3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2013; 4(2):152-164. doi:10.7150/jca.5702 This issue Cite
Research Paper
HtrA3 Is Downregulated in Cancer Cell Lines and Significantly Reduced in Primary Serous and Granulosa Cell Ovarian Tumors
Prince Henry's Institute of Medical Research, Clayton, Victoria 3168, Australia.
Received 2012-12-13; Accepted 2013-1-10; Published 2013-2-1
Abstract
Objective. The high temperature requirement factor A3 (HtrA3) is a serine protease homologous to bacterial HtrA. Four human HtrAs have been identified. HtrA1 and HtrA3 share a high degree of domain organization and are downregulated in a number of cancers, suggesting a widespread loss of these proteases in cancer. This study examined how extensively the HtrA (HtrA1-3) proteins are downregulated in commonly used cancer cell lines and primary ovarian tumors.
Methods. RT-PCR was applied to various cancer cell lines (n=17) derived from the ovary, endometrium, testes, breast, prostate, and colon, and different subtypes of primary ovarian tumors [granulosa cell tumors (n=19), mucinous cystadenocarcinomas (n=6), serous cystadenocarcinomas (n=8)] and normal ovary (n = 9). HtrA3 protein was localized by immunohistochemistry.
Results. HtrA3 was extensively downregulated in the cancer cell lines examined including the granulosa cell tumor-derived cell lines. In primary ovarian tumors, the HtrA3 was significantly lower in serous cystadenocarcinoma and granulosa cell tumors. In contrast, HtrA1 and HtrA2 were expressed in all samples with no significant differences between the control and tumors. In normal postmenopausal ovary, HtrA3 protein was localized to lutenizing stromal cells and corpus albicans. In serous cystadenocarcinoma, HtrA3 protein was absent in the papillae but detected in the mesenchymal cyst wall.
Conclusion. HtrA3 is more extensively downregulated than HtrA1-2 in cancer cell lines. HtrA3, but not HtrA1 or HtrA2, was decreased in primary ovarian serous cystadenocarcinoma and granulosa cell tumors. This study provides evidence that HtrA3 may be the most relevant HtrA associated with ovarian malignancy.
Keywords: HtrA3, ovarian cancer, protease, GCT, Serous cystadenocarcinoma.
Introduction
Ovarian cancer undergoes progressive “dedifferentiation” from a well to a poorly differentiated tumor, then spreads to the pelvic and abdominal cavities before metastasizing to distant sites. It represents one of the most aggressive and heterogeneous cancer types in women and is a leading cause of gynaecological deaths [1, 2]. The majority of ovarian cancer patients are diagnosed at late stages when conventional therapy is less effective, leading to high levels of morbidity and mortality [3]. It has therefore been proposed that survival for this highly lethal disease could be improved by developing screening methods that detect the disease when it is confined to the ovary. A number of biomarkers have been investigated which are involved in the development and progression of ovarian cancer [4]. Despite initial responses to surgery and chemotherapy, most of the affected women ultimately die from recurrence and development of chemoresistance [5]. These highlight the need for identifying new prognostic markers to better determine patients with more aggressive disease, appropriate for treatment.
The high temperature requirement (HtrA) family of serine proteases are well conserved from bacteria to humans. Bacterial HtrA is the most studied member of this family and is necessary for bacterial survival at elevated temperatures [6]. It acts as a chaperone at normal temperature and protease at elevated temperatures [7, 8]. To date, four human HtrAs have been identified: HtrA1 [9, 10], HtrA2 [11, 12], HtrA3 [13, 14] and HtrA4 [15, 16]. All HtrA proteins share a highly conserved serine protease domain and one PDZ domain at the C-terminus [17]. They appear to be involved in inflammation, apoptosis, ischemia/reperfusion, neurodegenerative conditions and neuromuscular disorders, and protection against stress conditions including heat shock [reviewed in [18]].
HtrA proteins, especially HtrA1 and HtrA3, are reported to be downregulated in a number of cancers [19-21]. Both HtrA1 and HtrA3 have been implicated as inhibitors of invasion and tumor suppression [22-24]. HtrA1 is downregulated in SV40 transformed fibroblasts [9, 10]. Several reports have indicated a tumor suppressive role for HtrA1 in breast, lymph node melanoma, ovarian and gastric cancers [21, 25-27].
HtrA2 is a stress-activated protease that is up-regulated in mammalian cells in response to cellular stress [12]. It is involved in apoptosis [28-30], a process which is impaired in cancer progression, and thus might play a role in malignancy.
HtrA3 was initially identified as a pregnancy-related serine protease [31]. Two variants of HtrA3 that arise from alternative mRNA splicing [long (HtrA3-L) and short (HtrA3-S)] have been identified [13]. HtrA3-S is identical to the long form except it lacks the terminal PDZ domain [13]. The HtrA3 isoforms may recognize different substrates and thus may have different functions. HtrA3 shares a high degree of sequence and domain organization with HtrA1 [10, 13], suggesting similar functions. HtrA3 mRNA levels are reported to be downregulated with increasing grades of human endometrial, ovarian and lung cancer [20, 32, 33]. The role of HtrA4, a newly identified member of the mammalian HtrA family, in cancer progression has not been reported.
The aims of the present study were to evaluate and compare the extent of the downregulation of HtrA1, HtrA2, and HtrA3 (HtrA3-L and HtrA3-S) mRNA levels in a number of commonly used human cancer cell lines and different sub-types of primary ovarian tumor tissues [serous, mucinous and granulosa cell tumor (GCT)]. We further used immunohistochemistry to investigate HtrA3 protein localization in primary ovarian cancers.
Materials and Methods
Cell Lines and total RNA
Human granulosa cell tumor-derived cancer cells KGN and COV434, endometrial ECC1, HEC-1A, HEC-1B, Ishikawa, RL95-2 and AN3CA, testicular Ntera-2D, breast MCF-7 and MDA-231, prostate PC3, DU145 and LNCap, and colon SW480 and WIDR cell lines were cultured in medium supplemented with 10% fetal calf serum (SAFC Biosciences, KS), 2mM L-glutamine, 100µg/ml streptomycin/100 IU/ml penicillin (both from Gibco, Carlsbad, CA). All cell lines were cultured in DMEM, except for HEC-1A in McCoy's, and RL95-2 cells in DMEM supplemented with 10µg/ml bovine insulin (Actrapid, Novo Nordisk, Denmark). RNA was isolated using RNeasy Minikit (Qiagen GmbH, Hilden, Germany) and ribonuclease-free deoxyribonuclease-1 (DNA-free kit, Ambion, TX). Total RNA from human breast, prostate and testes was from Ambion (Life Technologies). Approval was obtained from the Human Research Ethics Committee at Monash Medical Centre, Melbourne to isolate total RNA from ovarian, colon and endometrial tissues.
