Impact Factor
ISSN: 1837-9664
J Cancer 2013; 4(5):350-357. doi:10.7150/jca.6394 This issue Cite
Research Paper
Comprehensive microRNA Profiling of Prostate Cancer
1. Translational Surgical Pathology Section, Laboratory of Pathology, National Cancer Institute, National Institutes of Health, USA;
2. Urologic Oncology Branch, National Cancer Institute, National Institutes of Health, USA.
Received 2013-4-3; Accepted 2013-5-2; Published 2013-5-9
Abstract
MicroRNAs are small non-coding RNA molecules that have been shown to regulate the expression of genes linked to cancer. The relevance of microRNAs in the development, progression and prognosis of prostate cancer is not fully understood. It is also possible that these specific molecules may assist in the recognition of aggressive tumors and the development of new molecular targets. Our study investigated the importance of several microRNAs in cases of prostate cancer from 37 patients that were manually microdissected to obtain pure populations of tumor cells, normal epithelium and adjacent stroma. MicroRNA was extracted for PCR array profiling. Differentially expressed miRNAs for each case were used to compare tumor vs. normal epithelium and tumor-adjacent stroma samples.
Loss of 18 miRNAs (e.g.miR-34c, miR-29b, miR-212 and miR-10b) and upregulation of miR-143 and miR-146b were significantly found in all the tumors in comparison with normal epithelium and/or stroma (p≤ 0.001). A different signature was found in the high grade tumors (Gleason score ≥ 8) when compared with tumors Gleason score 6. Upregulation of miR-122, miR-335, miR-184, miR-193, miR-34, miR-138, miR-373, miR-9, miR-198, miR-144 and miR-215 and downregulation of miR-96, miR-222, miR-148, miR-92, miR-27, miR-125, miR-126, miR-27 were found in the high grade tumors.
MicroRNA profiling in prostate cancer appears to have unique expression patterns in comparison with normal tissue. These differential expressed miRNAs may provide novel diagnostic and prognostic tools that will assist in the recognition of prostate cancers with aggressive behavior.
Keywords: microRNA, Prostate Cancer, biomarkers.
Introduction
Prostate cancer is the most common non-skin malignancy in men. The American Cancer Society (ACS) 1 estimated that about 240,890 cases of prostate cancer will be newly diagnosed in 2011 and 33,720 men will die from prostate cancer in the United States.
Currently, prostate-specific antigen (PSA) testing, digital rectal examination and histopathological evaluation of prostate needle biopsies are all used for the detection and monitoring of prostate cancer progression. Prostate cancer Gleason score (Gleason grading system) is determined microscopically evaluating the degree of loss of normal glandular tissue architecture. It is used to help to evaluate patient's prognosis 2,3-4 and to guide the clinician's with the best suitable treatment option 5-7. However, the course of the disease is highly variable and occasionally some cases behave independently than their correspondent Gleason score group 8-9. Therefore, new and more specific biomarkers are needed to better predict cancer prognosis.
MicroRNAs (miRNAs) are small (<22 nt), non-coding RNA molecules that play a role in many biological processes. Actually more than 1,400 human miRNA sequences have been identified thus far and many of them linked to cancer pathogenesis. It has been shown that they modulate the expression levels of proteins based on sequence complementarity with their target mRNA. Interestingly, while coding sequences in one gene can be regulated by several miRNAs, one miRNA may target multiple mRNAs. Their relevance in cancer is related to the regulation of important cellular processes and pathways involving tumorigenesis, cell proliferation, differentiation and apoptosis among others 10.
