Impact Factor
ISSN: 1837-9664
J Cancer 2014; 5(3):253-259. doi:10.7150/jca.8085 This issue Cite
Research Paper
Association between APE1 Single Nucleotide Polymorphism (rs1760944) and Cancer Risk: a Meta-Analysis Based on 6,419 Cancer Cases and 6,781 Case-free Controls
1. Department of Oncology, the Second Affiliated Hospital of Xi'an Jiaotong University, Xi'an 710004, China;
2. Department of Pathology, the Second Affiliated Hospital of Xi'an Jiaotong University, Xi'an 710004, China;
* These authors contributed equally to this work.
Received 2013-11-8; Accepted 2014-2-1; Published 2014-3-13
Abstract
Apurinic/apyrimidinic endonuclease 1 (APE1) is an essential enzyme in the base excision repair pathway. Epidemiological studies have suggested associations between APE1 rs1760944 polymorphism and cancer risk. This study was aimed to evaluate the relationship between APE1 rs1760944 polymorphism and cancer risk. We searched Pubmed, ISI Web of Knowledge, Embase, Chinese National Knowledge Infrastructure (CNKI) databases until September 2013 to identify eligible studies. Odds ratios (ORs) and 95 % confidence intervals (CIs) were used to estimate the strength of the associations. 12 studies from 11 articles on APE1 rs1760944 genotypes and cancer risk were identified, including a total of 6,419 cancer cases and 6,781 case-free controls. Overall, APE1 rs1760944 polymorphism was significantly associated with the decreased risk of cancer in any genetic models (G vs. T: OR = 0.86, 95% CI = 0.82-0.90; homozygote comparison: OR = 0.74, 95% CI = 0.67-0.82; heterozygote comparison: OR =0.88, 95%CI = 0.81-0.95; dominant model TG+GG vs. TT: OR = 0.82, 95% CI = 0.76-0.89; recessive model GG vs. TT+TG: OR = 0.81, 95%CI = 0.75-0.88). In the stratified analysis by populations, the effect was remain in studies of Asian population (homozygote comparison: OR = 0.71, 95%CI = 0.63-0.79; heterozygote comparison: OR = 0.86, 95 %CI = 0.79- 0.94; dominant model: OR = 0.80, 95% CI = 0.74 -0.87 and recessive model: OR = 0.78, 95%CI = 0.71-0.86). Moreover, a significantly decreased risk was found in lung cancer studies (homozygote comparison: OR = 0.68, 95% CI = 0.59-0.79; heterozygote comparison: OR = 0.86, 95%CI = 0.77- 0.98; dominant model: OR = 0.80, 95%CI = 0.72-0.90 and recessive model: OR= 0.77, 95% CI= 0.68-0.87). These findings support that APE1 rs1760944 polymorphism has a possible protective effect on cancer susceptibility particularly among Asians. Further studies based on different ethnicity and various cancer types are warranted to verify our findings.
Keywords: APE1, single nucleotide polymorphism, cancer susceptibility, meta-analysis.
Introduction
Apurinic/apyrimidinic endonuclease 1 (APE1, also known as APEX, HAP1, and REF-1) is a multifunctional protein and plays a central role in the base excision repair (BER) pathway [1]. The human APE1 is located on chromosome 14q11.2 and consists of five exons, spans roughly 2.5 to 3 kb of DNA [2, 3]. Besides its role in DNA repair, APE1 is also known as a transcriptional coactivator for numerous transcription factors, such as AP-1, HIF-1α, p53 and NF-κB, which are involved in cancer promotion and progression [4]. APE1 represents a promising target for pharmacological treatment in some cancer types [3, 5].
Up to now, Epidemiologic studies suggested single nucleotide polymorphisms (SNP) in APE1 may confer individuals' susceptibility to cancer [6, 7]. A total of 18 SNPs in APE1 have been identified [2], of which, two functional SNPs (- 656 T> G in the promoter region, rs1760944 and 1349 T> G in the fifth Exon, rs1130409) have been wildly investigated [8]. It was reported that APE1 rs1760944 T>G polymorphism was associated with altered promoter activity in vitro [9]. Furthermore, the rs1760944 G allele was associated with the decreased risk of lung cancer by enhancing the transcriptional activity, compared with that of the T allele [9]. The results suggested that the rs1760944 G allele in the promoter region was associated with an increased transcriptional activity of the APE1 gene [10].
