by inducing invasion, migration and metastasis through regulation of the tumor microenvironment 29,30. Zou et al also stated that the CD133+CD29+ cellular fraction up-regulated self-renewal, proliferation and differentiation, and therefore reported CD29 to be a new stem cell marker for colon cancer,30 although its full biological function has yet to be elucidated. Here we present the first report of the possible association between CD29 expression and overall survival in CRC, which remains to some degree even after adjustment for stage in this limited size study.
CD44, the first stem cell marker to be identified in breast cancer, has long been thought to be a marker of tumor invasiveness and metastasis, and more recently has been described as a potential CRCSC marker21,23. CD44 is a transmembrane glycoprotein that can act as a receptor for extracellular matrices such as hyaluronic acid; it is a downstream target of the Wnt/B-catenin pathway21. High expression of CD44 in malignant colonic epithelial cells has been associated with tumor virulence; knockdown of CD44 in primary colon cancer cell lines reduced clonogenicity in vitro and tumorigenicity in vivo29. Currently, it is not known whether CD44 is a specific marker for stem cells, as a large population of CD44-exressing cells within a tumor has many splice variants21. Early reports of the CD44 gene and its splice variants describe decreased overall survival in patients with increased expression levels of the gene or its related protein. More recent results, however, are inconsistent; some suggest either no role for CD44, others demonstrate a worse clinical outcome with loss of protein expression 31,32,33,23. Further, some have identified increased expression of CD44 in colon along the progression from normal colonic epithelium to adenoma to carcinoma33,23. We did not find CD44 to be prognostic.
Epithelial cell adhesion molecule (EpCAM) is a transmembrane glycoprotein mediating epithelium-specific intercellular adhesion. It is also a molecule involved in cell signaling, migration, proliferation and differentiation34. As EpCAM is expressed wholly in epithelium-derived cancers, it has been reported as a diagnostic marker34. Enhanced EpCAM expression has been linked to advanced cancer stage and worse overall survival34, specifically for breast cancer and gallbladder cancer35,36. EpCAM expression has also been linked to higher primary tumor grade and inferior local and distant recurrence free survival in rectal cancer37. Lugli et al. also noted EpCAM to be a prognostic marker; however, they found a decreased expression of EpCAM to be prognostic. In addition, decreased EpCAM expression was found to be significantly associated with infiltrating tumor margin, tumor invasion, and presence of lymph node metastasis23. Similarly, we found that decreased EpCAM expression is associated with increasing lymph node stage (N0 vs. N1 vs. N2).
CD166 expression can be used to further select CSC's within the EpCAM(+)CD44(+) population 21. There appears to be heterogeneous expression of CD166 in CRC and the expression level is associated with poor clinical outcomes. It has been hypothesized that this effect is possibly mediated by CD166 regulation of cell-to-cell interactions29. In immunodeficient mice, CD44+CD166+ colon cancer cells display high tumor initiating potential29. Patel et al. found a significant increase in CD166 expression in adenomatous glands and an age-dependent increase in CD44 and CD166 expression. This was also associated with the number of colon polyps23,38. Weichert et al. described increased expression of CD166 in tumor tissue, and, in a group of 111 colorectal cancer cases, observed a between CD166 expression and shortened overall survival23,39. These findings suggest a role for both CD44 and CD166 in the colon adenoma to carcinoma sequence. It is possible that because of the small number of patients analyzed in this study an association between CD44, CD166 and outcomes could not be elucidated.
CD24 has also been implicated in CRCSC's. Choi et al examined 523 colorectal adenocarcinomas of various stages and found significant correlation between CD24 expression and degree of differentiation19,40. Other studies have shown that the degree of colorectal tumor CD24 expression is associated with lymph node metastasis and shortened patient survival 19,41,42. We chose not to use CD24 in this study. Although there may be a relationship between CD24 and oncologic outcome, the evidence for CD24 as CRCSC marker is less convincing.