Previous studies have suggested a role for miRNAs as diagnostic markers in tumors that show a specific profile 11, and more recently they have been used as therapeutic targets in several trials 12-13. Specific miRNA signatures in solid tumors have been found frequently in colon 14, breast15, bladder 16-17 and pancreatic cancer. In prostate cancer, screening of miRNAs profiles from tumor versus normal tissues has reported inconsistent patterns, showing occasionally up-regulation 18 and generally down-regulation of miRNAs 19. Although around 50 miRNAs have been linked to prostate cancer, few of them have been showed to be related with disease pathogenesis 20. MicroRNAs in prostate cancer have been suggested to have a relevant role as biomarkers; a more recent comparison of expression profile in prostate cancer versus benign hyperplastic prostate showed some miRNAs that may discriminate between the two groups 21.
In order to understand the role of miRNA in prostate cancer tumorigenesis, we performed a comprehensive differential miRNA expression analysis of a group of prostate tumors with different Gleason score compared to their corresponding normal epithelium and stromal tissue.
Material and Methods
MicroRNA expression profiling using RT2 PCR arrays: Extracted total RNA including miRNAs (10 ng/µl concentration) was first reverse transcribed into first strand cDNA using the RT2- miRNA First Strand Kit following manufacturer's recommendations (SA Biosciences, Rockville, MD). One µl cDNA per well was then mixed with SYBR Green qPCR Master Mix and placed into a 96-well PCR-array plate containing a panel of 88 mature miRNAs sequences. The arrays also contain appropriate small nucleolar RNA sequences that are used as housekeeping assays and quality controls. One µl was used in a 12 µl final volume reaction for Real-time PCR analysis on an Applied Biosystems Step-One Plus Real Time PCR system. Relative amounts were calculated by the ΔΔCT method and further normalized to the values of their corresponding normal tissue samples.
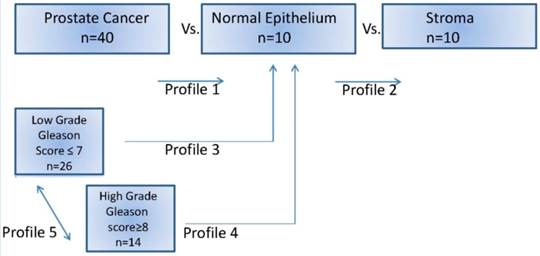
Differential miRNA expression profiling: Differentially expressed miRNAs for each case, and according Gleason score were compared with the normal epithelium and stromal tissue, defined as those with a two-fold change and a false discovery rate <5%. Low grade tumors were the presenting Gleason score 6 and 7 (3+3; 3+4) and High grade tumors were Gleason score 8 and 9 (4+4; 4+5). Unsupervised clustering was used to evaluate the role of significant expressed miRNAs according the Gleason grade. Different profiles were designed to compare prostate cancer (low and high grade) vs. normal epithelium and vs. stroma cells (Figure 1).
Statistical analysis: A two-fold change (Up or down) was determined as significant. MicroRNA expression was also correlated to the clinicopathologic features of the tumors. The relationship between these variables was evaluated by the Mann-Whitney or Kruskall-Wallis non-parametric tests and significance was defined as p<0.05.
MicroRNA prostate cancer profiles investigated in this study. After manual microdissection, separate groups of cells were preserved and then used for comparison.
Results
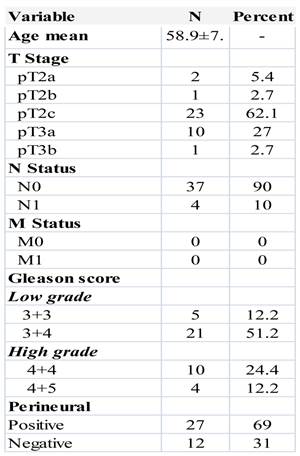
The clinicopathologic characteristics of the patients used for miRNA profiling are shown in Table 1. Patients mean age was 58.9 ±7.7. Twenty-six patients were pT2 stage and 11 pT3 stage. Three tumor samples from core biopsies were also included. Low grade cases included 5 cases with Gleason score 6 (3+3) and 21 with Gleason score 7 (3+4). The group of high grade cases were 10 with Gleason score 8 (4+4) and 4 cases with Gleason score 9 (4+5). We didn't have in our sample any case Gleason score 5, Gleason 7 (4+3) nor Gleason score 10 (5+5). Only 4 cases presented positive lymph nodes and none of them had metastasis.
