Impact Factor
ISSN: 1837-9664
J Cancer 2013; 4(6):468-472. doi:10.7150/jca.6568 This issue Cite
Research Paper
Low Frequency of the ERG Oncogene Alterations in Prostate Cancer Patients from India
1. Rajiv Gandhi Cancer Institute and Research Centre, New Delhi, India.
2. Center for Prostate Disease Research, Department of Surgery, Uniformed Services University of the Health Sciences, Rockville, Maryland, USA.
3. Urology Service, Walter Reed National Military Medical Center, Bethesda, Maryland, USA.
4. Bangalore Institute of Oncology - HCG, Bangalore, India.
5. Department of Genitourinary Pathology, Joint Pathology Center, Silver Spring, Maryland, USA.
Received 2013-4-26; Accepted 2013-5-28; Published 2013-7-5
Abstract
Introduction and Objective: ERG oncogene fusions (predominantly TMPRSS2-ERG) represent the most common (50-70% frequency) and validated prostate cancer (CaP) genome alteration in the Western countries. A common TMPRSS2-ERG fusion type leads to the androgen dependent tumor cell specific expression of the TMPRSS2-ERG fusion transcript and amino terminally truncated ERG oncoprotein. CaP prevalence and aggressiveness, as well as genomic alterations vary in different geographic locations in the world. Recent studies from our group highlighted significantly lower frequency of ERG alterations in prostate index tumors of African American men (~30%) in comparison to Caucasian Americans (~60%). Further, much lower frequencies (10 -25%) of ERG alterations have been reported in studies from China and Japan. There is no study on ERG alterations in CaP patients from India, representing a significant portion of the world male population. This study focuses on the frequency of ERG oncoprotein expression in CaP patients from India.
Methods: De-identified formalin-fixed paraffin-embedded (FFPE) specimens from radical prostatectomy (RP) specimens of 51 patients from the Rajiv Gandhi Cancer Institute and Research Centre (RGCI), New Delhi, India, were analyzed for ERG alterations. The ERG oncoprotein expression as a surrogate of ERG gene fusions was analyzed by using a highly specific ERG monoclonal antibody (9FY). TMPRSS2-ERG fusion was assessed by fluorescent in situ hybridization (FISH) assays using the break-apart ERG probes.
Results: Specimens reflecting prior hormonal treatment, or lacking any tumor content, were excluded from the analyses. Of the thirty evaluable specimens, ERG positive tumors were present in 8 cases (27%) and one tumor specimen exhibited rare ERG positive cells. None of the benign glands were positive for ERG supporting previous studies showing complete specificity of the ERG oncoprotein for detection of tumors cells in prostate.
Conclusions: Frequency of ERG oncoprotein expression is much lower in CaP patients from India in comparison to higher frequency of ERG alterations noted in Western countries. ERG frequency in Indian CaP is similar to observations from Japan and China. Since ERG oncogenic activation is a promising biomarker and therapeutic target for CaP, careful evaluation of ERG is needed in CaP patients from different parts of the world.
Keywords: prostate cancer, ERG oncogene, India.
Introduction
Prostate cancer (CaP) remains the most common non-skin cancer of men and the second leading cause of cancer deaths in the USA1. CaP is the fifth most common cancer in the world in both sexes and second most common in men after melanoma2. CaP prevalence and aggressiveness, as well as genomic alterations, vary in different geographic locations in the world with it being most prevalent in Caucasians and least prevalent among Asians3,4. According to the World Health Organization GLOBOCAN data from 2008, the highest regional incidence is in Australia and New Zealand (104.2 per 100,000) and the lowest in South-Central Asia (4.1 per 100,000)2. The incidence in India has been steadily increasing by approximately 10% from 1978 to 19974 and in 2008 was 3.7 per 100,000, with a mortality rate of 2.5 per 100,0002. It is also noted that a much higher percentage of the CaP is detected in the advanced stages which is likely due to the lack of wide spread use of CaP screening.