Lo et al. and Lu et al. reported that - 656 T > G polymorphism influenced the transcriptional activity of APE1 and contributed to lung cancer susceptibility [9, 11]. Some other studies in breast cancer, prostate cancer, and lung cancer also supported the association [12-14], however, it has not been replicated in one study in renal cell carcinoma [15].
Meta-analysis is a statistical technique for combining results from different studies to produce a single estimate of the major effect with enhanced precision [16]. Any single study is insufficient to confirm the association of the APE1 rs1760944 polymorphism with cancer risk [8]. It is important to summarize inconclusive results from different studies to provide evidence on the association of APE1 rs1760944 polymorphism with breast cancer risk. To clarify the effect of the APE1 rs1760944 polymorphism on cancer risk, we carried out a meta-analysis on all eligible case-control studies to estimate the overall cancer risk of the APE1 rs1760944. Furthermore, we conducted the subgroup analysis by stratification according to the ethnicity and cancer type.
Materials and methods
Publication Search
Computer searches were carried out independently by two authors, in PubMed, ISI Web of Knowledge, Embase, Chinese National Knowledge Infrastructure (CNKI) Data (the latest research was retrospected to September 2013) to collect articles with case-control or cohort studies related to the association of APE1 rs1760944 polymorphism and cancer risk.
The keywords were as follows: cancer/carcinoma, apurinic/apyrimidinic endonuclease-1/APE1/APEX/HAP1/REF-1, rs1760944 /656 T > G and polymorphism/genotype/SNP. Furthermore, reference lists of main reports and review articles were also reviewed by a manual search to identify additional relevant publications.
Selection Criteria
The following criteria were used to select studies for further meta-analysis: (1) case-control studies; (2) the studies evaluated the associations between APE1 rs1760944 polymorphism and cancer risk; (3) the studies contained at least two comparison groups (cancer group vs. control group); (4) the studies included detailed genotyping data.
Accordingly, the following exclusion criteria were also used: (1) the design of the experiments were not case-control studies; (2) the source of cases and controls, and other essential information were not provided; (3) the genotype distribution of the control population was departure from HardyeWeinberg equilibrium (HWE); (4) reviews and duplicated publications.
Data Extraction and Synthesis
Articles were performed independently by two reviewers and data with discrepancies in identification were discussed by all authors. For each included study, the following information was collected: first author, year of publication, country of origin, ethnicity, source of control, numbers of cases and controls, genotyping methods for APE1 rs1760944 T/G, total number of cases and controls as well as number of cases and controls with T/T, T/G and G/G genotypes. Different ethnicity descents were categorized as Caucasian, Asian, African, and “mixed”. All the case and control groups were well controlled. The non-cancer controls had no history of gynecologic disease, and there was no present evidence of any malignant disease. When studies included, subjects of more than one ethnicity, genotype data were extracted separately according to ethnicities for subgroup analyses.
Statistical analysis
The associations between APE1 rs1760944 polymorphism and cancer risk were measured by odds ratio (OR) with 95% confidence interval (CI). The significance of the pooled OR was determined by the Z test. Statistical heterogeneity among studies was assessed with the Q and I2 statistics. The Q test and I2 were claimed to test the variation which was due to heterogeneity or by random error. When P value of heterogeneity tests was no more than 0.1 (P≤0.1), we used random effects model. When P value of heterogeneity test was more than 0.1 (P≥0.1), we used fixed effects model. Sensitivity analysis was also tested by removing one study at a time to calculate the overall homogeneity and effect size. Publication bias were evaluated by the funnel plot and further assessed by the method of Egger's linear regression test. All statistical analyses were carried out with the review manager version 5.0 (Revman; The Cochrane Collaboration, Oxford, UK) and the Stata software version 10.0 (Stata Corporation, College Station, TX, USA). All P values in the meta-analysis were two-sided, and P value less than 0.05 were considered significant.