Clinicopathological features of prostate cancer patients.
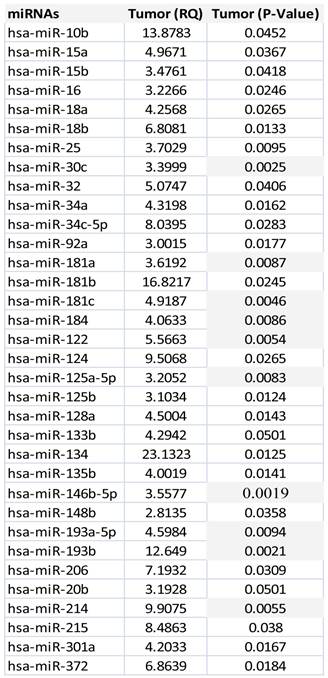
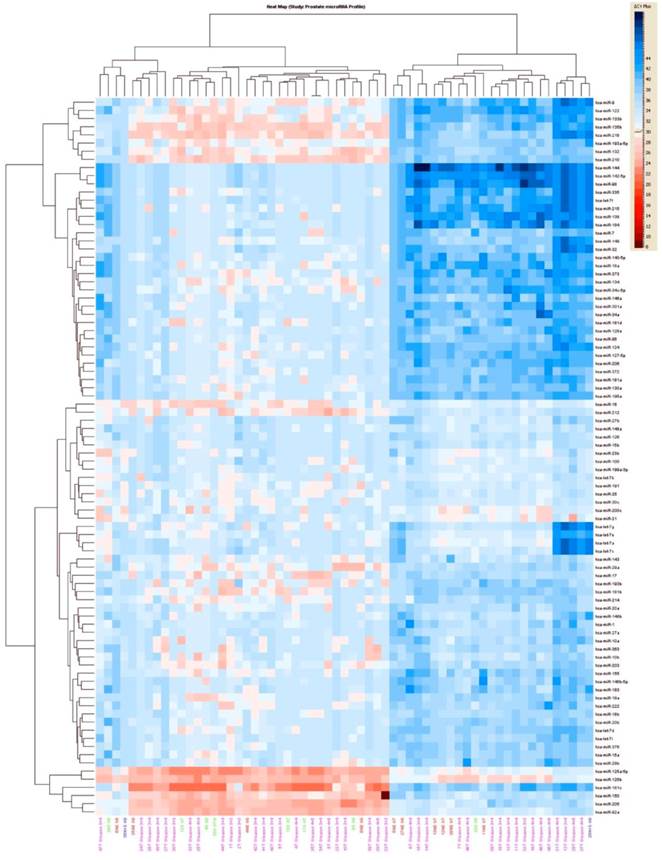
Unsupervised hierarchical clustering of all prostate tumors based on miRNA expression values showed that mainly two groups of miRNAs clusters separated from most of the normal epithelium samples (Figure 2). The first group of miRNAs is (miR-92a, miR-150, miR-181c, miR-205) and the second one include miR-9, miR-122, miR-133b, miR-135b, miR-210, miR -218, miR-193, miR-132). Expression values for dysregulated miRNAs ranged from 2.81 fold up to 23 fold change in all the tumors in comparison with normal epithelium. There were 34 statistically significant miRNAs that all of them were upregulated. MicroRNA-30c, miR-122, miR-125a, miR-181a, miR-181c miR-146b-5p, miR-184, miR193a, miR193b and miR-214 were the most significantly upregulated, (p<0.005), Table 2.
MicroRNA-193b and 181b, miR-20a and 10b, and miR-125a and 155 were differentially expressed in patients having perineural (PI), extracapsular extension and lymphatic invasion, respectively.