TMPRSS2-ERG gene fusion leading to ERG over expression represents a highly prevalent oncogenic alteration (50-70%) in CaP patients from Western countries5-11. ERG gene fusions often involve regulatory sequences of the androgen receptor (AR) responsive genes (predominantly TMPRSS2) and protein coding sequences of nuclear transcription factors of the ETS gene family (predominantly ERG)12-16. These gene fusions lead to unscheduled androgen dependent expression of ETS related transcription factors in tumor cell specific manner17. Extensive evaluations of ERG alterations at genome, transcript and protein levels demonstrate unprecedented specificity of ERG fusions for detecting prostate tumor cells14-17. Studies focusing on the oncogenic functions of ERG point to its involvement in: abrogating differentiation; facilitating cell invasion and epithelial to mesenchymal transition; and disrupting epigenetic, inflammatory and DNA damage control mechanisms 14-17. Therapeutic targeting of ERG or ERG interacting proteins, such as PARP, hold promise in developing new strategies for the treatment of CaP18,19. In summary multi-pronged evaluations of the ERG in CaP continue to reflect the critical causal role of this prevalent oncogenic activation in CaP.
Our recent report20 using a matched cohort of CaP cases showed a significantly lower frequency of ERG alterations in African Americans (28%) in comparison to Caucasian Americans (63%) 21,22. A much lower frequency of ERG (7.5-28%) alteration has also been reported in studies from China and Japan21,23,24,25. There is no study on ERG alterations in CaP patients from India, representing a significant portion of the world population. This study focused on the frequency of ERG oncoprotein expression in CaP patients from India.
Materials and Methods
Prostate Specimens
De-identified formalin-fixed, paraffin-embedded specimens from RP specimens of 51 patients from RGCI, New Delhi, India, were analyzed. The specimens were collected from 2003-2008 under an International Institutional Review Board-approved protocol.
Immunohistochemistry (IHC) Assay for ERG oncoprotein expression
Evaluation of the ERG oncoprotein expression in prostate tissues was performed as described previously26. Briefly, four µm sections were taken from the specimens that were deparaffinized, dehydrated and blocked in 0.6% hydrogen peroxide in methanol for 20 min. The sections were then microwaved in EDTA (pH 8.0) for 30 min. and cooled for 30 min. at room temperature in EDTA buffer. The sections were then blocked in 1% horse serum for 40 min. and were incubated with the ERG-MAb mouse monoclonal antibody (9FY, available from Biocare Medical Inc.) at a dilution of 1:1280 for 60 min. at room temperature. Sections were incubated with the biotinylated horse anti-mouse antibody at a dilution of 1:200 (Vector Laboratories, Burlingame, CA) for 30 min. followed by treatment with the ABC Kit (Vector Laboratories, Burlingame, CA) for 30 min. The color was developed by VIP (Vector Laboratories, Burlingame, CA) treatment for 5 min. and the sections were counterstained by hematoxylin. ERG expression was reported as positive or negative within the specimen. Positive staining of endothelial cells in specimens served as built-in control for the assay. ERG protein expression was also correlated with clinico-pathologic features.
Fluorescent In-situ Hybridization (FISH) Assay for detection of ERG gene rearrangements
Dual-color interphase FISH for detecting TMPRSS2-ERG fusion status or ERG rearrangement status of chromosome 21q22.2 was analyzed by FISH break-apart assay12. Biotin-14-dCTP labeled BAC clone RP11-24A11 (red avidin-rhodamine) and digoxigenindUTP-labeled BAC clone RP11-137J13 (green fluorescein-tagged anti-DIG antibody) were used in FISH assay as described previously26,27.