ALDH1 has been reported as a cancer stem cell marker in pancreatic, breast, prostate, and lung cancer, multiple myeloma and leukemia. More recently ALDH1 was identified as a CRCSC marker21. ALDH1 functions by catalyzing the irreversible oxidation of a range of aliphatic and aromatic aldehydes to their corresponding carboxylic acids27. High ALDH1 activity is detected in stem and progenitor cells of various lineages. Enhanced ALDH1 expression was found in areas where epithelial progenitor cells localize within normal breast, colon and stomach27. Various groups have reported that high ALDH1 expression is associated with reduced survival times in breast, lung, pancreas, bladder and prostate cancer patients27. Deng et al. found that ovarian cancer patients with high ALDH1 had shorter disease free and overall survival compared to those with low ALDH1 (p = 0.0036 and p = 0.0023, respectively)27. Lugli et al. also noted that increased ALDH1 expression is associated with higher tumor grade23. We similarly found an association between ALDH expression and disease behavior.
Although this study is the first to analyze the association between specific biomarkers within a panel of six CRCSC markers and overall survival, the study has its limitations, particularly a small overall sample size. This specifically reduced our ability to describe tumor characteristics and behaviors according to multi-marker phenotype of the investigated molecules, and required us to conduct the analysis according to each marker individually. This parsimonious analytical approach was done of necessity, as multi-marker phenotypes in tumor tissues are notably heterogeneous. The small number of matched normal controls and matched metastatic tissue cores is another limitation of this study. Finally, we were constrained by needing to use a retrospective study design. For this reason, we regard this study as hypothesis generating in nature, and report our findings in the context of a pilot study. Therefore, the data should be interpreted with caution. Further prospective investigation is needed to validate CRCSC as biomarkers.
The CSC hypothesis appears to have a promising role in CRC tumor biology, despite the remaining unanswered questions related to this novel premise. By focusing on the identification and treatment of tumor progenitor cells, we may ultimately be able to improve screening, early detection, treatment, and prognostication19. Further identification of novel cell surface or cytoplasmic markers associated with CRCSC, and validation of known CRCSC could be useful in identifying tumors with poor prognosis. This approach could further enhance our ability to assess response to therapy and optimize treatment selection and intensity of post-treatment surveillance and follow-up. In order to translate these findings into clinical practice, prospective comprehensive analysis of a panel of CRCSC expression in large groups of patients is imperative23. Moreover, there remains a paucity of data evaluating the prognostic significance of the co-expression of multiple CRCSC within the same tumor, nodal and distant metastases within the same patients 14,23.
Conclusion
In this study, we have evaluated the relationship between the expression of six CRCSC markers, clinically relevant features and outcomes. Our findings suggest that decreased CD29 expression, decreased EpCAM expression and increased ALDH1A1 and ALDH1B1 expression may represent suitable risk-stratifying worth exploring as prognostic biomarkers in CRC. These findings may lend themselves to a new strategy of individualized adjuvant therapy selection and post-treatment surveillance aimed at identifying patients with the highest likelihood of disease recurrence or progression based on CRCSC marker expression within the primary tumor.
The CSC hypothesis may herald a paradigm shift in oncologic diagnosis and treatment. This pilot study shows that putative CRC stem cell markers may have a role in predicting the behavior of CRC and estimating clinical outcomes. Prospective studies properly powered based on this study should be undertaken to determine the significance of these early findings.
Abbreviations
CRC: colorectal cancer; CRCSC: colorectal cancer stem cell; CSS: combined IHC staining score; DFS: disease-free survival; E5202: Eastern Cooperative Oncology Group Trial 5202; IHC: immunohistochemistry; MSI: microsatellite instability; RFS: recurrence-free survival; TMA: tissue microarray; OS: overall survival.
Contributing Author Declaration
We certify that all individuals who qualify as authors have been listed; each has participated in one or more of the following areas: conception and design of this work, the acquisition and/or analysis of data, the writing, and/or critical revision of the document, and supervision of this cooperative research effort. All contributing authors approve of the submission of this version of the manuscript and assert that the document represents valid work. If information derived from another source was used in this manuscript, we obtained all necessary approvals to use it and made appropriate acknowledgements in the document. All contributing authors take public responsibility for this work.
Disclaimer
The views expressed in this manuscript are those of the authors and does not necessarily reflect the views of policies of the Department of Health and Human Services, Department of the Army, the Department of Defense or the United States Government, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Funding
This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E.