Isolation of RNA from tissue specimens
Ovarian tumour tissues [granulosa cell tumors (n=19), mucinous cystadenocarcinomas (n=6) and serous cystadenocarcinomas (n=8)] were obtained from banked tissue samples which had previously been used in different studies [34-36]. Normal ovarian tissues were obtained from nine premenopausal women who had undergone elective hysterectomy with oophorectomy for a range of conditions not associated with ovarian malignancy. Details of individual patients are provided in Table 1. The tissues were snap-frozen and then stored at -80 0C prior to RNA extraction. The Human Research and Ethics Committee of Monash Medical Centre approved this study protocol.
Total RNA was extracted from tissues using guanidinium thiocyanate-cesium chloride as previously described [37]. All RNA samples were DNaseI-treated as per manufacturer's instructions (DNA-free kit; Ambion, Texas, USA). RNA concentration was determined at 260nm using an ND1000 Nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
Individual patient characteristics.
| Sample number | Sample Type | Patient Age | Menopausal Status | Stage |
|---|---|---|---|---|
| S1 | Serous | 56 | N/A | Unstaged |
| S2 | Serous | 71 | Post | 3C |
| S3 | Serous | 71 | Post | 3C |
| S4 | Serous | 75 | Post | 3C |
| S6 | Serous | 68 | Post | 3C |
| S7 | Serous | 83 | Post | 3C |
| S8 | Serous | 51 | Post | 3C |
| S9 | Serous | 72 | Post | 3C |
| M2 | Mucinous | 87 | Post | Unstaged |
| M5 | Mucinous | 65 | Post | 1 |
| M9 | Mucinous | 84 | Post | 1 |
| M10 | Mucinous | 64 | Post | 2 |
| M12 | Mucinous | N/A | N/A | N/A |
| M13 | Mucinous | N/A | N/A | N/A |
| N7 | Normal | 49 | Pre | Fibroid uterus |
| N8 | Normal | 33 | Pre | Intractable premenstrual tension |
| N9 | Normal | 30 | Pre | Endometrial carcinoma |
| N10 | Normal | 50 | Pre | Endometrial carcinoma |
| N11 | Normal | 48 | Pre | endometriosis/adenomyosis |
| N13 | Normal | 48 | Pre | N/A |
| 818 | Normal | N/A | N/A | N/A |
| 884 | Normal | N/A | N/A | N/A |
| 927 | Normal | N/A | N/A | N/A |
| G5 | GCT | 31 | Pre | 1 |
| G6 | GCT | 48 | Pre | Recurrent, metaststic |
| G7 | GCT | 43 | Pre | 1A |
| G9 | GCT | 85 | Post | Recurrent, metaststic |
| G10 | GCT | N/A | N/A | N/A |
| G11 | GCT | 66 | Post | Recurrent, metaststic |
| G12 | GCT | 58 | Post | Recurrent, metaststic |
| G13 | GCT | 17 | Pre | Juvenile, recurrent (metastatic) |
| G14 | GCT | 50 | Pre | Recurrent |
| G15 | GCT | 33 | Pre | Juvenile, recurrent |
| G17 | GCT | 53 | Pre | 1 |
| G18 | GCT | 54 | Pre | 1 |
| G19 | GCT | 45 | Pre | Recurrent |
| G20 | GCT | 71 | Post | Recurrent |
| G22 | GCT | 4 | Pre | Juvenile |
| G23 (137) | GCT | 81 | Post | High Grade |
| G24 (1342) | GCT | 50 | Post | 1c |
| G26 | GCT | 54 | Post | 1 |
| G27 | GCT | 79 | Post | 1 |
Pre: pre-menopausal; Post: post-menopausal; N/A: Not available. Samples from patients numbered - S1-S9 (n = 8) are serous; M2-M13 (n = 6) are mucinous; G5-G27 (n = 19) are from Granulosa cell tumor (GCT); N7-N13, 818, 884 and 927 (n = 9) are normal.
The primer sequences and reaction conditions for RT-PCR.
| Gene | Primer Sequences (5' - 3') F- Forward, R- Reverse | Annealing temperature (0C) and cycles (n) - Block PCR | PCR Product size (bp) |
|---|---|---|---|
| HTRA1 | F - AAA GCC ATC ACC AAG AAG AAG TAT R - TCC TCA TCC GTC ATC CAC | 58 (30 cycles) | 384 |
| HTRA2 | F - GCC GTG GTC TAT ATC GAG ATC CT R - TGC CGG ATG TGA TCG TGT | 58 (30 cycles) | 343 |
| HTRA3-L (Long) | F - ATG CGG ACG ATC ACA CCA AG R - CGC TGC CCT CCG TTG TCT G | 58 (30 cycles) | 337 |
| HTRA3-S (short) | F - GAG GGC TGG TCA CAT GAA GA R - GCT CCG CTA ATT TCC AGT | 53 (35 cycles) | 320 |
| 18S | F - CGG CTA CCA CAT CCA AGG AA R - GCT GGA ATT ACC GCG GCT | 64 (26 cycles) | 187 |
Reverse transcription polymerase chain reaction (RT-PCR) analysis
Total RNA (1µg) was reverse transcribed using AMV reverse transcriptase (Roche, Mannheim, Germany) as previously reported [24]. The cDNA product (1µl) was amplified using GoTag green master mix (Promega, Hawthorn, VIC, Australia) and 10 pmol of forward and reverse primers for HtrA1, HtrA2 and HtrA3 [long (L) and short (S) isoforms] and 18S (Table 2). All PCRs were run in duplicate and products were analysed by electrophoresis on 1% agarose gel (Roche), stained with Gel Red (Biotium, Stepney, SA, Australia) and imaged with Gel Doc-2000 (Bio-Rad, Hercules, CA). A non template control in which diethyl pyrocarbonate water was substituted for RT was included as a negative control. As a loading control, 18S was amplified for each RT reaction.
