Impact Factor
ISSN: 1837-9664
J Cancer 2014; 5(7):537-547. doi:10.7150/jca.7797 This issue Cite
Research Paper
FGF10: Type III Epithelial Mesenchymal Transition and Invasion in Breast Cancer Cell Lines
1. Group of Genetics, Cancer Research Center, Cancer Institute of Iran, Tehran University of Medical Sciences (TUMS), Tehran, Iran
2. Institute of Biochemistry and Biophysics (IBB), University of Tehran, Tehran, Iran
3. Group of experimental research in cancer, Cancer Research Center, Cancer Institute of Iran, Tehran University of Medical Sciences (TUMS), Tehran, Iran
4. Department of Genetics, Faculty of Medicine, Tehran University of Medical Sciences (TUMS), Tehran, Iran
* The first two authors have contributed equally to this work.
Received 2013-10-2; Accepted 2014-2-9; Published 2014-6-21
Abstract
Purpose: Fibroblastic growth factor-10 (FGF-10) has an important role in type I epithelial mesenchymal transition (EMT) during the embryonic period of life (gastrulation). Since EMT has a critical role during cancer cells invasion and metastasis (type III) this study sought to investigate the possible role of FGF-10 in type III EMT by monitoring breast cancer cell lines' behavior by FGF-10 regulation.
Methods: MCF-7 and MDA-MB-231 cell lines with different levels of FGF10 expression were treated with FGF-10 recombinant protein and FGF-10 siRNA, respectively.
Results: The cell viability, migration, colony formation and wound healing have a direct relationship with FGF-10 expression, while FGF-10 expression decreased apoptosis. All mesenchymal factors (such as vimentin, N cadherin, snail, slug, TGF-β) increased due to FGF-10 expression with contrary expression of epithelial markers (such as E-cadherin). Moreover, GSK3β phosphorylation (inactivation) increased with FGF-10 expression.
Conclusion: The important role of FGF-10 in type III EMT on cancer cells and initiation of metastasis via various kinds of signaling pathways has been suggested.
Keywords: FGF10, epithelial mesnchymal transition, invasion, breast cancer, metastasis.
Introduction
Metastasis is one of the important characteristics of a cancer cell which has a profound effect on the prognosis and destiny of the tumor. It is responsible for more than 90% of deaths among patients who are suffering from cancer (1). Many different factors may affect this process, from which a few are known. In this respect, the process called epithelial mesenchymal transition (EMT) is a known process related to the initiation of cell migration and metastasis (2).
The phenomenon called EMT is an orchestration of physiomorphological events that leads to a change in cell identity from an epithelial to mesenchymal (3). EMT is an orchestration which starts from cell-to-cell junction and cytoskeleton and leads to extracellular matrix (ECM) alteration and eventually cell migration (4). This is firstly defined in the process of embryogenesis and a period called gastrulation (5) and categorized into three types: type 1 in gastrulation, type 2 in wound healing and type 3 in metastasis. Type 3, the most interesting one for this study, happens during metastasis and invasion of cancer cells into the distant organs (6). Finding a way to regulate this phenomenon will have a practical importance for clinical outcome of cancer in life span and quality.
Many factors have different roles in the initiation and progression of this process, such as various types of growth factors. Among these factors, fibroblastic growth factor (FGF) has many important functions in the embryonic period and many diseases (7). It has 23 members which only 18 type of them are recognized as ligands for fibroblast growth factor receptor (FGFR) (8). FGF plays an important regulatory part in the main developmental pathways, such as mesodermal patterning in the initial step of embryonic life leading to organogenesis. Also, FGF signaling has a role in wound healing and angiogenesis (9).
Because of some similarities between cellular and molecular procedures during gestational life and cancer, investigations about the possible role of dysregulated molecules, which are responsible for organogenesis and rapid growth of cells in the fetus, on invasion and cancer metastasis is very important (10).
Fibroblastic growth factor-10 (FGF-10) is a well-known protein in type I EMT and organogenesis. It has important roles in the development of many organs such as kidney (11), prostate (12) and lung (13). There are some evidences showing that in some cancers, such as prostate cancer, there are correlations between the expressing levels of FGF-10and invasion of cancer cells (14). Moreover, other studies have demonstrated that targeting FGF-10 receptor cleavage or negative regulatory agents called polyoma enhancer activator 3 (PEA3) can affect the pattern of growth and behavior in breast cancer cell lines (15, 16). Hence, this study set out to show that the effect of these cell-line behaviors is directly related to FGF-10 through regulation of EMT mechanisms in which various types of signaling pathways are involved.
Materials and Methods
Cell lines and Primers
MCF-7 and MDA-MB-231 cell lines were prepared by Iran Genetic and Biologic Resource Center and Pasteur Institute of Iran, respectively. MCF-7 cell line was cultured in DMEM containing 10% FBS. MDA-MB-231 was cultured by RPMI 1640 with 10% FBS at 37˚C incubator with 5% CO2. Primers were designed by primer3 program (http://frodo.wi.mit.edu/) for real time PCR reaction (Table 1).
Gene regulation by siRNA and recombinant protein
FGF-10 and GFP siRNA (Santa Cruz, USAsc-39462) were used for FGF-10 silencing in MDA-MB-231 cell line based on the protocol manual with different concentrations of 10, 200, 400 nM for 106 cells. FGF-10 recombinant protein (R&D, USA 345-FG-025) was used for protein inducing in MCF-7 cell line in different concentrations of 50, 100 and 200 ng/ml to treat 106 cells.