Results
Characteristics of Studies
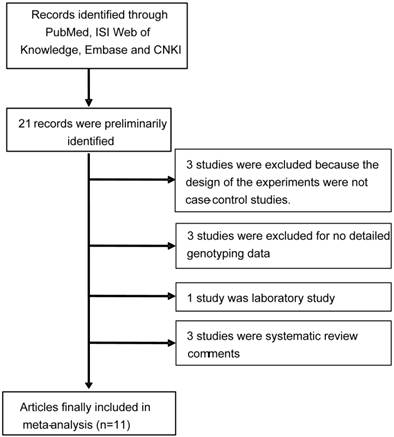
As shown in Figure 1, a total of 21 records that fulfilled our search criteria were preliminarily identified for further detailed evaluation, of which excluded 10 articles. Three studies were excluded because the designs of the experiments were not case-control studies. Two studies were not focused on APE1 rs1760944 polymorphism and breast cancer risk. Two studies was excluded because no detailed genotyping data. One was laboratory study, and the rest of the four studies were systematic review comments. Finally, 12 studies from 11 articles on APE1 rs1760944 genotypes and cancer risk were identified [9-15, 17-20], including a total of 6,419 cancer cases and 6,781 case-free controls. The characteristics of the included studies are listed in Table 1.
Among the eligible studies, only one study was based on Caucasian background which was carried out in America. Eleven were based on Asian background and carried out in China. All studies were case-control studies, including five lung cancer studies, one bladder cancer study, one renal cancer study, one advanced colorectal adenoma study, one cervical cancer study, one prostate cancer study, one glioblastoma study, and one breast cancer study. All cancers were confirmed by histology or pathology. Moreover, controls were mainly matched on age, of which four were population-based and eight were hospital-based.
Flow chart of study selection.
Characteristics of the studies included in the meta-analysis.
| First author | Year | Country | Ethnicity | Cancer type | Genotyping medthod | Source of control | Total sample size (case/control) |
|---|---|---|---|---|---|---|---|
| Berndt [17] | 2007 | America | Caucasian | ACA | TaqMan | PB | 767/720 |
| Lo [9] | 2009 | China | Asian | Lung cancer | Arrayed primer extension | HB | 725/728 |
| Lu [11] | 2009 | China | Asian | Lung cancer | Illumina | PB | 500/517 |
| Lu [11] | 2009 | China | Asian | Lung cancer | SNPscan | HB | 572/547 |
| Wang [18] | 2010 | China | Asian | BC | PCR-RELP | HB | 234/253 |
| Zhou [19] | 2011 | China | Asian | Glioblastoma | MassARRAY | HB | 766/824 |
| Li [14] | 2011 | China | Asian | Lung cancer | TaqMan | PB | 455/443 |
| Cao [15] | 2011 | China | Asian | RCC | TaqMan | HB | 612/632 |
| Wang [10] | 2013 | China | Asian | CC | PCR-RELP | HB | 306/306 |
| Kang [12] | 2013 | China | Asian | breast cancer | TaqMan | HB | 465/799 |
| Jing [13] | 2013 | China | Asian | PC | PCR-RELP | PB | 198/156 |
| Pan [20] | 2013 | China | Asian | Lung cancer | PCR-LDR | HB | 819/803 |
ACA: Advanced colorectal adenoma; RCC: Renal cell carcinoma; BC: Bladder cancer; CC: Cervical cancer; PC: Prostate cancer; PCR-RFLP: Polymerase Chain Reaction-restriction Fragment Length Polymorphism; PCR-LDR: Polymerase Chain Reaction-Ligation Detection Reaction; PB: Population Based; HB: Hospital Based.
Meta-analysis results
The frequency of the G allele varied widely across the twelve studies, ranging from 0.33 to 0.63 (Table 2). The average frequency of the G allele in Asian populations was 0.45, which was lower than that in European populations (0.52). There was no significantly different between Asians and Caucasians (P>0.05).