Immunohistochemistry
Normal ovarian tissues and serous epithelial ovarian cancer tissues from post-menopausal women were collected prospectively from women undergoing surgery for suspected gynaecological malignancies. All samples were collected from anaesthetised patients who had no prior treatment for their malignancies, and had no other history of malignant disease. Tissue samples were collected with prior approval from the Southern Health Human Research Ethics Committee and with prior informed written consent from participants. The tissues were formalin fixed and embedded in paraffin.
Sections of 5µm thickness on Superfrost plus slides were subjected to standard immunohistochemistry. After de-paraffinization and rehydration, antigen was retrieved by microwaving [5min 700W in 0.01mol/L sodium citrate buffer (pH 6.0)], endogenous peroxidase was quenched with 3% hydrogen peroxide in methanol for 10min, and the non-specific binding was blocked with pre-immune blocking serum [15% normal rabbit serum plus 2% normal human serum in high salt Tris buffered saline (HS-TBS) with 0.1% Tween-20] for 40min at room-temperature. The primary antibody [custom-made sheep anti-HtrA3 antibody [14]] or preimmune sheep IgG as a negative control (both 0.5µg/ml) was diluted in the blocking buffer (10% fetal calf serum in HB-TBS with 0.1% Tween-20) and sections were incubated at 37 0C for 1h and washed with high-salt TBS plus 0.6% Tween. The secondary antibody (biotinylated rabbit anti-sheep IgG, 1:200; Vector Laboratories, Burlingame, CA) was applied in the blocking buffer for 30min at room-temperature. Positive immunostaining was revealed by incubating the sections with an avidin-biotin-complex conjugated to horseradish peroxidase (1:100, Vectorstain elite ABC kit) for 30min at room-temperature, followed by the application of the peroxidase substrate 3,3N-diaminobenzidine (DAB, DakoCytomation) leading to a brown precipitate for positive staining. Sections were counterstained with hematoxylin and examined with Olympus BH2 microscope fitted with a Fujix HC-2000 high-resolution digital camera (Fujix, Tokyo, Japan). Sections from normal ovarian tissue were used as positive controls.
Densitometric and statistical analysis
Densitomety was performed using ImageJ software (Version 1.6, National Institutes of Health, Bethesda, MD). Selected bands were quantified based on their relative intensities. Fold change was calculated relative to 18S for each sample. Statistical analysis was performed using Prism for Windows (GraphPad 5.0 Software, San Diego, CA) using a non-parametric Kruskal-Wallis test, followed by Dunns post test. P <0.05 was considered significant for data acquired in arbitrary density units.
Results
HtrA3 is more extensively and drastically downregulated than HtrA1 or HtrA2 in commonly used cancer cell lines
Expression of HtrA1, HtrA2 and HtrA3 (HtrA3-L and HtrA3-S) in several well characterized cell lines derived from ovary, endometrium, testes, breast, prostrate, and colon was determined by conventional RT-PCR (Fig. 1A & B). Specific bands of expected size for each candidate gene were detected.
HtrA1 was consistently expressed in all control samples and in ovarian (n=2), endometrial (n=7), testicular (n=1) and prostate (n=3) cancer cell lines screened. It was downregulated in only one of the two breast and colon cancer cell lines examined (Fig. 1B).
Similarly, HtrA2 was also consistently expressed in most of the cancer cell lines screened. All endometrial (n=7), prostate (n=3) and colon (n=2) cancer cell lines and controls were positive for HtrA2 (Fig. 1A & B). Weak HtrA2 signal was observed in control ovary and in both GCT-derived cancer cell lines, and in control testes and the testicular cancer cell line. Very low levels of HtrA2 were detected in breast control and in both breast cancer cell lines (Fig. 1A & B).
In contrast, both HtrA3-L and HtrA3-S were downregulated in most of the cancer cell lines examined. In all control samples, positive HtrA3 expression was detected. HtrA3 expression was not detected in either GCT-derived cancer cell lines. Its expression was downregulated in five of the seven endometrial cancer cell lines, in both breast cancer cell lines, in two of the three prostate cancer cell lines, and one of the two colon cancer cell lines (Fig. 1A & B). However, positive HtrA3 mRNA was detected in the normal testis and testicular cancer cell lines (Fig. 1A & B).
HtrA3-L mRNA levels are significantly decreased in primary serous and granulosa ovarian tumors
We next determined the mRNA levels of HtrA1, HtrA2 and HtrA3 isoforms (HtrA3-L and HtrA3-S) in ovarian tissue samples. Different sub-types of ovarian tumor [serous tumors (n=8), mucinous tumors (n=6), granulosa cell tumors (GCT, n=19)] were compared to normal premenopausal ovarian tissue (n=9).
HtrA1 mRNA was clearly detected in all ovarian control and tumor samples (Fig. 2A). Densitometric analysis showed no significant differences between control and any ovarian tumor groups (Fig. 2B). HtrA1 mRNA was lower in serous tumors, but not significantly different from any other groups. HtrA1 expression was more variable across the GCT samples.
HtrA2 was also detected in all ovarian control and tumor samples screened with no significant differences between the groups (Fig. 3A & B).
Both HtrA3 isoforms (HtrA3-L and HtrA3-S) were expressed in normal and tumor samples (Fig. 4A). However, densitometric analysis corrected for 18S showed that the two isoforms were differentially expressed. Compared to the normal tissues, the mean level of HtrA3-L was significantly reduced in the serous tumors and GCT (Fig. 4B). HtrA3-L mRNA levels were variable in mucinous tumors and the mean level was not significantly different from the control.
In contrast, compared to the normal tissues, although the mean levels of HtrA3-S mRNA were also low in serous tumors (Fig. 4C), the levels were not significantly different from the control. No significant changes in HtrA3-S expression were observed in any cancer groups.