Copyright protection
One or more of the contributing authors are military service members (or employees of the U.S. Government: AS), and this work was prepared as part of their official duties. Title 17 U.S.C. 105 provides the “Copyright protection under this title is not available for any work of the United States Government.” Title 17 U.S.C. 101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person's official duties.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel R, Ward E, Brawley O. et al. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212-36
2. Bilchik AJ, DiNome M, Saha S. et al. Prospective multicenter trial of staging adequacy in colon cancer: preliminary results. Arch Surg. 2006;141:527-33
3. Manfredi S, Bouvier AM, Lepage C. et al. Incidence and patterns of recurrence after resection for cure of colonic cancer in a well defined population. Br J Surg. 2006;93:1115-22
4. Galizia G, Gemei M, Del Vecchio L. et al. Combined CD133/CD44 expression as a prognostic indicator of disease-free survival in patients with colorectal cancer. Arch Surg. 2012;147:18-24
5. Falcone A, Ricci S, Brunetti I. et al. Phase III trial of infusional fluorouracil, leucovorin, oxaliplatin, and irinotecan (FOLFOXIRI) compared with infusional fluorouracil, leucovorin, and irinotecan (FOLFIRI) as first-line treatment for metastatic colorectal cancer: the Gruppo Oncologico Nord Ovest. J Clin Oncol. 2007;25:1670-6
6. Wolpin BM, Meyerhardt JA, Mamon HJ. et al. Adjuvant treatment of colorectal cancer. CA Cancer J Clin. 2007;57:168-85
7. Tejpar S, Bertagnolli M, Bosman F. et al. Prognostic and predictive biomarkers in resected colon cancer: current status and future perspectives for integrating genomics into biomarker discovery. Oncologist. 2010;15:390-404
8. Gryfe R, Kim H, Hsieh ET. et al. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342:69-77
9. Ribic CM, Sargent DJ, Moore MJ. et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247-57
10. Sinicrope FA, Sargent DJ. Clinical implications of microsatellite instability in sporadic colon cancers. Curr Opin Oncol. 2009;21:369-73
11. Karapetis CS, Khambata-Ford S, Jonker DJ. et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757-65
12. Ahnen DJ, Feigl P, Quan G. et al. Ki-ras mutation and p53 overexpression predict the clinical behavior of colorectal cancer: a Southwest Oncology Group study. Cancer Res. 1998;58:1149-58
13. O'Brien CA, Pollett A, Gallinger S. et al. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106-10
14. Ricci-Vitiani L, Lombardi DG, Pilozzi E. et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111-5
15. Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755-68
16. Xin HW, Hari DM, Mullinax JE. et al. Tumor-initiating label-retaining cancer cells in human gastrointestinal cancers undergo asymmetric cell division. Stem Cells. 2012;30:591-8
17. Avital I, Moreira AL, Klimstra DS. et al. Donor-derived human bone marrow cells contribute to solid organ cancers developing after bone marrow transplantation. Stem Cells. 2007;25:2903-9
18. Hari D, Xin HW, Jaiswal K. et al. Isolation of live label-retaining cells and cells undergoing asymmetric cell division via nonrandom chromosomal cosegregation from human cancers. Stem Cells Dev. 2011;20:1649-58
19. Papailiou J, Bramis KJ, Gazouli M. et al. Stem cells in colon cancer. A new era in cancer theory begins. Int J Colorectal Dis. 2011;26:1-11
20. Vaiopoulos AG, Kostakis ID, Koutsilieris M. et al. Colorectal cancer stem cells. Stem Cells. 2012;30:363-71
21. Subramaniam D, Ramalingam S, Houchen CW. et al. Cancer stem cells: a novel paradigm for cancer prevention and treatment. Mini Rev Med Chem. 2010;10:359-71
22. Marquardt JU, Thorgeirsson SS. Stem cells in hepatocarcinogenesis: evidence from genomic data. Semin Liver Dis. 2010;30:26-34