Western blot analysis
Whole cell lysates were prepared from treated cells, in SDS lysis buffer (25 mM Tris-HCl pH 6.8, 0.8% SDS, 4% glycerol, 2% 2-mercaptoethanol, 0.008% bromophenol blue). Concerning MCF-7 cells, they were treated with FGF-10 for 48 hours and then the lysate was extracted. For MDA-MB cell lines, the extraction of protein was performed 48 hours after transfection with FGF-10 and GFP siRNA. For immunoblotation, the protein extracts were subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by transfer to PVDF western blotting membranes (Roche, USA). Blocking was performed with 5% nonfat dry milk in TBST (50 mmol/L Tris-HCl (pH7.5), 150 mmol/l NaCl, 0.1% Tween-20) for one hour. The membranes were rinsed 3 times with TBST and incubated overnight at 4°C with a monoclonal antibody against human FGF-10 (1:1000), a monoclonal anti-human GSK3β (total and p Ser -9) antibody (Santa cruz, USA) and an anti-human actin monoclonal antibody (1:5000) (Santa cruz, USA) in TBST. The next day, the membranes were washed several times with TBST and incubated with the secondary antibody: peroxidase conjugated affinity pure F(ab')2 fragment goat anti-mouse IgG + IgM (H+L)(Santa cruz, USA) in TBST for one hour at room temperature. Following another three washes with TBST, the membranes were incubated with Chemiluminescence Detection Kit (Pars Tous Biotechnology, Iran) for one minute exposure to the Kodak radiography transparent film and checking for the band intensity.
Fluorescent microscopy
The transfected cells with FGF-10 and GFP siRNA were cultured after 48 hours and observed for GFP signal for fluorescent microscopy (Leica, USA)
MTT assay
A number of 5x103 cells was seed in each well of 96 well plates and incubated at 37˚C for 24 hours. MDA-MB-231 and MCF-7 cells were treated with siRNA and FGF10 recombinant protein, respectively, in concentrations mentioned above. In different time periods (24, 48, 72 hours) 10 µl of MTT solution (Roche, USA, 11465007001) in concentration of 5 mg/ml was added to each well and the whole procedure was done based on the protocol manual. The absorbance was recorded at 550-570 nm by ELISA reader (BioTek, USA, Gen5 power wave xs2).
Colony agar assay
Equal amount of 1.2% agar and 2x culture medium containing 20% FBS was mixed and distributed in 6 well plates at final volume of 1.5 ml. Cells at concentration of 103 for each well (treated MDA-MB-231 cells with siRNA before use) were mixed with 0.6% agar and 2x culture medium containing 20% FBS and final volume of 1.5 ml was added to each well. The third layer (medium without serum containing FGF-10 recombinant protein for MCF-7 cells, for MDA-MB-231 cells medium without serum) was added after cooling of the rest and incubated at 37˚C in 5% CO2 incubator for colony growth in a period of 14-28 days. Colonies were counted in ten different fields.
Migration assay
The amount of 750 µl culture media containing 20µl/ml fibronectin without serum was added to the bottom wells of matrigel 24 well plates (SPL Life Sciences, Korea). Well inserts was applied on wells gently to prevent bubbling in the bottom. The treated cells were tripsynised and after a brief spin, 2x104 cells were counted and seed to each well insert of 24 well plates with 500 µl of media containing 10% FBS and incubated at 37˚C in 5% CO2 incubator for 24 hours. Inserts were removed and fixed with 3.7% paraformaldehyde for 20 minutes after rinsing with PBS. Fixed cells were stained with Geimsa (Sigma, USA) and noninvasive cells were removed with a cotton swab. The remaining migrated cells were observed by using light microscopy (Ziess, Germany) counting 10 different fields with 10x of magnification.
Wound healing assay
A number of 106 treated cells was seed in 6 well plates and incubated at 37˚C for 24-48 hours. A crash was made in each well by using sampler tip and the detached cells were removed by 1% serum washing. The cells were observed and the picture was captured using phase contrast microscopy (Ziess Germany Axiovert 25) and the free pixels were compared.
Apoptosis (Hoechst Acrydine orange)
The treated cells with FGF-10 siRNA and FGF-10 recombinant protein (using 1% FBS instead of 10% FBS for MCF-7 cells) were rinsed with PBS after 48 hours incubation at 37˚C. 100 µl of cell suspension was mixed with 1 µl of Hoechst 33258 (sigma, USA) in concentration of 1 mg/ml and incubated at room temperature for 5 minutes using 10 µl of solution for detecting apoptotic cells. For acridyne orange/ethidium bromide (sigma, USA) staining the solution in 1:1 volume 100µg /ml in PBS was added and incubated for 5 minutes at room temperature. Using a slide, 10 µl of cell suspension was applied for fluorescent microscopy (ziess, Germany Axiovert 25 plus). Ten fields were observed for apoptotic cell counting with 40x magnification.
Real Time PCR
RNA extraction was done from 106 treated cells using high pure RNA isolation kit (Roche, USA, 11828665001) based on the protocol manual. The synthesis of cDNA was made from 1-2 µg of extracted RNA using easy cDNA synthesis kit (Pars Tous, Iran) base on the protocol manual. Two µl of cDNA was added to primer mix (1µM) (Table 1) and syber green master mix (Takara, Japan) using real time PCR instrument (Qiagen 65 HO, USA). The program for reaction was a denatured at 95 for one minute followed by 40 cycles of initiation at 95 for 10 seconds, annealing at 56˚C for 15 seconds, elongating at 72˚C for 20 seconds and a single final step at 58˚C for 90 seconds. At the end of the program, the melting curve was checked and the data were analyzed by CT calculation using REST software (Qiagen, USA).