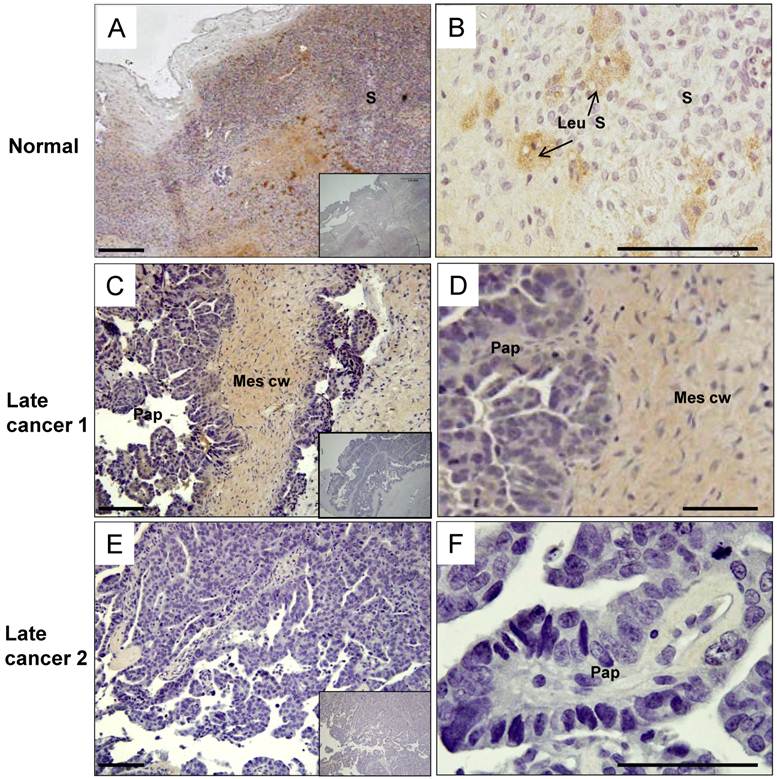
HtrA3 protein levels were decreased progressively in serous ovarian tumors
Since HtrA3-L mRNA was significantly downregulated in serous ovarian tumors, we next determined the HtrA3 cellular localisation in the post-menopausal ovarian control and late stage serous carcinoma by immunohistochemistry.
The post-menopausal ovary was dominated by a stroma composed of spindle shaped cells surrounded by abundant collagen bundles and blood vessels. In the post-menopausal ovary, ovarian follicles and the accompanying granulosa cell compartment are lost, as expected. In the control, moderate levels of HtrA3 (Fig. 5A & B) were detected predominantly in the stroma, particularly in the remaining luteinizing stromal cells and corpus albicans (Fig. 5B).
In the serous carcinoma tumors (grade-3, stages 2-4 - late cancer), papillae are lined by stratified cells of serous type with extensive cellular growth, cellular budding, and increased stromal invasion. In the papillae of the cancer, HtrA3 was significantly reduced or completely absent (Fig. 5C-F). On sections containing mesenchymal cyst wall and papillae (Fig. 5C & D), HtrA3 was detected in the mesenchymal cyst wall, but the papillae showed low HtrA3 staining. On sections containing no mesenchymal cyst wall (Fig. 5E & F), the papillae was almost negative for HtrA3.
RT-PCR analysis of the mRNA levels of human HtrA1, HtrA2 and HrA3 [long (HtrA3-L) and short (HtrA3-S)] in various cancer cell lines. (A) Representative semi-quantitative PCR results from testes, breast, prostate, and colon control and cancer cell lines. (B) Representative semi-quantitative PCR results from ovarian and endometrial control and cancer cell lines. 18S amplification was used as loading control. Pre-menopausal and post-menopausal ovary - normal controls for ovary; Mid-Proliferative and Mid-Secretory - normal controls for mid proliferative and secretory phase endometrium respectively. Cancer cell lines examined, testes - Ntera-2D; breast - MCK-7, MDA-231; prostate - PC3, DU145, and LNCap; colon - SW480 and WIDR; ovary - KGN and COV434; endometrium - ECC1, HEC1-A, HEC1-B, Ishikawa, RL95-2 and AN3CA.
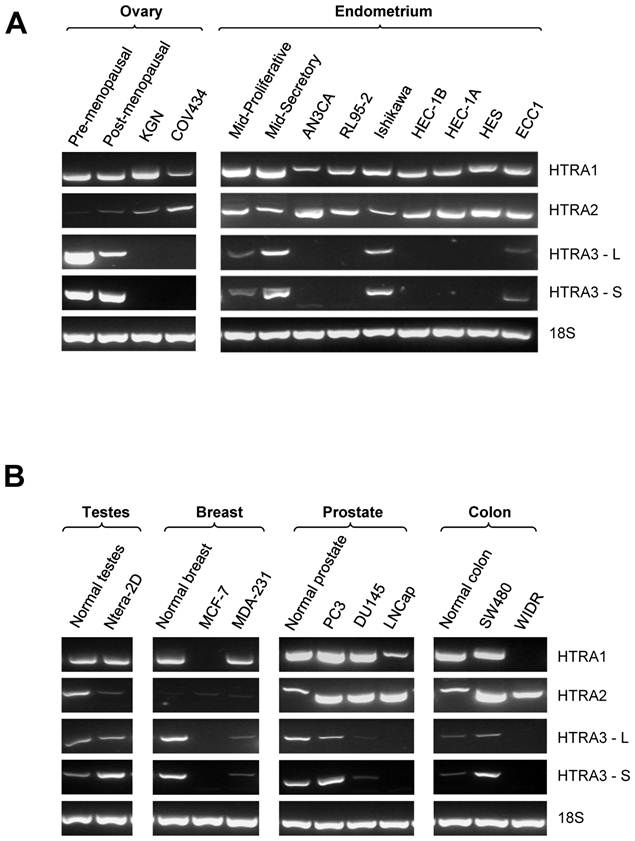
RT-PCR analysis of human HtrA1 expression in normal ovary and different types of ovarian tumors. (A) Representative semi-quantitative RT-PCR results from normal ovary and serous, mucinous and granulosa cell tumor (GCT) ovarian cancers. (B) Densitometric analysis showing the relative mRNA levels (corrected for 18S) in normal ovary and ovarian cancer samples. Patient details for normal and cancer groups are summarised in table 1.