MicroRNAs dysregulated in prostate tumor samples versus normal epithelium group.
Profile 1: All prostate tumor samples versus normal epithelium.
MicroRNA profile 2 was performed comparing the normal epithelium group versus stroma with all prostate tumors. Stroma versus normal epithelium miRNA (profile 2), showed some upregulated miRNAs ranging from 2.6 fold up 20.41 fold change, Table 3.
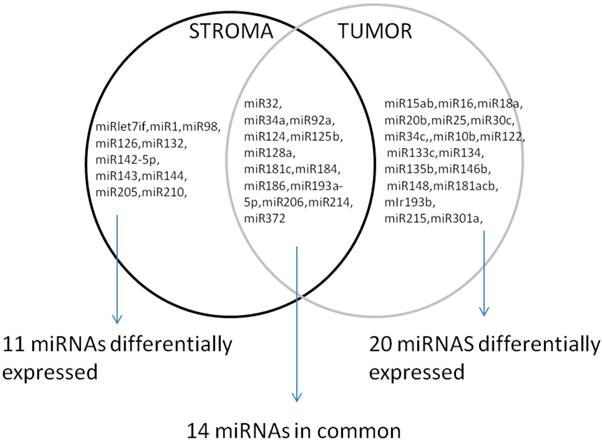
Interestingly, when the comparison between the normal epithelium versus stroma and tumor samples was done, 14 miRNAs were commonly present in all the three groups, but others were specific to discriminate between the normal epithelium, the stroma and the prostate tumor group, Figure 3.
Low grade prostate tumors, Gleason score 6 (3+3) and 7 (3+4) showed commonly upregulated miRNAs when compared with the normal epithelium (profile 3). The miRNA expression value that were most statistically significant (P<0.0001) had fold change ranging from 3 up to 1015-fold. They were let-7f, miR-1, miR-17, miR-98, miR-122, miR-125b, miR-125a-5p, miR-144, miR-142-5p, miR-146b-5p, miR-181a and miR-210. In high grade tumors that were Gleason score 8 (4+4) and 9 (4+9), (profile 4); they were commonly upregulated, fold change 3.7 up 1000-fold. The most significant were miR-32, miR-98, miR-125a-5p, miR-138, miR-142p, miR-144, miR-146-5p, miR-181a, miR-181c, miR-183, miR-184, miR-205, miR-206, miR-215, miR-272, and miR-301. However, dysregulated miRNAs expression profiles of low grade versus high grade did not showed any specific signatures to differentiate the two groups.
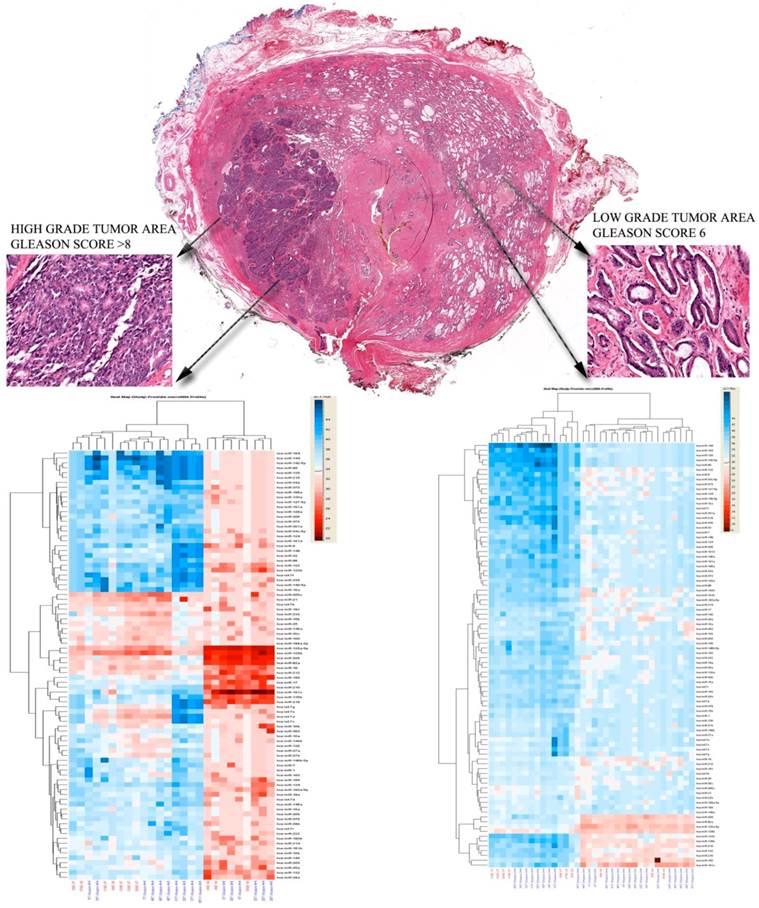
In some cases, we additionally investigated if prostate tumor heterogeneity was related with different miRNAs profiles, in the same case areas of low grade tumors and areas of high grade tumors were microdissected, and then compared with each other. Different profiles were seen in the two tumor areas, Figure 4.
Profile of upregulated miRNA found in stroma tissue in comparison with normal epithelium.
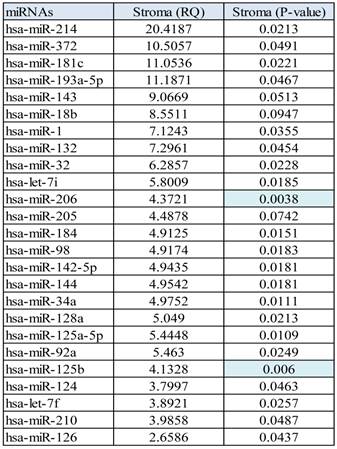
Differentially expressed miRNAs in tumor and stroma versus normal epithelium.