Primers used for quantitative RT-PCR.
| Right primer | Left primer | Gene Name |
|---|---|---|
| 5'-CCCCTTCTTGTTCATGGCTA-3' | 5'-ATGTCCGCTGGAGAAAGCTA-3' | FGF-10 |
| 5'-TTTGTCAGGGAGCTCAGGAT-3' | 5'-GAATGACAACAAGCCCGAAT-3' | E-CADHERIN |
| 5'-GGATTGCCTTCCATGTCTGT-3' | 5'-AACAGCAATTGATGCTGACG-3' | N-CADHERIN |
| 5'-GGACAGAGTCCCAGATGAGC-3' | 5'-CCCAATCGGAAGCCTAACTA-3' | SNAIL-1 |
| 5'-CACAGTGATGGGGCTGTATG-3' | 5'-CCTTCCTGGTCAAGAAGCAT-3' | SLUG |
| 5'-AGCCTCAGAGAGGTCAGCAA-3' | 5'-GGCTCAGATTCAGGAACAGC-3' | VIMENTIN |
| 5'-GACAAGCTTCCCGTTCTCAG-3' | 5'-TCACCAGGGCTGCTTTTAAC-3' | GAPDH |
| 5'-TACACGCGAGTGAAGGTGAG-3' | 5'-GAGACCGGTGAGCTGGATAG-3' | MMP9 |
| 5'-GTTGTGGGACCTGTGGAAGT-3' | 5'-GGGACACCAGAAGTCAACCA-3' | TIMP1 |
| 5'-CAACTCCGGTGACATCAAAA-3' | 5'-GTACCTGAACCCGTGTTGCT-3' | TGF-β1 |
| 5'-ATCATCGACGGTGGGTACAT-3' | 5'-TGGGGAAGTTGAAGTGGAAC-3' | CYCLIN D2 (CCND2) |
| 5'-TTTCAGGAGCTCGGTACCAC-3' | 5'-GCTTTCTGCCATTCTCATCG-3' | CDK2 |
Statistical Analysis
All data were elicited using triplicate samples. Standard error and p value were calculated by t-test using MS Excel 2010 software (Microsoft, USA).
Results
Confirmation of FGF-10 inducing and silencing through protein expression in dose-dependent manner
According to Figure 1a, the expression of FGF-10 in MCF-7 cells followed by recombinant protein induction was confirmed in a dose-dependent manner. Control cells have shown a weak expression of FGF-10, while the protein expression has increased to 4, 6, 7 fold during treatment of cells with 50, 100, 200 ng/ml of FGF-10 recombinant protein, respectively. Concerning MDA-MB-231 cell lines, the expression suppression was again a dose-dependent manner with the 52%, 75%, 97% in 100, 200, 400 nM concentration, respectively. The GFP siRNA transfection control and non-treated cells have shown the same amount of FGF-10 expression. However, using fluorescent microscopy, the expression of GFP was confirmed in GFP siRNA transfected cells, while other cells demonstrated no fluorescent signal.
Positive effect of FGF-10 on proliferation
MTT assay is a calorimetric technique for analyzing cell viability and drug cytotoxic on cells. It is a water soluble tetrasolium salt which appears as the yellow color in phenolic environment (17). Dehydrogenase enzyme activity in the living cells breaks the MTT ring and changes it into insoluble blue formazan crystals which could be recorded in 550-570 nm wavelength dissolved in isopropanol acid or DMSO. MCF-7 cells treated with FGF-10 recombinant protein in different dosages and time periods have shown an increase in absorbance (MTT activity) after 48 hours as it was normalized by untreated MCF-7 cells (Figure 1c). (P < 0.01)
Cell viability a) Western blot analysis for FGF-10 expression in MCF-7 cells induced by recombinant protein (50ng, 100ng, 200ng respectively). Also, silencing of FGF-10 in MDA-MB-231 using FGF-10 siRNA (10 nM, 200 nM, 400 nM) has shown a dosage-dependent pattern in FGF-10 regulation in both cell lines. C is the control cells with no treatment and GFP corresponds to GFP siRNA b) the expression of GFP was confirmed by using fluorescent microscopy only in GFP inducing MDA-MB-231 cell lines c) MCF-7 cell proliferation with FGF-10 recombinant protein treatment. d) MDA-MB-231 Cell proliferation after FGF-10 siRNA silencing.
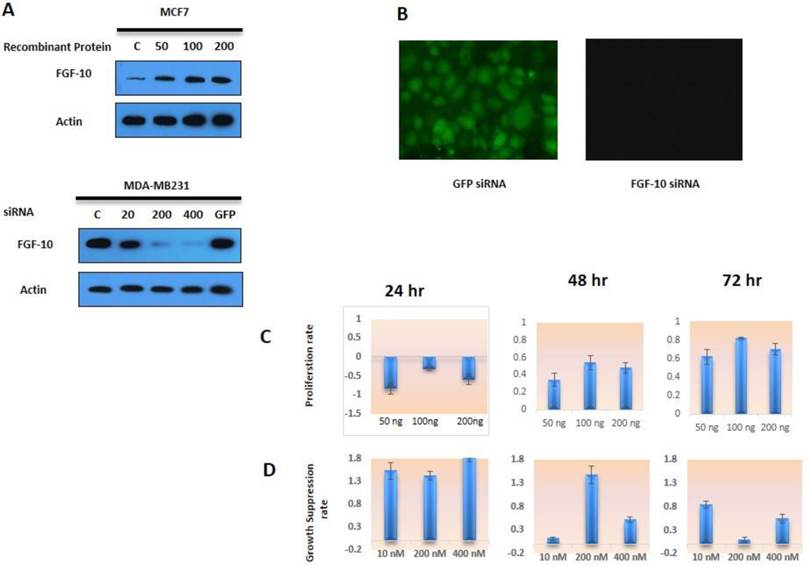
On the other hand, MDA-MB-231 cells treated with FGF-10 siRNA (Figure 1d) have also confirmed the direct effect of FGF-10 in growth suppression, especially after 24 hours (P < 0.05). On the whole, the data suggest that FGF-10 has a direct correlation with cell proliferation and viability. Reducing FGF-10 causes growth suppression while increasing induced proliferation acceleration.