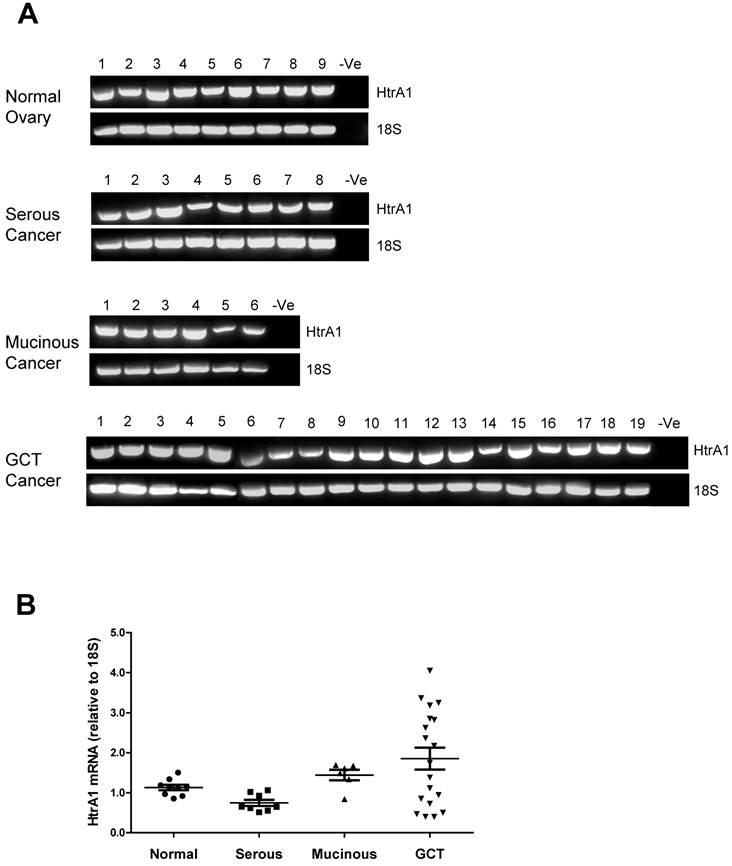
RT-PCR analysis of human HtrA2 expression in normal ovary and different types of ovarian tumors. (A) Representative semi-quantitative RT-PCR results from normal ovary and serous, mucinous and granulosa cell tumor (GCT) ovarian cancers. (B) Densitometric analysis showing relative mRNA levels (corrected for 18S) in normal ovary and ovarian cancer samples. Patient details for normal and cancer groups are summarised in table 1.
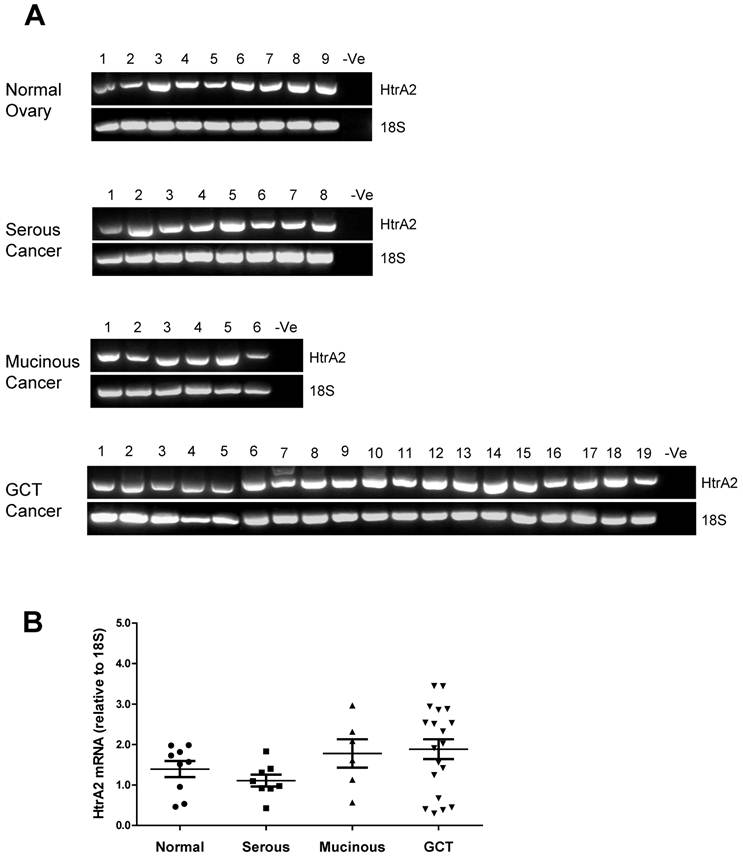
RT-PCR analysis of human HtrA3 (HtrA3-L and HtrA3-S) expression in normal ovary and different types of ovarian tumors. (A) Representative semi-quantitative RT-PCR results from normal ovary and serous, mucinous and granulosa cell tumor (GCT) ovarian cancers. (B & C) Densitometric analysis showing relative mRNA levels (corrected for 18S) in normal ovary and ovarian cancer samples. Patient details for normal and cancer groups are summarised in table 1. * P<0.05, ** P<0.01.
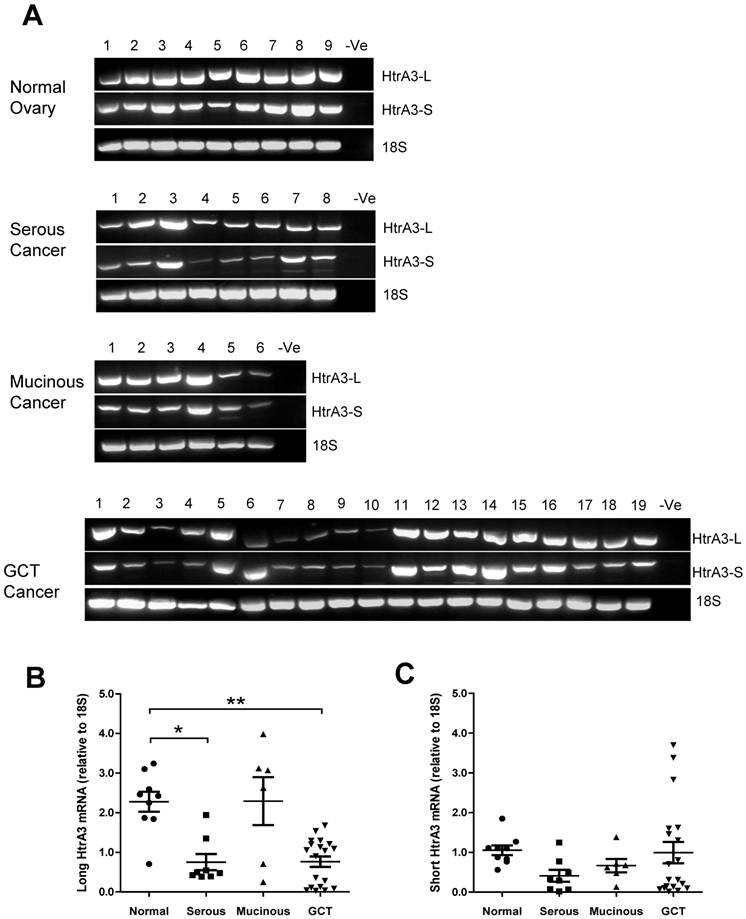
Human HtrA3 in normal and serous ovarian cancer tissues. Representative micrographs of post-menopausal control ovary and two serous (grade 3) cancer tissues immunostained for HtrA3. (A, B) Normal ovary, (C, D, E, F) Serous cancer - grade (G3) - from different patients. Right panel images are magnified view. Negative controls (preimmune sheep IgG) are in lower right inserts. Scale bar = 200 µm; applies to all panels. S - stromal cells; Leu S - lutenizing stromal cells; Mes cw - mesenchymal cell wall; Pap - papillae.
