Impact Factor
ISSN: 1837-9664
J Cancer 2014; 5(6):417-424. doi:10.7150/jca.9205 This issue Cite
Research Paper
Development of a Novel Anti-HIF-1α Screening System Coupled with Biochemical and Biological Validation for Rapidly Selecting Potent Anti-Cancer Compounds
Department of Pathology and Laboratory Medicine, University of Tennessee Health Science Center, Memphis, TN 38016, USA.
Received 2014-3-4; Accepted 2014-4-4; Published 2014-4-30
Abstract
Breast cancer (BCa) is the most diagnosed cancer and the second leading cause of cancer death in the American women. Adaptation to the hypoxic environment seen in solid tumors is critical for tumor cell survival and growth. The activation of hypoxia inducible factor-1 alpha (HIF-1α), an important master transcriptional factor that is induced and stabilized by intratumoral hypoxia, stimulates a group of HIF-1α-regulated genes including vascular endothelial growth factor (VEGF), leading tumor cells towards malignant progression. Therefore, a promising therapeutic approach to cancer treatment is to target HIF-1α. The goal of this project was to develop and validate a screening system coupled with secondary screen/validation process that has the capability to screen large numbers of potential anti-cancer small-molecule compounds based on their anti-HIF-1α activities. Breast cancer MDA-231 cells were used as the model to select potent anti-HIF-1α compounds by their abilities to inhibit transactivation of a VEGF promoter fused to a luciferase reporter gene under hypoxia. Positive compounds were then validated by a series of assays that confirm compounds' anti-HIF-1α activities including measurement of HIF-1α downstream VEGF gene expression and angiogenic ability of BCa cells. Results of our pilot screening demonstrate that this prototype screening coupled with validation system can effectively select highly potent anti-HIF-1α agents from the compound library, suggesting that this prototype screen system has the potential to be developed into a high-throughput screen (HTS) coupled with automated validation process for the screening and identification of novel and effective anti-cancer drugs based on anti-HIF-1α mechanism.
Keywords: HIF-1α, VEGF, Breast cancer
Introduction
One general characteristic of fast-growing solid tumors including breast cancer (BCa) is the development of intratumoral hypoxia, which activates hypoxia-inducible factor-1 (HIF-1), the transcription factor that stimulates a group of downstream genes including VEGF that are responsible for tumor malignant progression (1,2). HIF-1 is composed of an inducible subunit, HIF-1α and a constitutively expressed subunit, HIF-1ß (13). HIF-1 activity is mainly dependent on the level of HIF-1α protein. HIF-1α degrades quickly under normal oxygen tension (3) but it is inducible and stabilized under low oxygen tension (hypoxia) (4). Increased HIF-1α levels have been shown to correlate with decreased patient survival in many cancers including BCa (5).
Because HIF-1α is the transcriptional regulator for a group of malignant-pathway related genes that promote tumor progression and metastasis, targeting and inhibiting the activity of HIF-1α should provide an effective strategy to suppress tumor malignant progression. The goal of this project is to develop an efficient screening system coupled with validation processes for fast and efficient identification and selection of highly potent HIF-1α inhibitors. Our optimal objective is to establish an integrated screen/validation HTS system that allows massive and rapid selection of anti-HIF-1α small-molecule compounds for the best pre-therapeutic leads against BCa. The integrated screen and validation (based on molecular biology, biochemistry and biology) developed in this designed prototype system will greatly facilitate the drug discovery efforts in this important area to significantly benefit BCa and other cancer patients.
In this study, human BCa cells expressing chimeric construct containing a VEGF promoter fused to a reporter gene luciferase (VEGF-Luc) were treated with or without candidate compounds, followed by incubation in hypoxic conditions. As the HIF-1α-mediated VEGF transactivation is reflected by the increased luciferase activity under hypoxia, the potential anti- HIF-1α effect of the compounds was identified by their significant reduction of luciferase activity compared to the untreated control. The designed prototype anti-HIF-1α primary screen and coupled validation processes were tested for its ability to select the top anti-HIF-1α compounds from a small compound library based on their abilities to inhibit the HIF-1α/VEGF pathway including HIF-1α expression, VEGF secretion, and angiogenesis.