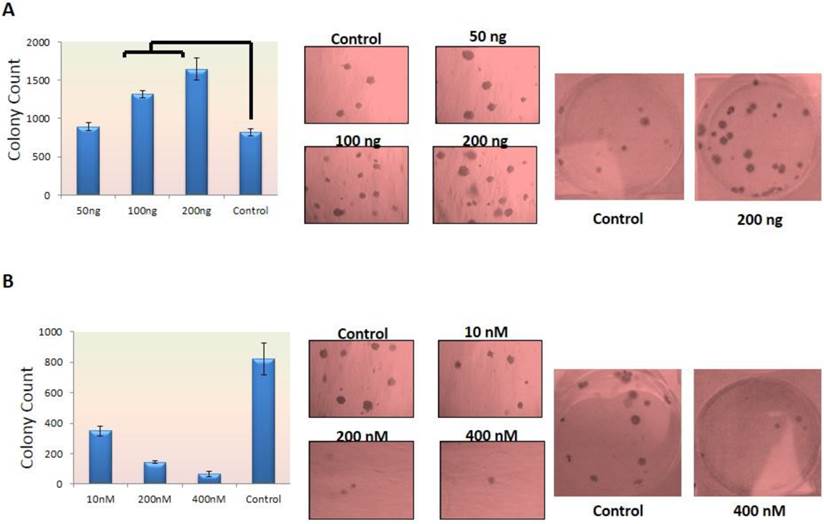
Tumorgenesis is increased under FGF-10 effect
Normal cells need signaling for growth and survival which is provided directly by ECM. If normal epithelial cells lose their interaction with ECM, this can lead to a program cell death called Anoikis (18). In cancer cells this correlation has been lost which means that cells have the capability of growth and survival without correlation with ECM. This ability also has a relationship with cell invasion and metastasis. Therefore, more invasive cells have capability of making more and bigger colonies (19). In this study the colony agar-assay method was selected to evaluate the ability of cells in tumorigenesis. In this method, two layers of agar were prepared and the cells were placed in the superior layer. Concerning MCF-7 cells, FGF-10 factor was placed with the medium in the superior part of two-layer agar but in the case of MDA-MB-231 cells, they were treated with siRNA before placement into the agar layer. The colony count was performed after 14-28 days. The colony formation depended on the number and capability of cells which could make tumor. When cells are induced by a factor, it is very important to know how this affects the cells during treatment. The tumor suppression effect of a factor can affect colony numbers even transiently without sustain treatment with FGF-10.
According to Figure 2, the colony formation increased 1.6 to 2 folds (P < 0.05) in MCF-7 cells treated with 100ng, 200ng FGF-10 recombinant protein, respectively (Figure 2a) while 2.4, 5.8 and 12.5 folds decrease were observed in colony formation (P < 0.001) when MDA-MB-231 cells were treated with 10nM, 200nM, 400nM FGF10 siRNA, respectively (Figure 2b). The results have demonstrated that the tumorogenesis ability of FGF-10 has direct relationship with the FGF-10 concentration.
Colony Formation in dosage-dependent manner a) colony counting after MCF-7 cell treatment with FGF-10 recombinant protein showing increase in colony count which is shown in graphs (left) and direct colony photography with two different magnifications (right). b) Colony number decreases after using FGF-10 siRNA for MDA-MB-231 which is illustrated by graph (left) and microscopic imaging with two different magnifications (right).
FGF-10 induces and increases wound healing ability of cells
The wound-healing assay is one of the earliest developed methods to study directional cell migration in vitro. It is mimicking cell migration during wound healing in vivo, by creating a "wound" in a cell monolayer, capturing the images at the beginning and at regular intervals during cell migration to close the wound, and comparing the images to quantify the migration rate of the cells. It is particularly suitable for studies on the effects of cell-matrix and cell-cell interactions on cell migration (20).
Concerning the MCF-7 cells, the FGF-10 induction with 50, 100, 200 ng/ml concentration was induced after crashing the cell layer while in the case of MDA-MB-231, the cells were treated with 10, 200, 400 nM concentration of siRNA and then the crashing process was performed. The image was taken 24 hours after the process and free area was calculated based on the free pixels by Photoshop software (Adobe Photoshop CS6, USA). As depicted in Figure 3a, the ability of cells to cover the wound has a direct correlation with FGF-10 and increasing the presence of FGF-10 inside the cell. The ability of MCF-7 cells in the presence of FGF-10 increased for filling in free area while in MDA-MB-231 cells this ability was decreased by inhibition of FGF-10.
Invasion and migration a) The ability of direct migration in MCF-7 cells after FGF-10 recombinant protein treatment (top) and in MDA-MB-231 after FGF-10 knock down by FGF-10 siRNA. Pictures have been taken using 10x magnification (bottom) demonstrating the ability of wound healing (arrows demonstrating the free area). b) Using the same method for gene regulation in matrigel, MCF-7 and FGF10 recombinant protein treatment (left), MDA-MB-231 and FGF10 siRNA treatment. Results for GFP siRNA and control cells were the same and were not mentioned in the pictures.