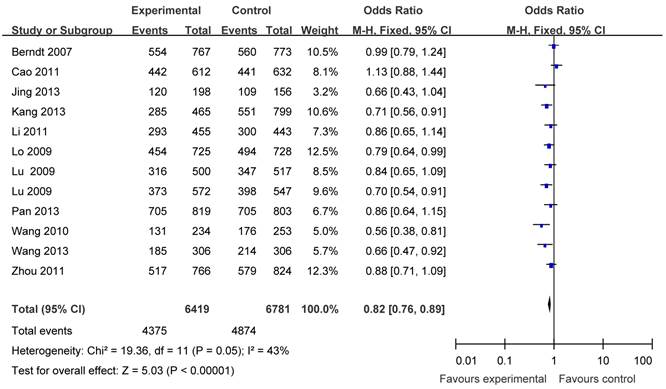
The main results of this meta-analysis were listed in Table 3. Overall, there was evidence of an association between cancer risk and the variant genotypes in different genetic models when all the eligible studies were pooled into the meta-analysis. As show in Table 3 and Figure 2, significant main effects were observed in any genetic models (G vs. T: OR = 0.86, 95% CI = 0.82-0.90, P <0.00001; homozygote comparison (GG vs. TT): OR = 0.74, 95% CI = 0.67-0.82, P <0.00001; heterozygote comparison (TG vs. TT): OR =0.88, 95%CI = 0.81-0.95, P = 0.002; dominant model TG+GG vs. TT: OR = 0.82, 95% CI = 0.76-0.89, P<0.00001; recessive model GG vs. TT+TG: OR = 0.81, 95%CI = 0.75-0.88, P<0.00001).
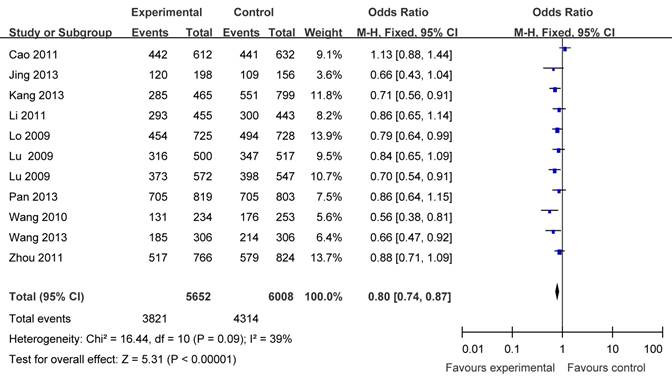
There were eleven articles including 5652 cases and 6008 controls based on Asians used to evaluate the relationship between APE1 rs1760944 polymorphism with cancer susceptibility. In the stratified analysis by populations, as shown in Table 3 and Figure 3, the effect was remain in studies of Asian population (homozygote comparison: OR = 0.71, 95%CI = 0.63-0.79; heterozygote comparison : OR = 0.86, 95 %CI = 0.79- 0.94; dominant model: OR = 0.80, 95% CI = 0.74 -0.87 and recessive model: OR = 0.78, 95%CI = 0.71-0.86).
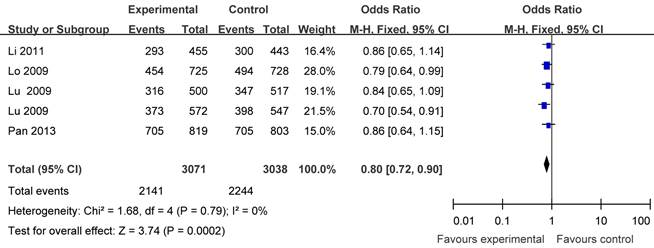
Five articles including 3071 cases and 3038 controls were used to evaluate the relationship between APE1 rs1760944 polymorphism with lung cancer risk. In the stratified analysis by cancer type, as shown in Table 3 and Figure 4, the protective effect was remain in lung cancer studies (homozygote comparison : OR = 0.68, 95% CI = 0.59-0.79; heterozygote comparison: OR = 0.86, 95%CI = 0.77- 0.98; dominant model: OR = 0.80, 95%CI = 0.72-0.90 and recessive model: OR= 0.77, 95% CI= 0.68-0.87). Further subgroup analyses were not performed because of limited data for this polymorphism.