Materials and methods
Cell lines, medium and culture conditions
Human breast cancer cell line MDA-MB-231 (ATCC) was cultured in RPMI-1640 medium (Gibco BRL, Gaithersburg, MD) containing10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 µg/ml streptomycin. Human microvascular endothelial cells (HMEC) were incubated in EBM-2 medium (Lonza, Walkersville, MD) containing 10% FBS. Both cell lines were incubated at 37oC under normoxic conditions (5% CO2, 21% O2). Whenever needed, MDA-MB-231 cells were also incubated at 37°C under hypoxic conditions (5% CO2, 0.5% O2, balanced with N2).
Compounds and anti-HIF-1α agents
By a contract between the two universities, we (UTHSC) are able to access a large small-molecule-compound library containing about 350,000 drug-like small molecule compounds that are housed in the Drug Discovery Center at University of Cincinnati (DDCUC). YC-1, the known anti-HIF-1α compound (6), was purchased from Alexis Biochemicals Corp (San Diego, CA). PG-928310 (benzamide, 2-iodo-N-[1-(phenyl methyl)-1H-benzimidazol-2-yl]), a compound which was originally identified from DDCUC compound library and subsequently purchased from Ryan Scientific, Inc (Mt. Pleasant, SC.). All compounds were dissolved in dimethyl sulfoxide (DMSO) to 100 mM stock solution and stored at -80 °C. The culture medium was used for all the subsequent dilution of compounds in the testing to reach the desired concentrations that usually range from 0-100 μM.
Screening for anti-HIF-1α compounds
MDA-MB-231 cells expressing VEGF Promoter-Luciferase (pVEGF-Luc) were established by transfecting cells with a chimeric construct containing a 2.4-kb VEGF promoter and a luciferase reporter gene (pVEGF-Luc) (7). Plasmids pMAP11WT and pMAP11Mutant, two VEGF upstream 5'-flanking sequence chimeric luciferase reporter gene constructs, are generous gifts from Dr. B.H. Jiang of West Virginia University (10).
MDA-MB-231 cells were seeded in 96-well tissue culture plate at 5x104 cells/well, and incubated overnight with candidate compound at 0, 1, 10 and 100 mM, respectively. The next day, a 20-ml mixture composed of 0.3 mg plasmid pVEGF-Luc, 1.5 ml Fugene 6 (Roche Applied Science, Indianapolis, IN), and serum-free medium was made and allowed to set at room temperature for 1 h to form the DNA-liposome complex. The mixture was then added onto the cells in the 96-well plate containing a serial dilution of the compounds. Practically, a mixture cocktail of DNA and Fugene 6 was made in a proportional increased amount, so the sample can be added into the 96-well simultaneously in an efficient manner by a multi-channel pipetman. After 30 h post-transfection, cells were incubated under hypoxic conditions (0.5% O2, 5% CO2, balanced by N2) for another 18 h prior to cell harvesting. The cells exposed to compounds were incubated with the same concentration of compounds throughout the experiment. After lysis, 2 μl of total 20 μl cell extracts per well was used to detect protein concentration for normalization (Coomassie Plus Protein Assay Reagent, Pierce, Rockford, IL), and the remaining 18 μl was used for measuring luciferase activity (Luciferase Assay System, Promega) by following the manufacturer's instructions. The luciferase activity was measured by a 96-well plate luminometer (Synergy HT Multi-Mode Microplate Reader, BioTek, Winooski, VT). The normalized luciderase activities (RLU per μg protein) were presented. The known anti-HIF-1α reagent, YC-1, was included in the assay as a positive control along with candidate compounds. Prior to the compound screen, the optimization of transfection has been performed in this system by transfecting pSV40-ß-gal (Promega) expression vector to MDA-MB-231 cells. We have consistently observed more than 40% transfected cells (blue-staining, ß-galactosidase-expressing cells after X-gal staining) under this procedure. For each compound screened, at least two independent experiments were done with each performed in duplicates or triplicates.