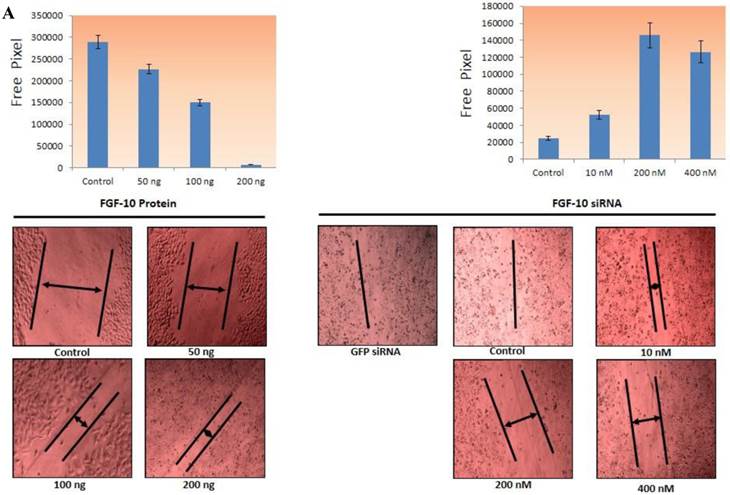

Invasion and migration ability of cells dependents on FGF-10 production
The ability of migration was investigated by Matrigel Migration Assay (Figure 3b). Cells with the ability of movement can migrate toward the fibronectin as a chemotactic reagent while cells without movement ability cannot show any reaction. Therefore the method demonstrates the ability of cells response to the migration stimulants.
Based on the results from wound healing assay, 100ng/ml FGF-10 recombinant protein and 200 nM FGF-10 siRNA were used for MCF-7 and MDA-MB-231, respectively. In MCF-7, the treated cells with 100ng/ml FGF-10 recombinant protein have shown 4.1 folds increase in migration (P < 0.001). However, FGF-10 down regulation by using 200 nM of FGF-10 siRNA after 48 hours has shown 3.1 folds decrease in migration in MDA-MB-231 (P < 0.001). Therefore, it was suggested that FGF-10 has a direct relationship with the migration.
Apoptosis is decreased under FGF-10 protein
In Hoechst 33258 staining, normal cells appear as uniform blue while apoptotic nucleus resembles as irregular and bright blue caused by chromatin aggregation and nucleus fragmentation (21). On the other hands, Acridyne orange is a vital stain which stains both living and death cells and binds to cell DNA, having lower affinity to RNA. However, ethidium bromide stains only cells which have lost their membrane totality (dead cells) (22). Living cells usually stain in green color by acridyne orange staining.
In MCF-7 cell lines, the cells were cultured in 1% FBS containing medium to induce a cellular stress and apoptosis. As it was observed in control cells image, apoptotic cells had increased compared to FGF-10 treated cells. This means that these cells could tolerate the stress condition and apoptosis rate was reduced, suggesting the anti-apoptotic effect of FGF-10. Still, MDA-MB-231 cell lines become apoptotic after FGF-10 silencing.
Apoptotic cells in MCF-7 by Hoechst 33258 staining (Figure 4a) decreased by 10 to 14 folds using 50ng/ml, 100ng/ml and 200ng/ml FGF-10 recombinant protein, respectively (P < 0.05), even in stressful condition, while siRNA down regulation of FGF-10 in MDA-MB-231 has shown 7, 23 and 34 folds increase in apoptosis in 10nM, 200nM and 400 nM FGF10 siRNA treated cells, respectively (P < 0.01).
Apoptosis. a) Hoechst staining has shown a deceased apoptosis induced by stress in MCF-7 cell lines treated with FGF-10 recombinant protein (left graph) while down regulation of FGF-10 using FGF-10 siRNA in MDA-MB-231 enhanced the apoptosis (right graph). b) The same experiment has been done by using acridine orange as mentioned in the text also demonstrated the number of ethidium bromide stained nucleus (apoptosis) decreased by FGF-10 treatment in MCF-7 cells (left graph) while down regulation of FGF10 in MDA-MB-231 cells caused an increase in apoptotic cells (right graph). (microscopic images by 10x magnification)
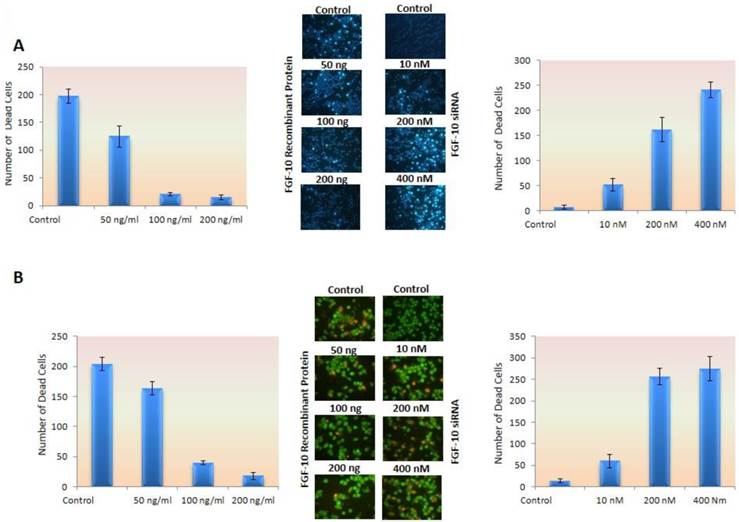
Concerning acridyne orange staining, apoptotic cells showed 5.2 and 11.3 folds decrease after (50 ng/ml) 100ng/ml and 200ng/ml FGF0-10 recombinant protein treatment of MCF-7 (P < 0.05), while there was 4.6, 19.7 and 21.2 folds increase after 10 nM, 200 nM and 400 nM FGF-10 siRNA treatment of MDA-MB-231 cells, respectively (P < 0.01) (Figure 4b).