Results
Of the original 51 specimens, 30 (Table 1) were evaluable for identifying the incidence of ERG positivity (Figure 1). Specimens were not included if they had prior hormonal treatment (13 patients), or the specimen did not contain any tumor (8 patients). ERG positive tumors were present in 8 of 30 cases (27%) and one tumor specimen exhibited rare ERG positive cells. None of the benign glands were positive for ERG supporting previous studies on complete specificity of ERG oncoprotein for detection of tumors cells in prostate. There were no primary Gleason 5 patterns in the ERG positive cohort (Table 2). All of the tumors that were positive for ERG were confirmed to have ERG rearrangement by FISH. Three specimens demonstrated collision tumors where there was a distinct margin within a tumor of glands staining positive for ERG and those that did not. Of 27 evaluable specimens with clinical data, ERG expression had no significant association with any of the clinicopathological parameters (data not shown).
Baseline characteristics of the 30 evaluable prostate tumor specimens.
| Variable | Number of prostate tumor specimens |
|---|---|
| N total | 30 (100 %) |
| Gleason sum | N (%) |
| 6 | 11 (37) |
| 3+4 | 8 (27) |
| 4+3 | 4 (13) |
| 8 | 7(23) |
| 9 and 10 | 0 |
| ERG IHC positive | 8 (27) |
| ERG IHC negative | 22 (73) |
Heat map illustrating the correlation of ERG positive specimens with primary Gleason score.
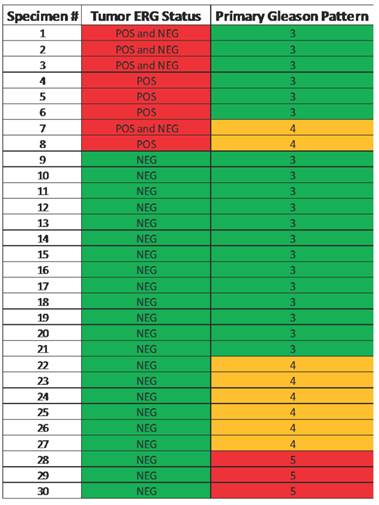
Note that none of the primary pattern Gleason 5 lesions were ERG positive.
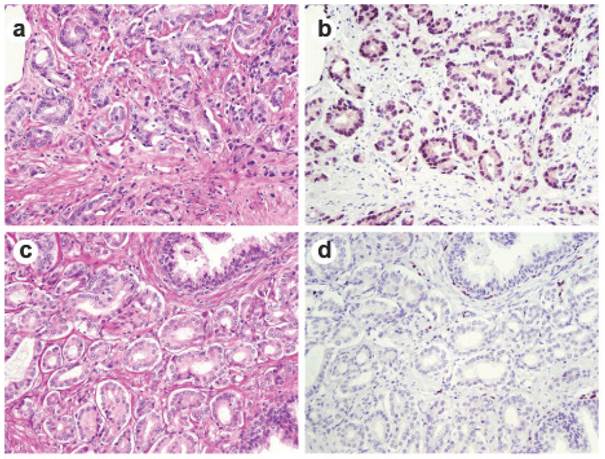
ERG immunohistochemical staining in prostate tissue. Representative consecutive prostate tumor sections were stained by H&E (a and c) and by ERG-MAb mouse monoclonal antibody (b and d). Nuclear staining of the tumor epithelium is apparent in ERG positive tumors (b), and missing in ERG negative tumors (d).
Discussion
This study provides first glimpse into the ERG gene and protein alterations in CaP from India. As noted before CaP associated ERG alterations are highest in men of European ancestry and lowest in Asians men with intermediate frequencies noted for African Americans. Although larger studies are needed, ERG frequencies in Indian CaP are similar to Asian frequencies. Further, there are no studies of ERG in CaP from Africa till date. Clearly there is need for careful evaluations of the CaP associated ERG alterations in the global context as this cancer gene is the most common CaP gene defect in Western countries and it is a promising therapeutic and biomarker target.