Tests of Heterogeneity
Statistically significant heterogeneity was observed between trials of the following analyses using Q statistic (G vs. T: P = 0.005, I2 = 59%; GG vs. TT: P = 0.008, I2 = 57%; dominant model TG+GG vs. TT: P = 0.05, I2 = 43%); recessive model GG vs. TT+TG (P = 0.04, I2 = 47%), and the random-effects model was performed in these studies. There was no significant heterogeneity in heterozygote comparison (TG vs.TT: P = 0.33, I2 = 12%), and a fixed-effects model was performed.
Publication Bias
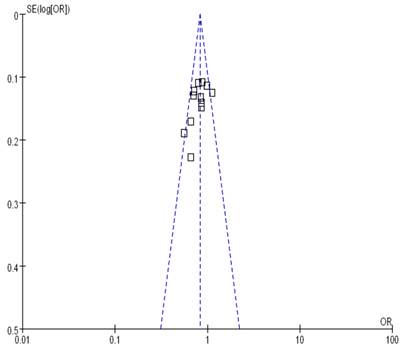
Begg's funnel plot and Egger's test were performed to assess the publication bias. As show in Figure 5, the funnel plots did not reveal any obvious asymmetry in all genotypes in overall population, and the results of Begg's test revealed no publication bias (P>0.05).
Forest plots of APE1 rs1760944 polymorphism and cancer risk in the overall population (TG+GG vs TT). The squares and horizontal lines correspond to the study specific OR and 95% CI. The area of the squares reflects the weight (inverse of the variance). The diamond represents the summary OR and 95% CI.
Forest plots showing the relationship between APE1 rs1760944 polymorphism and cancer risk in Asians subgroup (TG+GG vs. TT).
Forest plot showing the relationship between APE1 rs1760944 polymorphism and lung cancer risk (GG+TG vs. TT).
APE1 rs1760944 polymorphism Genotype Distribution and Allele Frequency in Cases and Controls.
| First author | Year | Genotype (N) | Allele frequency (N, %) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | ||||||||||
| total | TT | TG | GG | total | TT | TG | GG | T | G | T | G | ||
| Berndt[17] | 2007 | 767 | 106 | 310 | 244 | 773 | 114 | 317 | 243 | 736(48) | 798(52) | 743(48) | 803(52) |
| Lo [9] | 2009 | 725 | 271 | 332 | 122 | 728 | 234 | 341 | 153 | 874(60) | 576(40) | 809(56) | 647(44) |
| Lu [11] | 2009 | 500 | 184 | 241 | 75 | 517 | 170 | 238 | 109 | 609(61) | 391(39) | 578(56) | 456(44) |
| Lu [11] | 2009 | 572 | 199 | 288 | 85 | 547 | 149 | 293 | 105 | 686(60) | 458(40) | 591(54) | 503(46) |
| Wang [18] | 2010 | 234 | 92 | 108 | 34 | 253 | 77 | 124 | 52 | 314(67) | 154(33) | 278(55) | 228(45) |
| Zhou [19] | 2011 | 766 | 233 | 392 | 125 | 824 | 237 | 424 | 155 | 890(58) | 642(42) | 914(55) | 734(45) |
| Li [14] | 2011 | 455 | 162 | 227 | 66 | 443 | 143 | 206 | 94 | 551(61) | 359(39) | 492(56) | 394(44) |
| Cao [15] | 2011 | 612 | 170 | 307 | 135 | 632 | 191 | 307 | 134 | 647(53) | 577(47) | 689(55) | 575(45) |
| Wang [10] | 2013 | 306 | 121 | 139 | 46 | 306 | 92 | 154 | 60 | 381(62) | 231(38) | 338(55) | 274(45) |
| Kang [12] | 2013 | 465 | 180 | 207 | 78 | 799 | 248 | 381 | 170 | 567(61) | 363(39) | 877(55) | 721(45) |
| Jing [13] | 2013 | 198 | 78 | 93 | 27 | 156 | 47 | 76 | 33 | 249(63) | 147(37) | 170(55) | 142(45) |
| Pan [20] | 2013 | 819 | 114 | 384 | 321 | 803 | 98 | 369 | 336 | 612(37) | 1026(63) | 565(35) | 1041(65) |
Meta-analysis results.