EMT induction factors change under FGF-10 effects and initiate EMT program
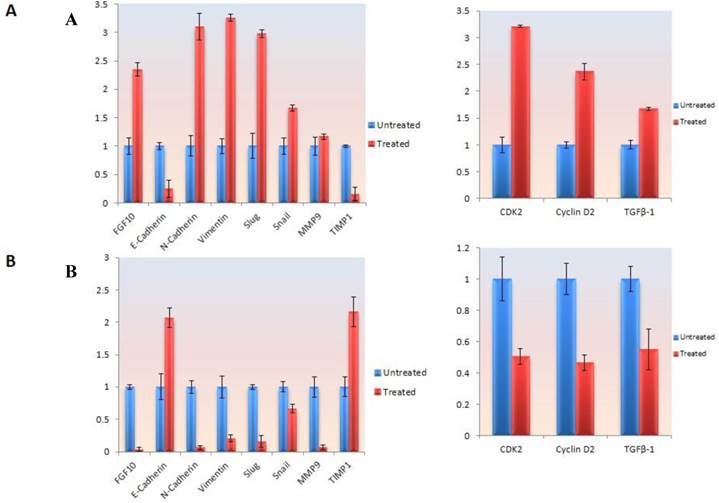
In MCF-7 cell lines, treatment with FGF-10 recombinant protein enhances the FGF-10 expression to 2.3 times, which is because of self-inducing effect by FGF-10. The expression of N-cadherin and vimentin (two characteristic factors of mesenchymal cells) has increased 3.2 and 3.1 folds, respectively. Slug transcription factor was affected more than snail (3 times compared with 1.6 times). Both of these transcription factors were the main agents in EMT which could regulate the expression of EMT-involved genes. While there was 5 folds decrease in E-cadherin expression (epithelial characteristics' factors), MMP9 demonstrated no change after treatment except TIMP1 showing 5 folds decease in expression. Transformation growth factor (TGF-β1) expression was activated by 1.6 fold after FGF-10 recombinant protein treatment. This enhancement was 3.2 and 2.4 for CDK2 and cyclin D2 expression (cell cycle regulatory factors), respectively (Figure 5a).
Gene expression and protein modification. a) Quantitative Real time PCR for gene expression analysis in different genes involved in EMT and cell cycle in MCF-7 cells treated by FGF-10 recombinant protein. b) Same analysis in MDA-MB-231 cells treated with FGF-10 siRNA. c) Western Blot analysis has shown the phosphorylation (inactivation) of GSK3β with the same direction of FGF-10 expression in both cell lines, demonstrating that the phosphorylation of GSK3β is increasing when FGF10 is up regulated (in MCF-7, left) and decreasing during FGF-10 down regulation (in MDA-MB-231, right).
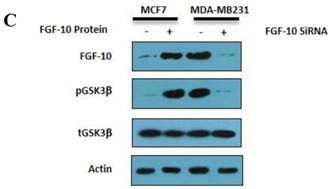
On the other hand, FGF-10-silenced MDA-MB-231 lead to a conflicting finding which indicated a complete suppression of FGF-10 and N-cadherin expression which was followed by 5 and 6.7 times down-regulation of vimentin and slug, respectively, while snail had a 1.4 fold down-regulation. Interestingly, FGF-10 down-regulation had effects on both MMP9 (complete suppression) and TIMP1 (2.2 times up-regulation). E-cadherin epithelial marker has shown a 2.1 folds increase in expression. Also, CDK2, Cyclin D2 and TGFβ-1 expression data demonstrated 2 times down-regulation (Figure 5b).
FGF-10 induced GSK3β phosphorylation in breast cancer cells
As it is illustrated in Figure 5c, treating MCF-7 cells with FGF-10 recombinant protein will cause enhancement of FGF-10 expression by 7.1 folds compared with untreated cells as well as 44.6 folds increase in the amount of phosphorylated (inactivated) GSK3β due to FGF-10 treatment. The results of FGF-10 silencing by FGF-10 siRNA is contradictory by means of 98% silencing of FGF-10 and 96% decrease in the amount of phosphorylated (inactivated) GSK3β. These phosphorylation alteration in the presence or absence of FGF-10 could be related to the relationship between the signaling pathways which they are involved in. GSK3β has a critical role in EMT phenomenon through phosphorylation in WNT signaling pathway and data suggests the role of FGF-10 in the phosphorylation regulation of GSK3β.
Discussion
To investigate the role of FGF-10, two cell lines with conflicting characteristics were used. The MCF-7 cell line has an epithelial appearance with low level of FGF-10 expression (23). Also, this cell line has low ability of movement and invasion (15). On the other hand, EMT markers are down-regulated in this cell line (24). However, MDA-MB-231 has mesenchymal appearance with high level of FGF-10 expression and high ability of invasion and up-regulation of EMT markers (25).
According to these findings, we can suggest that FGF-10 can follow all steps and prepare cells to invade as it is shown in the migration assay. Other findings have demonstrated that FGF-10 regulation has a direct role on cell viability and proliferation, immortality and apoptosis the same as its role in early stages of life.
The ability of colony making in soft agar and tumor formation after inoculation in mouse are two main components of invasion and tumorigenesis in cancer cells (26). In this study the ability of cell invasion was determined by colony formation rates. As we know, more colony formation ability and colony size in a cancer cell indicates greater invasiveness and tumorigenesis (27). Our study has shown a direct correlation between FGF-10 and colony formation.