While there is hardly any information available on CaP associated somatic gene alterations in Indian population, there are numerous studies of association of single nucleotide polymorphisms (SNP). Kesarwani et al. demonstrated single nucleotide polymorphisms in tumor necrosis factor alpha may influence CaP risk, stage and progression in the North Indian population28. Another genetic factor that has been studied among several populations is the glutathione S-transferase gene (GSTM1, GSTT1 and GSTP1). In the North Indian population, there was a higher risk of CaP in men with the GSTP1-313 G, GSTM1 and GSTT1 alleles. A combination of the GST genotypes further increased risk of CaP29. The results found here are similar to those in Japan, but differ from prior studies looking at Austrian, German and American populations30-33. In contrast to these studies, the widely studied SNPs on chromosome 8q24 showing association with risk of CaP in Western countries were also found in Indian population34.
Genetics, diet, lifestyle and environmental conditions are all likely affecting the incidence of any cancer including CaP among different demographics. The multifactorial influences involved in the development and progression of CaP represent a complex puzzle. The CaP associated ERG gene alteration is a potentially important piece in this puzzle that may guide us to differences of underlying biology of CaP among different demographics. These observations will have a significant impact in the management of CaP in different populations as we begin to develop the biological stratification of CaP.
Acknowledgements
A research grant from National Cancer Institute R01CA162383 (S. S.) and CPDR program fund HU0001-10-2-0002 supported this work. The authors would like to thank Yongmei Chen for statistical support.
Disclaimer
The views expressed in this paper are those of the authors and do not reflect the official policy of the Department of the Army, Department of Defense or the U.S. Government.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11-30
2. Ferlay JSH, Bray F, Forman D, Mathers C and Parkin DM. GLOBOCAN 2008 v1.2, Cancer Incidence and Mortality Worldwide: IARC CancerBase No 10. http://globocan.iarc.fr
3. Gronberg H. Prostate cancer epidemiology. Lancet. 2003;361(9360):859-864
4. Sim HG, Cheng CW. Changing demography of prostate cancer in Asia. Eur J Cancer. 2005;41(6):834-845
5. Mosquera JM, Mehra R, Regan MM. et al. Prevalence of TMPRSS2-ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin Cancer Res. 2009;15(14):4706-4711
6. Gopalan A, Leversha MA, Satagopan JM. et al. TMPRSS2-ERG gene fusion is not associated with outcome in patients treated by prostatectomy. Cancer Res. 2009;69(4):1400-1406
7. Lapointe J, Kim YH, Miller MA. et al. A variant TMPRSS2 isoform and ERG fusion product in prostate cancer with implications for molecular diagnosis. Modern Pathology. 2007;20(4):467-473
8. Perner S, Demichelis F, Beroukhim R. et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66(17):8337-8341
9. Rajput AB, Miller MA, De Luca A. et al. Frequency of the TMPRSS2:ERG gene fusion is increased in moderate to poorly differentiated prostate cancers. J Clin Pathol. 2007;60(11):1238-1243
10. Tu JJ, Rohan S, Kao J, Kitabayashi N, Mathew S, Chen YT. Gene fusions between TMPRSS2 and ETS family genes in prostate cancer: frequency and transcript variant analysis by RT-PCR and FISH on paraffin-embedded tissues. Modern Pathology. 2007;20(9):921-928
11. Yoshimoto M, Joshua AM, Cunha IW. et al. Absence of TMPRSS2:ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Modern Pathology. 2008;21(12):1451-1460
12. Tomlins SA, Rhodes DR, Perner S. et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644-648
13. Petrovics G, Liu A, Shaheduzzaman S. et al. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene. 2005;24:3847-3852