Discussion
This study describes the comparison of the expression of HtrA serine proteases HtrA1, HtrA2 and HtrA3 (HtrA3-L and HtrA3-S) in various cancer cell lines and in primary ovarian tumors. Our study demonstrates that both HtrA3-L and HtrA3-S are more widely lost than HtrA1 or HtrA2 in various cancer cell lines. In addition, we present evidence that HtrA3-L mRNA is significantly downregulated in primary serous and GCTs compared to normal controls. Our results also confirmed a downregulation of HtrA3 protein in late stage serous carcinoma. These findings provide further evidence that HtrA3-L may play an active role as a tumor suppressor in human GCTs and serous carcinomas.
Our results are in agreement with the previous studies showing the downregulation of human HtrA3 in ovarian, endometrial and lung cancers. Both HtrA1 and HtrA3 are secreted proteins with similar domain organisation, suggesting they have similar functions. Though previous studies have reported the downregulation of HtrA1 mRNA in different types of ovarian tumors [21, 33], our study revealed that the downregulation of HtrA3 is more widespread and extensive. In this study, HtrA1 was not found to be significantly reduced in various cancer cell lines or in primary ovarian tumors compared to normal controls. A previous study by Chien et al [21] has also shown that HtrA3 expression was lost in ovarian cell lines expressing HtrA1. Our study found differential expression for HtrA1 and HtrA3, not only in cancer cell lines but also in primary ovarian tumors. However, no direct correlation was observed between HtrA1 and HtrA3 levels in any of the sample group. The differential expression between HtrA1 and HtrA3 in ovarian cancer cells is intriguing; its significance warrants further investigation.
Both HtrA3-L and HtrA3-S are downregulated in human GCT-derived cell lines, while only HtrA3-L was significantly downregulated in human primary GCT and serous carcinoma. Though levels of HtrA3-S also showed a similar downward pattern in serous carcinoma, it was not significantly different. Higher expression levels of HtrA3-L have been previously reported in rat ovaries and also in primary granulosa cells treated with follicle-stimulating hormone [38]. It is therefore possible that HtrA3 levels are decreased or non-detectable in the GCTs and GCT derived cell lines which cannot undergo luteinization. Since both HtrA3-L and HtrA3-S isoforms are proteolytically active [39], further work needs to resolve the role of HtrA3-L in ovarian tumorigenesis.
HtrA3 can bind to TGF-β family members and inhibit TGF-β signalling, and its proteolytic activity is essential for the inhibition [40]. It has also been shown to regulate the IGF/IGFBP system [13, 41] A number of ovarian processes require the action of growth factors including IGF-1 and TGF-β superfamily members [42]. Studies have shown that TGF-β1 can act as a tumor suppressor at the early stages of tumorigenesis, when the tumor is still benign, and as a stimulator of tumor progression, invasion and metastasis at the late stages of tumorigenesis [43]. Furthermore, HtrA3 exhibits substrate specificity toward certain extracellular matrix proteoglycans like decorin, biglycan and fibronectin [40], facilitates cell migration and invasion [24, 44], and hence HtrA3 may also modulate the ECM environment of the ovary. It is therefore possible that HtrA3 plays a role in ovarian tumorigenesis by regulating the actions and signalling of these growth factor systems in the ovary.
Additional support for HtrA3 as a putative tumor suppressor came from a study in lung cancer. Beleford et al [19] showed that down-regulation of HtrA3 expression in smoking related lung cancers was associated with the cigarette smoke-induced methylation of HtrA3. As evidence exists showing that HtrA3 can be a predictive factor of clinical response to chemotherapy; a higher response rate was found for patients with lung cancers expressing higher levels of HtrA3 [32]. Similarly, due to epigenetic inactivation of HtrA1, lower levels of HtrA1 have been reported in lymph node melanoma, ovary, lung and breast cancers [21, 25, 27, 45]. A previous study by Chien et al has indicated that HtrA1 expression enhanced sensitivity to cisplatin and paclitaxel, whereas down-regulation attenuated cytotoxicity in ovarian and gastric cancers [46]. A better understanding of the molecular mechanisms involved in ovarian tumor development may help to distinguish between the high- and low-risk patients and improve their clinical management.
Proteases have been implicated in the development and progression of cancer, by degrading ECM and facilitating cell migration and invasion [47]. However, a new concept is emerging demonstrating that proteases can act as tumour-suppressors. Specific serine proteases in the normal breast, prostate and testis are downregulated in cancers and function as tumour suppressors either by a loss of the gene or methylation in the promoter [48, 49]. Our data suggests that HtrA3 may represent another tumor suppressor in this group.
Moreover, the epithelial-to-mesenchymal transition (EMT) which is critical during embryonic development has a parallel role in tumorigenesis and contributes to tumor invasion, metastasis, and acquisition of therapeutic resistance [50-52]. We have previously demonstrated that HtrA3 expression levels are inversely correlated with motility and invasion [24, 44], and acquisition of increased motility is one of the typical features of EMT. Similar to HtrA1 [53], HtrA3 may function as a tumor suppressor by controlling the EMT, and may function in chemotherapeutic responsiveness by mediating DNA damage response pathways.
In summary, we have shown that both HtrA3-L and HtrA3-S are more consistently downregulated than HtrA1 or HtrA2 in various commonly used cancer cell lines. Importantly, HtrA3-L mRNA and protein was significantly downregulated in primary serous and GCTs compared to controls. Collectively this study highlights the potential role of HtrA3 as a tumor suppressor. Further studies are needed to enhance our understanding of the role of HtrA3 in normal and malignant cells.
Acknowledgements
The authors would like to thank Maria Alexiadis for assistance with primary ovarian cancer RNA extraction and RT preparation.