Western blot analysis of HIF-1α and VEGF expression
For detecting time-course HIF-1α induction in MDA-MB-231 cells by hypoxia, cells were incubated under hypoxic conditions and harvested at various indicated time points. For detecting inhibition of HIF-1α expression by compound, MDA-MB-231 cells were cultured with or without indicated compounds at various concentrations for 72 h with the last 18 hours under hypoxic conditions. The cells were lysed in lysis buffer containing 10 mM Tris-HCl (pH 8.0), 0.25 M Sucrose, 0.05 mM CaCl2, 0.02% Azide, 0.5% NP-40, 1x protease inhibitor cocktail (Sigma, St. Louis, MO), and 1x phosphatase inhibitor cocktail I and II (Sigma), and processed for gel electrophoresis. Protein concentration was determined by using Coomassie Plus Protein Assay Reagent (Pierce). Protein extracts were loaded on a precast 4-12% gradient polyacrylamide gel (NuPAGE gel, Invitrogen, Carlsbad, CA) (100 µg/per lane) and subject to SDS-gel electrophoresis, followed by transferring to a polyvinylidene fluoride (PVDF) membrane (Immobilon-P Transfer Membrane, Millipore). The membrane was incubated with blocking solution (5% nonfat milk and 0.02% sodium azide in phosphate-buffered saline) overnight at 40C. The membrane was incubated for 16 h at 40C with mouse anti-human HIF-1α antibody (1:1,000 dilution, cat#610958, BD Biosciences). The membrane was then incubated for 1 h at room temperature with corresponding secondary antibody coupled to peroxidase, followed by detection with chemiluminescent reagents (ECL kit, Amersham). For detecting VEGF induction by hypoxia, BCa cells incubated under normoxic and hypoxic conditions for 18 h were extracted and processed to immunoblot as described above with anti-VEGF as the primary antibody (Santa Cruz Biotech, Santa Cruz, CA). The same blots were also immunoblotted with anti-actin antibody (Millipore, Temecula, CA) as the internal control.
ELISA assay to detect secreted VEGF concentration
Cells were plated in six-well plates and cultured in medium with or without compound for 48 h under normoxic conditions, followed by incubation for another 18 h under hypoxic conditions. Secreted VEGF amounts in extracellular medium were measured by VEGF ELISA Kit (Quantikine Human VEGF Immunoassay, R&D Systems, Minneapolis, MN) by following the manufacturer's instructions. Two independent experiments were conducted with each performed in triplicates.
In vitro angiogenesis assay
Preparation of conditioned medium: MDA-MB-231 cells in medium with or without compound were incubated for 48 h under normoxic conditions and then incubated for additional 24 h under hypoxic conditions. Culture supernatants were collected, and sequentially centrifuged at 1,000 rpm for 10 min, and stored at -800C. The final supernatant was used as conditioned medium for the following tubular formation assay.
We used HMEC as a model for tubular formation assay as previously described (7). Briefly, 150 µl matrigel (BD Biosciences) was pipetted into a prechilled 48-well plate and polymerized for 1 hour at 37°C. HMECs incubated in EBM-2 medium (Lonza, Walkersville, MD) containing 10% FBS were harvested after trypsin treatment, resuspended in the above-mentioned conditioned medium, and then replated onto the layer of matrigel at a density of 3x104 cells/1000 µl per well. Matrigel cultures were incubated at 37°C. At time points of 1, 5, and 8 h after HMEC planting onto the matrigel, the tube-like structure of the cells was photographed. To compare tubular connections (i.e., tubular network formation) in a quantitative manner, five randomly selected fields at each time point were counted for the vessel crossing points.