14. Rubin MA, Maher CA, Chinnaiyan AM. Common gene rearrangements in prostate cancer. J Clinic Oncol. 2011;29:3659-3668
15. Sreenath TL, Dobi A, Petrovics G. et al. Oncogenic activation of ERG: A predominant mechanism in prostate cancer. J Carcinogen. 2011;10:37
16. Dobi A, Sreenath T, Srivastava S. Androgen dependent oncogenic activation of ETS transcription factors by recurrent gene fusions in prostate cancer: Biological and Clinical Implications. In: (ed.) Wang Z. Androgen-Responsive Genes in Prostate Cancer. New York, USA: Springer. 2013
17. Sun C, Dobi A, Mohamed A. et al. TMPRSS2-ERG fusion, a common genomic alteration in prostate cancer activates C-MYC and abrogates prostate epithelial differentiation. Oncogene. 2008;27:5348-5353
18. Brenner JC, Ateeq B, Li Y. et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19:664-78
19. Shao L, Tekedereli I, Wang J. et al. Highly specific targeting of the TMPRSS2/ERG fusion gene using liposomal nanovectors. Clin Cancer Res. 2012;18:6648-57
20. Rosen P, Pfister D, Young D. et al. Differences in frequency of ERG oncoprotein expression between index tumors of Caucasian and African American patients with prostate cancer. Urology. 2012Oct;80(4):749-53
21. Magi-Galluzzi C, Tsusuki T, Elson P. et al. TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. The Prostate. 2011;71(5):489-497
22. Petrovics G, Liu A, Shaheduzzaman S. et al. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene. 2005;24(23):3847-852
23. Miyagi Y, Sasaki T, Fujinami K. et al. ETS family-associated gene fusions in Japanese prostate cancer: analysis of 194 radical prostatectomy samples. Modern Pathology. 2010;23(11):1492-1498
24. Mao X, Yu Y, Boyd LK. et al. Distinct genomic alterations in prostate cancers in Chinese and Western populations suggest alternative pathways of prostate carcinogenesis. Cancer Res. 2010;70(13):5207-5212
25. Furusato B, van Leenders GJ, Trapman J. et al. Immunohistochemical ETS-related gene detection in a Japanese prostate cancer cohort: diagnostic use in Japanese prostate cancer patients. Pathol Int. 2011;61:409-14
26. Furusato B, Tan SH, Young D. et al. ERG oncoprotein expression in prostate cancer: clonal progression of ERG-positive tumor cells and potential for ERG-based stratification. Prostate Cancer Prostatic Dis. 2010Sep;13(3):228-37
27. Perner S, Demichelis F, Beroukhim R. et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66(17):8337-41
28. Kesarwani P, Mandhani A, Mittal RD. Polymorphisms in tumor necrosis factor-A gene and prostate cancer risk in North Indian cohort. J Urol. 2009;182(6):2938-2943
29. Srivastava DS, Mandhani A, Mittal B, Mittal RD. Genetic polymorphism of glutathione S-transferase genes (GSTM1, GSTT1 and GSTP1) and susceptibility to prostate cancer in Northern India. BJU Int. 2005;95(1):170-173
30. Gsur A, Haidinger G, Hinteregger S. et al. Polymorphisms of glutathione-S-transferase genes (GSTP1, GSTM1 and GSTT1) and prostate-cancer risk. Int J Cancer. 2001;95(3):152-155
31. Murata M, Shiraishi T, Fukutome K. et al. Cytochrome P4501A1 and glutathione S-transferase M1 genotypes as risk factors for prostate cancer in Japan. Jpn J Clin Oncol. 1998;28(11):657-660
32. Rebbeck TR, Walker AH, Jaffe JM, White DL, Wein AJ, Malkowicz SB. Glutathione S-transferase-mu (GSTM1) and -theta (GSTT1) genotypes in the etiology of prostate cancer. Cancer Epidemiol Biomarkers Prev. 1999;8(4 Pt 1):283-287
33. Steinhoff C, Franke KH, Golka K. et al. Glutathione transferase isozyme genotypes in patients with prostate and bladder carcinoma. Arch Toxicol. 2000;74(9):521-526
34. Tan YC, Zeigler-Johnson C, Mittal RD. et al. Common 8q24 sequence variations are associated with Asian Indian advanced prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17(9):2431-5
Author contact
![]() Corresponding author: Shiv Srivastava, PhD; ssrivastavaorg.
Corresponding author: Shiv Srivastava, PhD; ssrivastavaorg.

 Global reach, higher impact
Global reach, higher impact