There are many studies showing that FGF-10 expression has an important role on progenitor epithelial cell maintenance. For example, FGF-10 inhibits terminal differentiation in the embryonic lung (28, 29) and by b-catenin signaling, through embryonic WNT/FGF-10 signaling for maintenance of parabronchial SMC (PSMC) progenitors, and also by FGF-10 secretion which keeps notch signaling activated and induces snail expression in surviving Clara cells (30). On the other hand, it is demonstrated that FGF-10 signaling is required for expanding pancreatic progenitor cells (31, 32). In a study it was demonstrated that FGFR1/2 deficient keratinocytes had a reduced migration velocity which can explain our results in wound healing assay (33). Since many studies have suggested the role of FGF-10 on delay in epithelial differentiation and further marker presentation, in this study we had investigated E-cadherin as an epithelial marker and also N-cadherin, vimentin, fibronectin as mesenchymal markers.
CDK2 and also Cyclin D2-G1/S transition proteins-regulation followed by FGF-10 treatment in the same direction, clarify the activating role of FGF-10 in cancer cell proliferation by promoting G1 to S phase transition and preparation of cells for synthesis and mitosis (34, 35). It can also explain why during down-regulation of FGF-10, cell growth suppressing and apoptosis mechanisms are activated. Apoptosis and immortality are two opposite sides of growth spectrum (36). More invasiveness has direct relationship with immortality. In aggressive cancer cells, apoptotic pathways are inhibited and cells are resistant to stress conditions (37).
EMT has an orchestrated alteration in different components of cells and even in their extracellular microenvironment (4). It is a wide spectrum from nucleus and transcription factor alterations through cell junction and ECM. MMP9 and TIMP1 are ECM proteins altered by FGF-10 induction. This study shows that FGF-10 induction enhances MMP9 expression while it decreases the level of TIMP1 expression which has a reverse function. It enhanced the mesenchymal N-cadherin expression while the expression of E-cadherin gene, which is responsible for epithelial adherence junction, was reduced. However, even in the transcriptional level, the snail and slug expressions (two EMT-inducer transcription factors) enhanced by the FGF-10 induction. Data for FGF-10 suppression in MDA-MB-231 has confirmed the reverse effect of FGF-10. On the whole, it has suggested that FGF-10 could be a strong inducer of type III EMT by affecting different components at intra-cellular and extra-cellular level
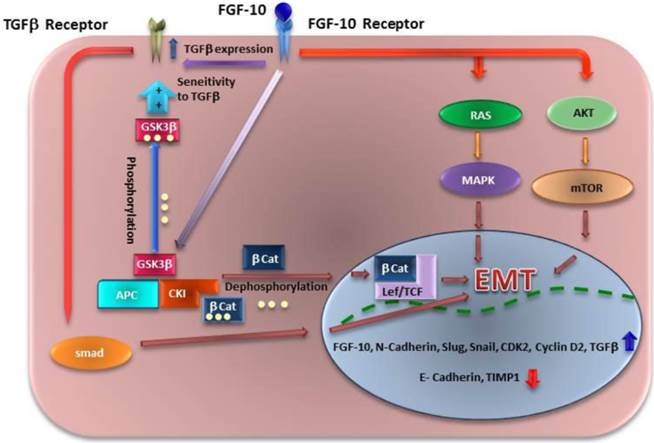
Until now, the main signaling pathways which are described for FGF-10 are through AKT- mTOR and RAS-MAPK dependent pathways (38, 39). In this study we have shown that FGF-10 has an activating role on TGF-β expression. On the other hand, GSK3β is phosphorylated during up-regulation of FGF-10 which means it is another possible pathway through WNT signaling activation as it was mentioned before by cross talk between FGF and WNT signaling (40). Also, based on another finding on ELM (The Eukaryotic Linear Motif Resource: http://elm.eu.org/) (41) shows a GSK3βSer/Thr phosphorylation recognition site which is confirmed by western blot analysis suggesting a direct effect of FGF-10 on GSK3β Ser-9 phosphorylation. GSK3β inactivation also has shown to have a relationship with cell sensitivity to TGF-β due to Smad3 stability (42). Taken together, all the data suggest that FGF-10 has a possible cumulative role in EMT signaling through AKT, RAS, WNT, and TGFβ (Figure 6). Regarding TGF-β, it is more likely that FGF-10 up-regulates the expression of this protein followed by its enhanced sensitivity to cells which may increase the effect of TGF-β in cells.
Conclusion
Our data suggested that if FGF-10, a protein with function in early life organogenesis and developmental roles, is dysregulated in cancer, it can act as a wild agent involving cell proliferation, immortality, and tumorigenesis, plus EMT followed by invasion and metastasis. Yet there are more unanswered questions about the possible key molecules in apoptosis or other details in FGF-10 and downstream signaling.
Different mechanisms of signaling for EMT which might be induced by FGF10. FGF10 has a direct effect on its expression, followed by TGF-β up regulation. FGF10 may have inducing effect on EMT through Ras/MAPK and AKT/Pi3K/mTOR pathways. Another finding has shown that WNT signaling activity though GSK3β phosphorylation leads to βCatenin (βCat) localization into the nucleus and Lef/TCF activation. Also GSK3β phosphorylation (inactivation) leads to stabilization of Smad 3 and sensitivity of cells to TGF-β which induces another way of EMT through TGF-β/Smad pathway.