| Comparisons | OR | 95%CI | P value | Heterogeneity | Effects model | |
|---|---|---|---|---|---|---|
| I2 | P value | |||||
| G vs T | 0.86 | 0.82-0.90 | <0.00001 | 59% | 0.005 | Random |
| Asian | 0.84 | 0.80-0.88 | <0.00001 | 54% | 0.02 | Random |
| Lung cancer | 0.83 | 0.78-0.90 | <0.00001 | 0% | 0.72 | Fixed |
| GG vs TT | 0.74 | 0.67-0.82 | <0.00001 | 57% | 0.008 | Random |
| Asian | 0.71 | 0.63-0.79 | <0.00001 | 48% | 0.04 | Random |
| Lung cancer | 0.68 | 0.59-0.79 | <0.00001 | 0% | 0.70 | Fixed |
| TG vs TT | 0.88 | 0.81-0.95 | 0.002 | 12% | 0.33 | Fixed |
| Asian | 0.86 | 0.79-0.94 | 0.0007 | 8% | 0.37 | Fixed |
| Lung cancer | 0.86 | 0.77-0.98 | 0.02 | 0% | 0.66 | Fixed |
| TG+GG vs TT | 0.82 | 0.76-0.89 | <0.00001 | 43% | 0.05 | Random |
| Asian | 0.80 | 0.74-0.87 | <0.00001 | 39% | 0.09 | Random |
| Lung cancer | 0.80 | 0.72-0.90 | 0.0002 | 0% | 0.79 | Fixed |
| GG vs TT+TG | 0.81 | 0.75-0.88 | <0.00001 | 47% | 0.04 | Random |
| Asian | 0.78 | 0.71-0.86 | <0.00001 | 37% | 0.10 | Random |
| Lung cancer | 0.77 | 0.68-0.87 | <0.0001 | 11% | 0.34 | Fixed |
Funnel plot assessing evidence of publication bias from 12 studies (GG+TG vs. TT).
Discussion
Human DNA repair systems play an important role in protecting the genome from DNA damage caused by endogenous and environmental agents [21]. The base excision repair (BER) pathway removes DNA damage caused by ionizing radiation, reactive oxidative species and methylating agents [22]. APE1 is a multifunctional protein and plays a central role in the BER pathway [3]. Polymorphisms in the DNA repair genes may contribute to the DNA repair capacity variations in the general population. Previous studies suggested the APE1 polymorphisms could influence the sensitivity to ionizing radiation [23].
Aberrant expression of APE1 gene may lead to defects in repairing these lesions and confer individuals' susceptibility to the cancer. For instance, there are some studies that suggested polymorphisms in APE1 may regulate its expression and influence individual's susceptibility to cancer [9-13]. APE1 rs1760944 might act as an important role in the prediction of gastric cancer survival [24]. Epidemiological studies have now linked genetic polymorphisms in APE1 with the risk various types of cancer including lung cancer, breast cancer, colorectal cancer and bladder cancer [7].
The relationship between APE1 rs1760944 polymorphism and cancer was inconsistent [12-15]. In this meta-analysis, we pooled all 12 eligible case-control studies to estimate the overall cancer risk of the APE1 rs1760944. We found that APE1 rs1760944 T>G variant had a significantly decreased risk of cancer in overall population.