Results
Proof of principle of the designed screening system
HIF-1α is an important transcriptional factor for a group of malignant-pathway related genes (including VEGF) and it promotes tumor angiogenesis and metastasis (8, 9). Human BCa MDA-MB-231 line is a suitable model for the anti-HIF-1α screen because the cells express a very low basal level of endogenous HIF-1α protein in normoxia but a significantly higher induced HIF-1α expression in hypoxia with a peak at 18 h (Fig. 4). To screen for effective anti- HIF-1α compounds, MDA-MB-231 cells expressing a 2.4-kb full-length VEGF promoter fused to a reporter gene luciferase (VEGF-Luc) were treated with or without compounds, followed by incubation for 18 h in hypoxia. As the HIF-1α-mediated VEGF transactivation is reflected by the increased luciferase activity, the potential anti-HIF-1α compound is identified by its significant reduction of luciferase activity compared to the untreated control under hypoxia.
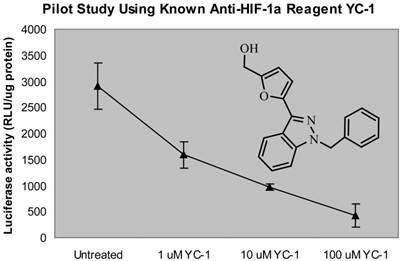
First, we used a known anti-HIF-1α compound, YC-1 (6) to test the proof of principle of the designed screening system. Indeed, we observed that YC-1 has a dose-dependent inhibition of VEGF-Luc transactivity as detected by this screen system (Fig. 1), confirming that this screening system has the ability to identify the anti-HIF-1α compound in MDA-MB-231 cells expressing VEGF-Luc under hypoxic conditions.
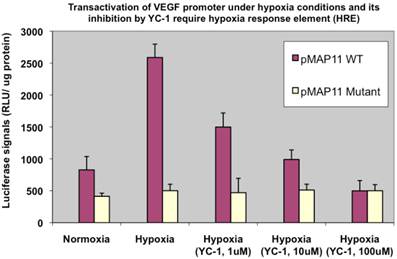
Secondly, to ensure it is indeed the HIF-1α binding that mainly affects VEGF transactivation and YC-1-involved inhibition is via HIF-1α pathway, we used two other chimeric luciferase reporter gene constructs, pMAP11WT (1,10), which contains only 47 bp of VEGF upstream 5'-flanking sequence encompassing a hypoxia response element (HRE) (the DNA region where HIF-1α binds to); and pMAP11Mutant (1,10), which is exactly the same as pMAP11WT except it contains a mutant HRE sequence (so that HIF-1α binding site is abolished), to transfect MDA-MB-231 cells and examined the luciferase activity, respectively. The results (Fig. 2) demonstrated that it is indeed HIF-1α, which activated pMAP11WT via binding to the HRE but not mutant pMAP11Mutant, mainly responsible for the transactivation of VEGF promoter and YC-1 mediated inhibition of transactivation under hypoxic conditions.
Proof of concept of the “proto-type” throughput screening system for anti-HIF-1α compounds. MDA-MB-231 cells expressing VEGF-Luc in 96-well platform were treated with a serial dilution of YC-1 (a known HIF-1α inhibitor) at 370C for 24 h, with the last 18 h at hypoxic conditions (0.5% O2, 5% CO2, and balanced with N2). The cells were then harvested for analysis of luciferase activity. The normalized data (as luciferase activity unit per ug cell extracts) is presented. RLU stands for “relative light units”. Calculated IC50 of YC-1 is 2.8 µM.
YC-1 inhibition involves HRE, the HIF-1α binding region of the promoter. MDA-MB-231 cells transfected with pMAP11WT and pMAP11mut, respectively, and under either normoxic or hypoxic conditions, were examined for their luciferase activity. The normalized data is presented.