23. Lugli A, Iezzi G, Hostettler I. et al. Prognostic impact of the expression of putative cancer stem cell markers CD133, CD166, CD44s, EpCAM, and ALDH1 in colorectal cancer. Br J Cancer. 2010;103:382-90
24. Pilati P, Mocellin S, Bertazza L. et al. Prognostic value of putative circulating cancer stem cells in patients undergoing hepatic resection for colorectal liver metastasis. Ann Surg Oncol. 2012;19:402-8
25. Zlobec I, Gunthert U, Tornillo L. et al. Systematic assessment of the prognostic impact of membranous CD44v6 protein expression in colorectal cancer. Histopathology. 2009;55:564-75
26. Willis ND, Przyborski SA, Hutchison CJ. et al. Colonic and colorectal cancer stem cells: progress in the search for putative biomarkers. J Anat. 2008;213:59-65
27. Deng S, Yang X, Lassus H. et al. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PLoS One. 2010;5:e10277
28. Dalerba P, Dylla SJ, Park IK. et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104:10158-63
29. Kemper K, Grandela C, Medema JP. Molecular identification and targeting of colorectal cancer stem cells. Oncotarget. 2010;1:387-95
30. Zou J, Yu XF, Bao ZJ. et al. Proteome of human colon cancer stem cells: a comparative analysis. World J Gastroenterol. 2011;17:1276-85
31. Mulder JW, Kruyt PM, Sewnath M. et al. Colorectal cancer prognosis and expression of exon-v6-containing CD44 proteins. Lancet. 1994;344:1470-2
32. Wielenga VJ, van der Voort R, Mulder JW. et al. CD44 splice variants as prognostic markers in colorectal cancer. Scand J Gastroenterol. 1998;33:82-7
33. Coppola D, Hyacinthe M, Fu L. et al. CD44V6 expression in human colorectal carcinoma. Hum Pathol. 1998;29:627-35
34. Patriarca C, Macchi RM, Marschner AK. et al. Epithelial cell adhesion molecule expression (CD326) in cancer: a short review. Cancer Treat Rev. 2012;38:68-75
35. Gastl G, Spizzo G, Obrist P. et al. Ep-CAM overexpression in breast cancer as a predictor of survival. Lancet. 2000;356:1981-2
36. Prince S, Zeidman A, Dekel Y. et al. Expression of epithelial cell adhesion molecule in gallbladder carcinoma and its correlation with clinicopathologic variables. Am J Clin Pathol. 2008;129:424-9
37. Gosens MJ, van Kempen LC, van de Velde CJ. et al. Loss of membranous Ep-CAM in budding colorectal carcinoma cells. Mod Pathol. 2007;20:221-32
38. Patel BB, Yu Y, Du J. et al. Age-related increase in colorectal cancer stem cells in macroscopically normal mucosa of patients with adenomas: a risk factor for colon cancer. Biochem Biophys Res Commun. 2009;378:344-7
39. Weichert W, Knosel T, Bellach J. et al. ALCAM/CD166 is overexpressed in colorectal carcinoma and correlates with shortened patient survival. J Clin Pathol. 2004;57:1160-4
40. Choi D, Lee HW, Hur KY. et al. Cancer stem cell markers CD133 and CD24 correlate with invasiveness and differentiation in colorectal adenocarcinoma. World J Gastroenterol. 2009;15:2258-64
41. Lim SC, Oh SH. The role of CD24 in various human epithelial neoplasias. Pathol Res Pract. 2005;201:479-86
42. Weichert W, Denkert C, Burkhardt M. et al. Cytoplasmic CD24 expression in colorectal cancer independently correlates with shortened patient survival. Clin Cancer Res. 2005;11:6574-81
Author contact
![]() Corresponding author: Udo Rudloff, MD, PhD. National Cancer Institute, NIH, 10 Center Drive, CRC3-5950Phone: (301) 496-3098, rudloffunih.gov; or Itzhak Avital, MD FACS, Medical Director, Bon Secours Cancer Institute, Richmond VA, Itzhak_Avitalcom; Itzhak.avitalcom.
Corresponding author: Udo Rudloff, MD, PhD. National Cancer Institute, NIH, 10 Center Drive, CRC3-5950Phone: (301) 496-3098, rudloffunih.gov; or Itzhak Avital, MD FACS, Medical Director, Bon Secours Cancer Institute, Richmond VA, Itzhak_Avitalcom; Itzhak.avitalcom.

 Global reach, higher impact
Global reach, higher impact