Histological appearance of a selected case analyzed. A) Low power slide scan demonstrating the heterogeneity of the disease. Note the occurrence of malignant transformation in different parts of the gland, areas of high grade disease (Gleason >8) and small areas of low grade cancer are both occurring in the peripheral zone. B) High magnification of the high grade area. Note the complete loss of normal gland pattern with solid tumor cell clustering. C) Low Gleason score areas also in the peripheral zone. The micrographs highlight the need for cell enrichment for the analysis of prostate cancer samples.
Discussion
Our data show consistent dysregulation of miRNAs in human prostate cancer, some of which are potentially involved in tumor formation and progression. We had determined initially genome-wide expression changes of miRNAs in a group of prostate tumors representing different Gleason score, which were classified as low grade tumors (Gleason score ≤7) and high grade tumors (Gleason score≥8), and compared for the first time the profile with normal epithelium and stromal tissue. The carefully needle microdisecction by a specialized pathologist of whole mount FFPE prostatectomy samples support the specificity of the results for each cells group with less chance to contamination as can be happen in other studies. The analysis revealed an important role of miRNAs as key regulators in the pathogenesis of prostate cancer.
There were 34 statistically significant miRNAs that all of them were upregulated. They ranged from 2.81 fold up to 23 fold change and the ones that shown more significance (p≤0.001) were the following: miR-30c, miR-122, miR-125a-b, miR-181a, miR-181c miR-146b-5p, miR-184, miR193a, miR193b and miR-214. According Ozen et al, miR-125, miR-145 and lect-7c were found dysregulated in tumors versus benign hyperplasia tissue, supporting the relevance of miR-125 in prostate cancer 21. Tong et al, found downregulated miR-23, miR-100,miR-145, miR-221 and miR-22 in tumors versus normal tissues, keeping consistency with Schaefer et al with the miR-125b, miR-145, miR-221 and miR-222 in prostate tumor samples 22-23. This upregulated miRNA profile is also found in our study (see Table 2).