In the subgroup meta-analysis based on ethnicity, compared with T allele, a significantly decreased cancer risk is associated with G allele in Asian. Furthermore, compared with TT genotype, a significantly decreased risk of cancer is associated with TG genotype, GG genotype and the combined TG/GG genotypes subgroup. However, there were only one study based on Caucasian background and no study based on African among the eligible studies. Further investigations on large scale on Caucasian and African populations are needed to verify this result.
In the stratified analysis by cancer type, the protective effect was remain in lung cancer studies (homozygote comparison: OR = 0.68, 95% CI = 0.59-0.79; heterozygote comparison: OR = 0.86, 95%CI = 0.77- 0.98; dominant model: OR = 0.80, 95%CI = 0.72-0.90 and recessive model: OR= 0.77, 95% CI= 0.68-0.87). However, there were only one study in the other cancers such as bladder cancer, renal cancer, cervical cancer and prostate cancer. Further subgroup analyses were not performed because of limited data for this polymorphism.
Some limitations of this meta-analysis should be noted. Firstly, this meta-analysis was based on pooled data and no individual data was available; thus, we could not assess the risk of cancer according to stratification of age, environment factors, and other risk factors of cancer. Secondly, small study effect, in which effects reported in small studies are larger, could not be avoided in that some studies were of a relative small size (<5,00 ). Moreover, further large scale multicenter studies with more detailed individual data, with different environmental background are warranted to further validated gene-gene and gene-environment interactions on APE1 rs1760944 polymorphism and cancer risk.
Conclusion
In summary, our present meta-analysis provides evidence of the association between APE1 rs1760944 polymorphism and cancer risk. APE1 rs1760944 polymorphism plays a possible protective effect in cancer in Asians. Further studies based on different ethnicity and various cancer types are warranted to verify our findings.
Acknowledgements
This study was supported by National Natural Science Foundation, People's Republic of China (No.81102711), the International Cooperative Project of Shaanxi province, People's Republic of China (No. 2013KW-32-01) and Specialized Research Fund of the Second Affiliated Hospital of Xi'an Jiaotong University, China [RC (GG) 201203].
Competing Interests
The authors have declared that no competing interest exists.
References
1. Krokan HE, Nilsen H, Skorpen F, Otterlei M, Slupphaug G. Base excision repair of DNA in mammalian cells. FEBS Lett. 2000;476:73-7
2. Xi T, Jones IM, Mohrenweiser HW. Many amino acid substitution variants identified in DNA repair genes during human population screenings are predicted to impact protein function. Genomics. 2004;83(6):970-9
3. Li M, Wilson DM 3rd. Human Apurinic/Apyrimidinic Endonuclease 1. Antioxid Redox Signal. 2013 Aug 20. [Epub ahead of print] PubMed. PMID: 23834463
4. Tell G, Damante G, Caldwell D, Kelley MR. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxid. Redox Signal. 2005;7:367-84
5. Tell G, Fantini D, Quadrifoglio F. Understanding different functions of mammalian AP endonuclease (APE1) as a promising tool for cancer treatment. Cell Mol Life Sci. 2010;67(21):3589-608
6. Wu B, Liu HL, Zhang S, Dong XR, Wu G. Lack of an association between two BER gene polymorphisms and breast cancer risk: a meta-analysis. PLoS One. 2012;7(12):e50857
7. Gu D, Wang M, Zhang Z, Chen J. The DNA repair gene APE1 T1349G polymorphism and cancer risk: a meta-analysis of 27 case-control studies. Mutagenesis. 2009;24(6):507-12