Grant/Funding support
This project was supported by NHMRC Research Fellowships GNT0494808 (SRF to GN), GNT01002559 (SPRF to PJF), Program Grant GNT0494802, GNT1013533 (CDF to CH), the Victorian Government's Operational Infrastructure Support Program, and the Ovarian Cancer Research Foundation (JR and ANS). The data submitted were audited (PHI Data Audit 12-26).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Greenlee RT, Murray T, Bolden S, Wingo PA. Cancer statistics, 2000. CA Cancer J Clin. 2000;50:7-33
2. Hough CD, Sherman-Baust CA, Pizer ES, Montz FJ, Im DD, Rosenshein NB, Cho KR, Riggins GJ, Morin PJ. Large-scale serial analysis of gene expression reveals genes differentially expressed in ovarian cancer. Cancer Res. 2000;60:6281-7
3. Brun JL, Feyler A, Chene G, Saurel J, Brun G, Hocke C. Long-term results and prognostic factors in patients with epithelial ovarian cancer. Gynecol Oncol. 2000;78:21-7
4. Coticchia CM, Yang J, Moses MA. Ovarian cancer biomarkers: current options and future promise. J Natl Compr Canc Netw. 2008;6:795-802
5. Herzog TJ. Recurrent ovarian cancer: how important is it to treat to disease progression? Clin Cancer Res. 2004;10:7439-49
6. Lipinska B, Zylicz M, Georgopoulos C. The HtrA (DegP) protein, essential for Escherichia coli survival at high temperatures, is an endopeptidase. J Bacteriol. 1990;172:1791-7
7. Krojer T, Garrido-Franco M, Huber R, Ehrmann M, Clausen T. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature. 2002;416:455-9
8. Spiess C, Beil A, Ehrmann M. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell. 1999;97:339-47
9. Hu SI, Carozza M, Klein M, Nantermet P, Luk D, Crowl RM. Human HtrA, an evolutionarily conserved serine protease identified as a differentially expressed gene product in osteoarthritic cartilage. J Biol Chem. 1998;273:34406-12
10. Zumbrunn J, Trueb B. Primary structure of a putative serine protease specific for IGF-binding proteins. FEBS Lett. 1996;398:187-92
11. Faccio L, Fusco C, Chen A, Martinotti S, Bonventre JV, Zervos AS. Characterization of a novel human serine protease that has extensive homology to bacterial heat shock endoprotease HtrA and is regulated by kidney ischemia. J Biol Chem. 2000;275:2581-8
12. Gray CW, Ward RV, Karran E, Turconi S, Rowles A, Viglienghi D, Southan C, Barton A, Fantom KG, West A, Savopoulos J, Hassan NJ, Clinkenbeard H, Hanning C, Amegadzie B, Davis JB, Dingwall C, Livi GP, Creasy CL. Characterization of human HtrA2, a novel serine protease involved in the mammalian cellular stress response. Eur J Biochem. 2000;267:5699-710
13. Nie GY, Hampton A, Li Y, Findlay JK, Salamonsen LA. Identification and cloning of two isoforms of human high-temperature requirement factor A3 (HtrA3), characterization of its genomic structure and comparison of its tissue distribution with HtrA1 and HtrA2. Biochem J. 2003;371:39-48
14. Nie G, Li Y, Hale K, Okada H, Manuelpillai U, Wallace EM, Salamonsen LA. Serine peptidase HTRA3 is closely associated with human placental development and is elevated in pregnancy serum. Biol Reprod. 2006;74:366-74
15. Inagaki A, Nishizawa H, Ota S, Suzuki M, Inuzuka H, Miyamura H, Sekiya T, Kurahashi H, Udagawa Y. Upregulation of HtrA4 in the placentas of patients with severe pre-eclampsia. Placenta. 2012;33:919-26
16. Wang LJ, Cheong ML, Lee YS, Lee MT, Chen H. High-temperature requirement protein A4 (HtrA4) suppresses the fusogenic activity of syncytin-1 and promotes trophoblast invasion. Mol Cell Biol. 2012;32:3707-17
17. Oka C, Tsujimoto R, Kajikawa M, Koshiba-Takeuchi K, Ina J, Yano M, Tsuchiya A, Ueta Y, Soma A, Kanda H, Matsumoto M, Kawaichi M. HtrA1 serine protease inhibits signaling mediated by Tgfbeta family proteins. Development. 2004;131:1041-53
18. Clausen T, Southan C, Ehrmann M. The HtrA family of proteases: implications for protein composition and cell fate. Mol Cell. 2002;10:443-55
19. Beleford D, Liu Z, Rattan R, Quagliuolo L, Boccellino M, Baldi A, Maguire J, Staub J, Molina J, Shridhar V. Methylation induced gene silencing of HtrA3 in smoking-related lung cancer. Clin Cancer Res. 2010;16:398-409
20. Bowden MA, Di Nezza-Cossens LA, Jobling T, Salamonsen LA, Nie G. Serine proteases HTRA1 and HTRA3 are down-regulated with increasing grades of human endometrial cancer. Gynecol Oncol. 2006;103:253-60
21. Chien J, Staub J, Hu SI, Erickson-Johnson MR, Couch FJ, Smith DI, Crowl RM, Kaufmann SH, Shridhar V. A candidate tumor suppressor HtrA1 is downregulated in ovarian cancer. Oncogene. 2004;23:1636-44
22. Ajayi F, Kongoasa N, Gaffey T, Asmann YW, Watson WJ, Baldi A, Lala P, Shridhar V, Brost B, Chien J. Elevated expression of serine protease HtrA1 in preeclampsia and its role in trophoblast cell migration and invasion. Am J Obstet Gynecol. 2008;199:557.e1-10
23. Chien J, Campioni M, Shridhar V, Baldi A. HtrA serine proteases as potential therapeutic targets in cancer. Curr Cancer Drug Targets. 2009;9:451-68
24. Singh H, Makino SI, Endo Y, Nie G. Inhibition of HTRA3 stimulates trophoblast invasion during human placental development. Placenta. 2010;31:1085-92
25. Baldi A, De Luca A, Morini M, Battista T, Felsani A, Baldi F, Catricala C, Amantea A, Noonan DM, Albini A, Natali PG, Lombardi D, Paggi MG. The HtrA1 serine protease is down-regulated during human melanoma progression and represses growth of metastatic melanoma cells. Oncogene. 2002;21:6684-8
26. Shridhar V, Sen A, Chien J, Staub J, Avula R, Kovats S, Lee J, Lillie J, Smith DI. Identification of underexpressed genes in early- and late-stage primary ovarian tumors by suppression subtraction hybridization. Cancer Res. 2002;62:262-70