Some of these miRNAs may regulate the expression of cancer-related genes in prostate cancer affecting the phenotype of these cells, because in same patients expression profiles were different according the most common morphologic pattern evaluated by a pathologist.
Consistent with previous studies, our data supported the hypothesized involvement of miRNAs in prostate tumors. A different signature was found in the high grade tumors (Gleason score ≥ 8) when compared with tumors Gleason score 6. Upregulation of miR-122, miR-335, miR-184, miR-193, miR-34, miR-138, miR-373, miR-9, miR-198, miR-144 and miR-215 and downregulation of miR-96, miR-222, miR-148, miR-92, miR-27, miR-125, miR-126, miR-27 were found in the high grade tumors.
Interestingly, stroma and normal epithelium showed different pattern of expression, been more dysregulated in some tumors suggesting a potential previous alteration than in the rest of the cases.
Differently expressed miRNAs have been suggested to have an impact as a diagnostic, prognosis and predictor biomarkers in prostate cancer. Previous studies were focused in blood-based circulating miRNAs profile in prostate cancer supported by their tissue specificity feature 24.
Within the group of miRNAs that were specific for stromal prostate tissue that discriminated between tumor cells when were compared with normal epithelium were the following:, miR-let7, miR-1,miR-98, miR-126, miR-132, miR-142, miR-143, miR-144, miR-205, miR-210. Interestingly, a recent study performed in prostate cancer suggested that miR-205 was linked with basement membrane located in the extracellular matrix of the normal epithelial glands in the prostate. The lack of the basement membrane integrity may potentially be related with tumor cell invasion and metastasis formation and the therapeutic role of this miRNA is highlighted 25.
Our data are consistent with the results from independent groups analyzing prostate cancer demonstrating expression of miRNAs cluster at significantly higher levels than normal tissue.
Conclusions
MicroRNA profiling in prostate tumors demonstrates distinctive expression patterns for tumor cells. While the miRNA signature can reflect differences in the underlying and unique molecular changes for tumors with high or low grade Gleason score, it can be used to support the diagnosis but also to detect some cases that will have a poor prognosis.
MicroRNA expression seems to be involved in some steps of pathogenesis: both as an element for tumor development as well as a consequence of or in response to the initial malignant transformation and part of tumor progression.
In summary, the results of this study uncover an etiological contribution of miRNAs in prostate cancer and validate their use for future studies concentrating on the role of individual miRNAs in disease progression that could be used for development of miRNA-based diagnostic and therapeutic strategies. A more detailed understanding of the molecular mechanisms and regulatory pathways at work in prostate cancer will enormously assist in improving the design and target selection of therapeutic strategies.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Brawley OW. Prostate cancer epidemiology in the United States. World J Urol. 2012;30:195-200
2. Stamey TA, McNeal JE, Yemoto CM, Sigal BM, Johnstone IM. Biological determinants of cancer progression in men with prostate cancer. JAMA. 1999;281:1395-400
3. Lilleby W, Torlakovic G, Torlakovic E, Skovlund E, Fossa SD. Prognostic significance of histologic grading in patients with prostate carcinoma who are assessed by the Gleason and World Health Organization grading systems in needle biopsies obtained prior to radiotherapy. Cancer. 2001;92:311-9