Screening and validation of potential anti-HIF-1α compounds
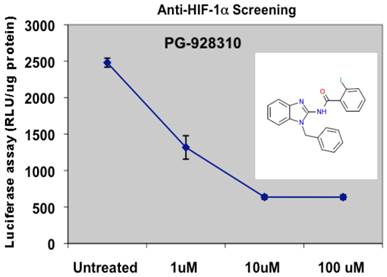
The prototype screening was performed at the 96-well platform. So far we have screened several hundreds of compounds from the DDCUC compound library, and we categorized and ranked the compounds based on their anti-HIF-1α abilities. For those compounds in the category that have IC50 lower than 10 µM, we performed a subsequent screening by biochemical validation assays for confirmation of their anti-HIF-1α and anti-VEGF abilities: that is, HIF-1α protein analysis and VEGF ELISA assays, respectively. Those compounds showing active (inhibitory effects) in both validation assays were further analyzed by in vitro angiogenic assay for their anti-angiogenesis (biological) effects. The compounds that were scrutinized through all these selection criteria and showed the overall best potency was selected as lead compounds. One such compound, PG-928310, stood out as a potential lead compound from our initial screenings: it showed more potent anti-HIF-1α ability than YC-1: PG-928310 has an IC50 concentration at 0.8 µM (Fig. 3), lower than YC-1's IC50 at 2.8 µM (Fig. 1).
Confirmation of the identified compound for its anti-HIF-1α expression by Western
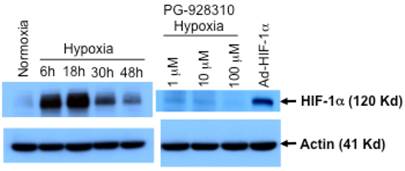
As another proof of principle of this screening system that the identified compound indeed inhibits HIF-1α expression, we analyzed HIF-1α expression at the protein levels of cells incubated with and without the identified lead compound PG-928310. As shown in Fig. 4, MDA-MB-231 cells had a low basal level of HIF-1α protein at normoxia, and an apparent hypoxic induction of endogenous HIF-1α with a peak time at 18 h after hypoxic incubation, justifying that MDA-MB-231 line is a suitable model for screening anti-HIF-1α compounds. We also observed that this hypoxic induction of HIF-1α (at 18 h after hypoxic incubation) was effectively inhibited by the compound PG-928310 at a dose-dependent manner (Fig. 4). These results validated the anti-HIF-1α capability of the compounds identified by the designed screening.
Screening for potential anti-HIF-1α compounds. MDA-MB-231 cells expressing VEGF-Luc in 96-well plate were treated with a serial dilution of compound PG-928310 and processed as described in M&M section for hypoxia treatment and luciferase detection.
Confirmation of the identified compound for its effective inhibition of HIF-1α-regulated downstream gene
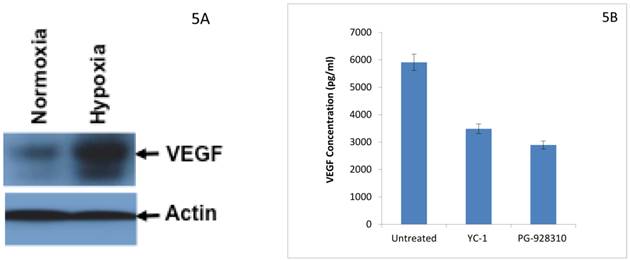
Hypoxia induces HIF-1α expression in BCa cells (Fig. 4), which in turn leads to increased expression of its downstream gene VEGF in BCa cells (Fig. 5A). As confirmed by the VEGF ELISA assay (a biochemical validation), the lead compound PG-928310 indeed inhibited HIF-1α-regulated VEGF signaling, and had more potent (51%) inhibition of VEGF secretion from BCa cells than YC-1 (41% inhibition) (Fig. 5B). These results indicate that active compounds identified by the prototype screening system can effectively inhibit HIF-1α downstream gene VEGF, demonstrating that this HIF-1α-targeting prototype screening is a logically designed, scientifically sound and feasible system to identify compounds that block VEGF-signaling/angiogenesis pathways.