8. Zhou B, Shan H, Su Y, Xia K, Shao X, Mao W. et al. The association of APE1 -656 T>G and 1349 T>G polymorphisms and cancer risk: a meta-analysis based on 37 case-control studies. BMC Cancer. 2011;11:521
9. Lo YL, Jou YS, Hsiao CF, Chang GC, Tsai YH, Su WC. et al. A polymorphism in the APE1 gene promoter is associated with lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2009;18(1):223-9
10. Wang M, Chu H, Wang S, Wang M, Wang W, Han S. et al. Genetic variant in APE1 gene promoter contributes to cervical cancer risk. Am J Obstet Gynecol. 2013;209(4):360.e1-7
11. Lu J, Zhang S, Chen D, Wang H, Wu W, Wang X. et al. Functional characterization of a promoter polymorphism in APE1/Ref-1 that contributes to reduced lung cancer susceptibility. FASEB J. 2009;23(10):3459-69
12. Kang H, Dai Z, Ma X, Ma L, Jin Y, Liu X. et al. A genetic variant in the promoter of APE1 gene (-656 T>G) is associated with breast cancer risk and progression in a Chinese population. Gene. 2013;531(1):97-100
13. Jing B, Wang J, Chang WL, Li B, Chen J, Niu YJ. Association of the polymorphism of APE1 gene with the risk of prostate cancer in Chinese Han population. Clin Lab. 2013;59(1-2):163-8
14. Li Z, Guan W, Li MX, Zhong ZY, Qian CY, Yang XQ. et al. Genetic polymorphism of DNA base-excision repair genes (APE1, OGG1 and XRCC1) and their correlation with risk of lung cancer in a Chinese population. Arch Med Res. 2011;42(3):226-34
15. Cao Q, Qin C, Meng X, Ju X, Ding Q, Wang M. et al. Genetic polymorphisms in APE1 are associated with renal cell carcinoma risk in a chinese population. Mol Carcinog. 2011;50(11):863-70
16. Zhao Y, Han Y, Zhang L, Wang Y, Ma Y, Zhang F. et al. Quantitative assessment of the effect of cytochrome P450 2C9 gene polymorphism and colorectal cancer. PLoS One. 2013;8(4):e60607
17. Berndt SI, Huang WY, Fallin MD, Helzlsouer KJ, Platz EA, Weissfeld JL. et al. Genetic variation in base excision repair genes and the prevalence of advanced colorectal adenoma. Cancer Res. 2007;67(3):1395-404
18. Wang M, Qin C, Zhu J, Yuan L, Fu G, Zhang Z. et al. Genetic variants of XRCC1, APE1, and ADPRT genes and risk of bladder cancer. DNA Cell Biol. 2010;29(6):303-311
19. Zhou K, Hu D, Lu J, Fan W, Liu H, Chen H. et al. A genetic variant in the APE1/Ref-1 gene promoter -141 T/G may modulate risk of glioblastoma in a Chinese Han population. BMC Cancer. 2011;11:104
20. Pan H, Niu W, He L, Wang B, Cao J, Zhao F. et al. Contributory Role of Five Common Polymorphisms of RAGE and APE1 Genes in Lung Cancer among Han Chinese. PLoS One. 2013;8(7):e69018
21. Wood RD, Mitchell M, Lindahl T. Human DNA repair genes, 2005. Mutat Res. 2005;577(1):275-83
22. Mohrenweiser HW, Wilson DM 3rd, Jones IM. Challenges and complexities in estimating both the functional impact and the disease risk associated with the extensive genetic variation in human DNA repair genes. Mutat Res. 2003;526:93-125
23. Hu JJ, Smith TR, Miller MS, Lohman K, Case LD. Genetic regulation of ionizing radiation sensitivity and breast cancer risk. Environ Mol Mutagen. 2002;39(2-3):208-15
24. Zhao Q, Wang W, Zhang Z, Wang S, Wang M, Zhou J. et al. A genetic variation in APE1 is associated with gastric cancer survival in a Chinese population. Cancer Sci. 2011;102(7):1293-7
Author contact
![]() Corresponding author: Email: dzj0911com (Z-J. D.) or kanghf73com (H.-F. K.) Tel.: +86-29-8767-9226 (Z-J. D.) Fax: +86-29-8767-9282(Z-J. D.).
Corresponding author: Email: dzj0911com (Z-J. D.) or kanghf73com (H.-F. K.) Tel.: +86-29-8767-9226 (Z-J. D.) Fax: +86-29-8767-9282(Z-J. D.).

 Global reach, higher impact
Global reach, higher impact