4. Spires SE, Cibull ML, Wood DP Jr, Miller S, Spires SM, Banks ER. Gleason histologic grading in prostatic carcinoma. Correlation of 18-gauge core biopsy with prostatectomy. Arch Pathol Lab Med. 1994;118:705-8
5. Roach M 3rd, Marquez C, Yuo HS. et al. Predicting the risk of lymph node involvement using the pre-treatment prostate specific antigen and Gleason score in men with clinically localized prostate cancer. Int J Radiat Oncol Biol Phys. 1994;28:33-7
6. Lughezzani G, Gallina A, Larcher A. et al. Radical prostatectomy represents an effective treatment in patients with specimen-confined high pathological Gleason score prostate cancer. BJU Int. 2012
7. Cerovic S, Brajuskovic G. [Forty years' use of Gleason grading in the diagnostics and treatment of prostate cancer]. Vojnosanit Pregl. 2006;63:1049-53
8. Arora R, Koch MO, Eble JN, Ulbright TM, Li L, Cheng L. Heterogeneity of Gleason grade in multifocal adenocarcinoma of the prostate. Cancer. 2004;100:2362-6
9. Bazinet M, Hamdy SM, Begin LR, Stephenson RA, Fair WR. Prognostic significance of antigenic heterogeneity, Gleason grade, and ploidy of lymph node metastases in patients with prostate cancer. Prostate. 1992;20:311-26
10. Lovat F, Valeri N, Croce CM. MicroRNAs in the pathogenesis of cancer. Semin Oncol. 2011;38:724-33
11. Sempere LF. Integrating contextual miRNA and protein signatures for diagnostic and treatment decisions in cancer. Expert Rev Mol Diagn. 2011;11:813-27
12. Hassan O, Ahmad A, Sethi S, Sarkar FH. Recent updates on the role of microRNAs in prostate cancer. J Hematol Oncol. 2012;5:9
13. Gordanpour A, Nam RK, Sugar L, Seth A. MicroRNAs in prostate cancer: from biomarkers to molecularly-based therapeutics. Prostate Cancer Prostatic Dis. 2012
14. Oberg AL, French AJ, Sarver AL. et al. miRNA expression in colon polyps provides evidence for a multihit model of colon cancer. PLoS One. 2011;6:e20465
15. Ferracin M, Querzoli P, Calin GA, Negrini M. MicroRNAs: toward the clinic for breast cancer patients. Semin Oncol. 2011;38:764-75
16. Song T, Xia W, Shao N. et al. Differential miRNA expression profiles in bladder urothelial carcinomas. Asian Pac J Cancer Prev. 2010;11:905-11
17. Enkelmann A, Heinzelmann J, von Eggeling F. et al. Specific protein and miRNA patterns characterise tumour-associated fibroblasts in bladder cancer. J Cancer Res Clin Oncol. 2011;137:751-9
18. Lu J, Getz G, Miska EA. et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-8
19. Volinia S, Calin GA, Liu CG. et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257-61
20. Pang Y, Young CY, Yuan H. MicroRNAs and prostate cancer. Acta Biochim Biophys Sin (Shanghai). 2010;42:363-9
21. Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788-93
22. Tong AW, Fulgham P, Jay C. et al. MicroRNA profile analysis of human prostate cancers. Cancer Gene Ther. 2009;16:206-16
23. Schaefer A, Stephan C, Busch J, Yousef GM, Jung K. Diagnostic, prognostic and therapeutic implications of microRNAs in urologic tumors. Nat Rev Urol. 2010;7:286-97
24. Selth LA, Tilley WD, Butler LM. Circulating microRNAs - macro-utility as markers of prostate cancer? Endocr Relat Cancer. 2012
25. Gandellini P, Profumo V, Casamichele A. et al. miR-205 regulates basement membrane deposition in human prostate: implications for cancer development. Cell Death Differ. 2012
Author contact
![]() Corresponding author: Maria J. Merino M.D. Address: 10 Center Drive, Building 10 MSC Room 2B44, Bethesda MD 20892. Phone: (301) 496-2441 Fax: (301) 480-1458. E-mail: mjmerinonih.gov.
Corresponding author: Maria J. Merino M.D. Address: 10 Center Drive, Building 10 MSC Room 2B44, Bethesda MD 20892. Phone: (301) 496-2441 Fax: (301) 480-1458. E-mail: mjmerinonih.gov.

 Global reach, higher impact
Global reach, higher impact