Inhibition of HIF-1α protein by lead compound PG-928310. MDA-MB-231 cells were grown either at normoxic (21% O2), or hypoxic conditions (0.5% O2) and harvested at different indicated times after hypoxic incubation. MDA-MB-231 cells treated with compound PG-928310 at different indicated concentrations for 18 h hypoxic incubation were harvested. The cell extracts were proceeded for the Western blot analysis using anti-HIF-1α antibody as the primary antobody. Mouse fibroblast MEF cells transduced by adenoviral vector expressing HIF-1α (Ad-HIF-1α) were used as the positive control. The same blots were immunoblotted with anti-actin antibody as the internal controls.
Identified anti-HIF-1α compound from prototype screen inhibits HIF-1α-regulated downstream gene VEGF. 5A. Hypoxia induced expression of HIF-1α-regulated downstream gene VEGF in BCa cells. Western analysis using anti-VEGF as the primary antibody. 5B. YC-1 and other anti-HIF-1α compounds decreased VEGF secretion from BCa cells. MDA-MB-231 cells were cultured with or without 10 µM YC-1 and 10 µM PG-928310 for 72 h at 37 0C with the last 24 h under hypoxia. The cell medium (supernatant) was collected for VEGF determination by ELISA assay. The normalized results based on equal amount of cells are presented here.
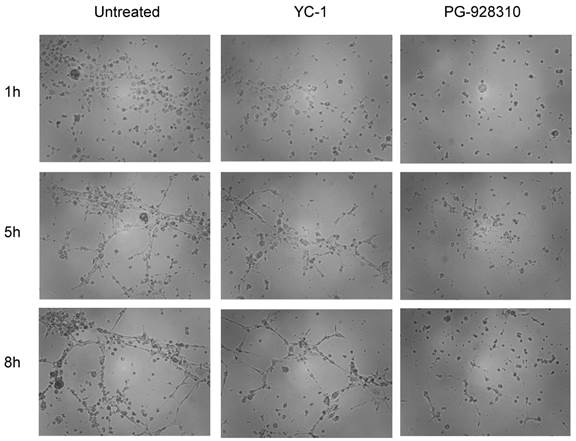
Compounds selected from the anti-HIF-1α screen system effectively inhibit angiogenesis
When the human microvascular endothelial cells (HMEC) grew in conditioned medium isolated from the untreated MDA-MB-231 cells, the cells tended to form tubular structures very quickly on the matrigel; even at 1 h, the cells started to aggregate together to form a pre-tubular structure (Fig. 6, left column). The apparent tubular structure was observed at 5 h, and reached the mature, well-developed tubular network by 8 h. These results suggested that there were some growth factors (including VEGF) secreted from MDA-MB-231 cells that were able to stimulate angiogenesis of HMEC cells. (Our previous publication demonstrated that VEGF secreted from MBA-MB-231 cells is the major growth factor stimulating the tubular network of HMEC, as HMEC in conditioned medium preincubated with anti-VEGF antibody had significantly abolished the tubular structure (7). However, when HMEC cells grew in conditioned medium isolated from YC-1- or PG928310-treated MDA-MB-231 cells, their abilities to form the tubular network were significantly reduced. For example, at 5 h, YC-1 and PG-928310 had 26.5% and 69.9% inhibition, respectively, compared to the untreated (Fig. 6, middle row). These results from biological validation assay demonstrate that the identified compounds by our prototype screen system indeed inhibit HIF-1α/VEGF signaling pathway and block BCa cell-induced angiogenesis.