27. Loss LA, Sadanandam A, Durinck S, Nautiyal S, Flaucher D, Carlton VE, Moorhead M, Lu Y, Gray JW, Faham M, Spellman P, Parvin B. Prediction of epigenetically regulated genes in breast cancer cell lines. BMC Bioinformatics. 2010;11:305
28. Srinivasula SM, Gupta S, Datta P, Zhang Z, Hegde R, Cheong N, Fernandes-Alnemri T, Alnemri ES. Inhibitor of apoptosis proteins are substrates for the mitochondrial serine protease Omi/HtrA2. J Biol Chem. 2003;278:31469-72
29. Trencia A, Fiory F, Maitan MA, Vito P, Barbagallo AP, Perfetti A, Miele C, Ungaro P, Oriente F, Cilenti L, Zervos AS, Formisano P, Beguinot F. Omi/HtrA2 promotes cell death by binding and degrading the anti-apoptotic protein ped/pea-15. J Biol Chem. 2004;279:46566-72
30. Yang QH, Church-Hajduk R, Ren J, Newton ML, Du C. Omi/HtrA2 catalytic cleavage of inhibitor of apoptosis (IAP) irreversibly inactivates IAPs and facilitates caspase activity in apoptosis. Genes Dev. 2003;17:1487-96
31. Nie GY, Li Y, Minoura H, Batten L, Ooi GT, Findlay JK, Salamonsen LA. A novel serine protease of the mammalian HtrA family is up-regulated in mouse uterus coinciding with placentation. Mol Hum Reprod. 2003;9:279-90
32. Beleford D, Rattan R, Chien J, Shridhar V. High temperature requirement A3 (HtrA3) promotes etoposide- and cisplatin-induced cytotoxicity in lung cancer cell lines. J Biol Chem. 2010;285:12011-27
33. Narkiewicz J, Klasa-Mazurkiewicz D, Zurawa-Janicka D, Skorko-Glonek J, Emerich J, Lipinska B. Changes in mRNA and protein levels of human HtrA1, HtrA2 and HtrA3 in ovarian cancer. Clin Biochem. 2008;41:561-9
34. Fuller PJ, Alexiadis M, Jobling T, McNeilage J. Seladin-1/DHCR24 expression in normal ovary, ovarian epithelial and granulosa tumours. Clin Endocrinol (Oxf). 2005;63:111-5
35. Jamieson S, Alexiadis M, Fuller PJ. Expression status and mutational analysis of the ras and B-raf genes in ovarian granulosa cell and epithelial tumors. Gynecol Oncol. 2004;95:603-9
36. Jamieson S, Butzow R, Andersson N, Alexiadis M, Unkila-Kallio L, Heikinheimo M, Fuller PJ, Anttonen M. The FOXL2 C134W mutation is characteristic of adult granulosa cell tumors of the ovary. Mod Pathol. 2010;23:1477-85
37. Fuller PJ, Chu S, Jobling T, Mamers P, Healy DL, Burger HG. Inhibin subunit gene expression in ovarian cancer. Gynecol Oncol. 1999;73:273-9
38. Bowden MA, Drummond AE, Fuller PJ, Salamonsen LA, Findlay JK, Nie G. High-temperature requirement factor A3 (Htra3): a novel serine protease and its potential role in ovarian function and ovarian cancers. Mol Cell Endocrinol. 2010;327:13-8
39. Singh H, Makino S, Endo Y, Li Y, Stephens AN, Nie G. Application of the wheat-germ cell-free translation system to produce high temperature requirement A3 (HtrA3) proteases. Biotechniques. 2012;52:23-8
40. Tocharus J, Tsuchiya A, Kajikawa M, Ueta Y, Oka C, Kawaichi M. Developmentally regulated expression of mouse HtrA3 and its role as an inhibitor of TGF-beta signaling. Dev Growth Differ. 2004;46:257-74
41. Hou J, Clemmons DR, Smeekens S. Expression and characterization of a serine protease that preferentially cleaves insulin-like growth factor binding protein-5. J Cell Biochem. 2005;94:470-84
42. Richards JS, Russell DL, Ochsner S, Hsieh M, Doyle KH, Falender AE, Lo YK, Sharma SC. Novel signaling pathways that control ovarian follicular development, ovulation, and luteinization. Recent Prog Horm Res. 2002;57:195-220
43. Akhurst RJ, Derynck R. TGF-beta signaling in cancer--a double-edged sword. Trends Cell Biol. 2001;11:S44-51
44. Singh H, Endo Y, Nie G. Decidual HtrA3 negatively regulates trophoblast invasion during human placentation. Hum Reprod. 2011;26:748-57
45. Esposito V, Campioni M, De Luca A, Spugnini EP, Baldi F, Cassandro R, Mancini A, Vincenzi B, Groeger A, Caputi M, Baldi A. Analysis of HtrA1 serine protease expression in human lung cancer. Anticancer Res. 2006;26:3455-9
46. Chien J, Aletti G, Baldi A, Catalano V, Muretto P, Keeney GL, Kalli KR, Staub J, Ehrmann M, Cliby WA, Lee YK, Bible KC, Hartmann LC, Kaufmann SH, Shridhar V. Serine protease HtrA1 modulates chemotherapy-induced cytotoxicity. J Clin Invest. 2006;116:1994-2004
47. Turk B. Targeting proteases: successes, failures and future prospects. Nat Rev Drug Discov. 2006;5:785-99
48. Goyal J, Smith KM, Cowan JM, Wazer DE, Lee SW, Band V. The role for NES1 serine protease as a novel tumor suppressor. Cancer Res. 1998;58:4782-6
49. Lopez-Otin C, Matrisian LM. Emerging roles of proteases in tumour suppression. Nat Rev Cancer. 2007;7:800-8
50. Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315:1650-9
51. Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15:740-6
52. Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265-73
53. Wang N, Eckert KA, Zomorrodi AR, Xin P, Pan W, Shearer DA, Weisz J, Maranus CD, Clawson GA. Down-regulation of HtrA1 activates the epithelial-mesenchymal transition and ATM DNA damage response pathways. PLoS One. 2012;7:e39446
Author contact
![]() Corresponding author: Harmeet Singh, Prince Henry's Institute of Medical Research, PO Box 5152, 246 Clayton Road, Clayton, VIC 3168, Australia. Fax: +61 3 9594 6125; email: harmeet.singhorg.
Corresponding author: Harmeet Singh, Prince Henry's Institute of Medical Research, PO Box 5152, 246 Clayton Road, Clayton, VIC 3168, Australia. Fax: +61 3 9594 6125; email: harmeet.singhorg.

 Global reach, higher impact
Global reach, higher impact