Discussion
The current clinical chemotherapy agents are not ideal for BCa as they are not curative and only provide a modest extension of survival with sometimes a severely adverse effect on the patient's quality of life. Therefore, there is an urgent need to search for new and more effective anti-BCa drugs. There is an extremely large amount of small molecule compounds in the database currently available, however, the existing screening system is inefficient and time-consuming; thus, the effort for identifying a new generation of effective anti-cancer drugs is hindered. The goal of this project is to develop an efficient screen system coupled with validation steps that is capable of screening large numbers of potential anti-cancer drugs (small molecule compounds) based on their anti-HIF-1α activity. Although HTS screen using promoter constructs for assaying activities of transcription factors has been proposed before, this project is the first to attempt linking a prototype HTS screening system, which allows highly efficient screening for new anti-cancer drug based on the anti-HIF-1α mechanism, with the processes of validation on the compounds' inhibitory effect on target HIF-1α's expression and its downstream target gene (VEGF) and relevant biological effect (angiogenesis).
Conditioned medium of MDA-MB-231 cells treated with anti-HIF-1α compound inhibited HMEC tubular network formation. HMEC cells were planted onto matrigel-coated wells in the conditioned medium derived from untreated, YC-1- or PG-928310-treated MDA-MB-231 cells, respectively. The development of the tubular network of HMEC cells was recorded at 1 h (top row), 5 h (middle), and 8 h (bottom row) after HMEC cells seeding onto the matrigel.
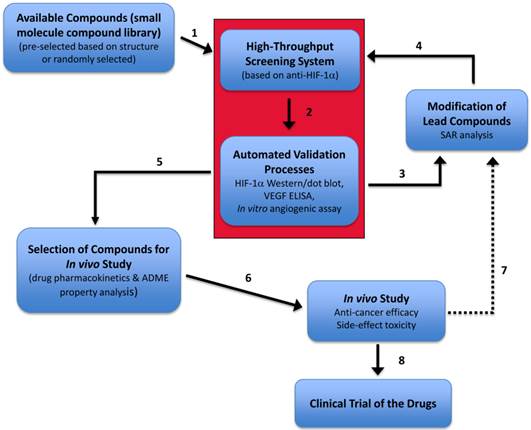
Screening and development of novel anti-HIF-1α drugs. A planned complete drug development diagram is illustrated. The red box represents the prototype screen system (the focus of this study) that is composed of a multiple-well platform screening of compounds and the coupled validation assays, with an optimal goal to become a high-throughput screening and automation validation processes. SAR, structure-activity relationship. ADME, (absorption, distribution, metabolism, and excretion).
Searches for anti-HIF-1α compounds have been attempted in order to use HIF-1 as the therapeutic target for cancer treatment. Although several small molecule inhibitors of HIF-1α (4, 6) were identified including YC-1 (6), these identified inhibitors either do not have sufficient potency to effectively inhibit tumor angiogenesis or have some severe side effects that exclude their potentials for clinical use (for example, YC-1 prolongs bleeding time due to its inhibition of platelet aggregation (11); at present, few small molecule HIF-1α inhibitors have completed evaluation in the clinical setting and none have proven particularly effective in BCa clinical trial (12). Therefore, more specific small-molecule inhibitors of HIF-1α activity need to be identified in order to select the best next generation of anti-BCa drugs that give the maximal anti-cancer efficacy with minimal or no side-effect cytotoxicity.
In addition to the goal of establishing this rapid HTS screening system coupled with automated validation processes which is the essential core of this project (red box, Fig. 7), our optimal objective of this drug screen/validation/development plan (Fig. 7) include: (i) the identified lead compounds can be further chemically modified in order to improve the drug property; and (ii) the complete in vitro and further in vivo validation systems. This drug screening and development plan (Fig. 7) will allow fast screening and selection of effective anti- HIF-1α compounds and prompt the identified lead compounds to quickly go through the entire preclinical evaluation cycle and be ready for clinical trial in a timely manner.
Consistent with the observation from others, we found that pVEGF-Luc stably transfected cells expressed much lower luciferase activity under hypoxia than that of transient transfected cells (not shown), therefore reducing the screen sensitivity of finding the potential anti-HIF-1α compounds. While our proposed study using transient transfection may add some extra steps for the screen procedure, it can significantly enhance screen sensitivity. As the proposed 96-well platform in this study can be easily adapted to 384-well or higher-well platform, this pilot study would provide a prototype of an optimal HTS system coupled with automated validation processes, as in a sequential “assembly line” manner in the future that can rapidly identify/filter and validate anti-HIF-1α compounds in a highly efficient screening manner. In summary, by establishing this “proof of concept” pilot study that employs a HTS system coupled with effective validation processes, we are in the process attempting to build up a novel screen system model that can screen and validate effective anti-HIF-1α compounds in an optimal automation manner from a large amount of compounds database. This new system may facilitate identification of some new (hitherto unknown) lead compounds that exhibit better therapeutic index compared to existing chemotherapy agents with minimal cytotoxicity.
Acknowledgements
This research project was partially supported by NIH grant CA107162 (YL). We thank Dr. Wei Li for his help selecting some potential compounds from DDCUC library for testing our prototype screening system.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Forsythe JA, Jiang BH, Iyer NV. et al. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604-13
2. Dvorak HF, Nagy JA, Feng D. et al. Vascular permeability factor/vascular endothelial growth factor and the significance of microvascular hyperpermeability in angiogenesis. Curr Top Microbiol Immunol. 1999;237:97-132
3. Iliopoulos O, Levy AP, Jiang C. et al. Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc Natl Acad Sci USA. 1996;93:10595-9
4. Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721-32
5. Chan DA, Giaccia AJ. Hypoxia, gene expression, and metastasis. Cancer Metastasis Rev. 2007;26:333-9
6. Yeo EJ, Chun Y-S, Cho Y-S. et al. YC-1: a potential anticancer drug targeting hypoxia-inducible factor 1. J Natl Cancer Inst. 2003;95:516-25
7. Zhang J, Lu A, Beech D, Jiang B, Lu Y. Suppression of breast cancer metastasis through the inhibition of VEGF-mediated tumor angiogenesis. Cancer Ther. 2007;5:273-86
8. Liao D, Corle C, Seagroves TN, Johnson RS. Hypoxia-inducible factor-1alpha is a key regulator of metastasis in a transgenic model of cancer initiation and progression. Cancer Res. 2007;67:563-72
9. Hiraga T, Kizaka-Kondoh S, Hirota K. et al. Hypoxia and hypoxia-inducible factor-1 expression enhance osteolytic bone metastases of breast cancer. Cancer Res. 2007;67:4157-63
10. Fang J, Xia C, Cao Z. et al. Apigenin inhibits VEGF and HIF-1 expression via PI3K/AKT/p70S6K1 and HDM2/p53 pathways. FASEB J. 2005;19:342-53
11. Ko FN, Wu CC, Kuo SC. et al. YC-1, a novel activator of platelet guanylate cyclase. Blood. 1994;84:4226-33
12. Onnis B, Rapisarda A, Melillo G. Development of HIF-1 inhibitors for cancer therapy. J Cell Mol Med. 2009;13:2780-6
13. Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510-4
Author contact
![]() Corresponding author: Yi Lu, Ph.D. Department of Pathology and Laboratory Medicine, University of Tennessee Health Science Center, Cancer Research Building, Room 218, 19 South Manassas Street, Memphis, TN 38163, USA. Tel: (901) 448-5436 Fax: (901) 448-5496 e-mail: yluedu.
Corresponding author: Yi Lu, Ph.D. Department of Pathology and Laboratory Medicine, University of Tennessee Health Science Center, Cancer Research Building, Room 218, 19 South Manassas Street, Memphis, TN 38163, USA. Tel: (901) 448-5436 Fax: (901) 448-5496 e-mail: yluedu.

 Global reach, higher impact
Global reach, higher impact