Acknowledgements
The authors would like to thank Seyed Muhammed Hussein Mousavinasab for his sincere cooperation in editing this text. Also, we appreciate Tayebeh Sabokbar and Elnaz Aghdami for their technical assistance and comments. This study was supported by cancer research center (CRC) of Tehran University of Medical Sciences and Iran National Science Foundation (INSF), grant number #90000937.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Mehlen P, Puisieux A. Metastasis: a question of life or death. Nature Reviews Cancer. 2006;6:449-58
2. Hance M, Dole K, Gopal U, Bohonowych J, Jezierska-Drutel A, Neumann C. et al. Secreted Hsp90 is a novel regulator of the epithelial to mesenchymal transition (EMT) in prostate cancer. J Biol Chem. 2012;287(45):37732-44
3. Radisky D. Epithelial-mesenchymal transition. Journal of Cell Science. 2005;118:4325
4. Shirkoohi R. Epithelial Mesenchymal Transition from a natural gestational orchestration to a bizarre cancer disturbance. Cancer Sci. 2013;104(1):28-35
5. Kong D, Li Y, Wang Z, Sarkar F. Cancer Stem Cells and Epithelial-to-Mesenchymal Transition (EMT)-Phenotypic Cells: Are They Cousins or Twins? Cancers. 2011;3:716-29
6. Keshamouni V, Schiemann W. EMT in Tumor Metastasis: A Method to the Madness. Future Oncol. 2009;5(8):1109-11
7. Gann P, Morrow M. Combined hormone therapy and breast cancer. A single-edged sword (editorial). JAMA. 2003;289:3304-6
8. Hogan B, Kolodziej P. Organogenesis:molecular mechanisms of tubulogenesis. Nat Rev Genet. 2002;3:513-23
9. Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow M. Sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophilaairways. Cell. 1998;92:253-63
10. Ma Y, Zhang P, Wang F, Yang J, Yang Z, Qin H. The relationship between early embryo development and tumourigenesis. J Cell Mol Med. 2010;14(12):2697-701
11. Michos O, Cebrian C, Hyink D, Grieshammer U, William sL, D'Agati V. et al. Kidney development in the absence of Gdnf and Spry1 requires Fgf10. PLoS Genet. 2010;6(1):e1000809
12. Meeks J, Schaeffer E. Genetic regulation of prostate development. J Androl. 2011;32(3):210-7
13. Li C, Hu L, Xiao J, Chen H, Li J, Bellusci S. et al. Wnt5a regulates Shh and Fgf10 signaling during lung development. Dev Biol. 2005;287(1):86-97
14. Rosli S, Shintani T, Hayashido Y, Toratani S, Usui E, Okamoto T. 1α,25(OH)2D3 down-regulates HBp17/FGFBP-1 expression via NF-κB pathway. J Steroid Biochem Mol Biol. 2012 Epub ahead of print
15. Chioni A, Grose R. FGFR1 cleavage and nuclear translocation regulates breast cancer cell behavior. J Cell Biol. 2012;197(6):801-17
16. Chioni A, Grose R. Negative regulation of fibroblast growth factor 10 (FGF-10) by polyoma enhancer activator 3 (PEA3). European Journal of Cell Biology. 2009;88:371-84
17. Sylvester P. Optimization of the tetrazolium dye (MTT) colorimetric assay for cellular growth and viability. Methods Mol Biol. 2011;716:157-68
18. Taddei M, Giannoni E, Fiaschi T, Chiarugi P. Anoikis: an emerging hallmark in health and diseases. J Pathol. 2012;226(2):380-93
19. Weihua Z, Lin Q, Ramoth A, Fan D, Fidler I. Formation of solid tumors by a single multinucleated cancer cell. Cancer. 2011;117(17):4092-9
20. Rodriguez L, Wu X, Guan JL. Wound-healing assay. Methods Mol Biol. 2005;294:23-9
21. Vardevanyan P, Antonyan A, Parsadanyan M, Pirumyan K, Muradyan A, Karapetian A. Influence of ionic strength on Hoechst 33258 binding with DNA. J Biomol Struct Dyn. 2008;25(6):641-6
22. Landex N, Kayser L. Optimization of an Acridine Orange-bisbenzimide procedure for the detection of apoptosis-associated fluorescence colour changes in etoposide-treated cell cultures. J Mol Histol. 2004;35(2):133-9
23. Altomare D, Testa J. Perturbations of the AKT signaling pathway in human cancer. Oncogene. 2005;24:7455-64
24. Ahmad I, Iwata T, Leung H. Mechanisms of FGFR-mediated carcinogenesis. Biochim Biophys Acta. 2012;1823(4):850-60
25. Hunakova L, Sedlakova O, Cholujova D, Gronesova P, Duraj J, Sedlak J. Modulation of markers associated with aggressive phenotype in MDA-MB-231 breast carcinoma cells by sulforaphane. Neoplasma 2009. 2009;56(6):548-56
26. Kang S, Elf S, Dong S, Hitosugi T, Lythgoe K, Guo A. et al. Fibroblast growth factor receptor 3 associates with and tyrosine phosphorylates p90 RSK2, leading to RSK2 activation that mediates hematopoietic transformation. Mol Cell Biol. 2009;29:2105-17
27. Choe C, Chen N, Sawadogo M. Decreased tumorigenicity of c-Myc-transformed fibroblasts expressing active USF2. Exp Cell Res. 2005;302(1):1-10
28. Nyeng P, Norgaard G, Kobberup S, Jensen J. FGF10 maintains distal lung bud epithelium and excessive signaling leads to progenitor state arrest, distalization, and goblet cell metaplasia. BMC Dev Biol. 2008 8(2)
29. Hyatt B, Shangguan X, Shannon J. FGF-10 induces SP-C and Bmp4 and regulates proximal-distal patterning in embryonic tracheal epithelium. Am J Physiol Lung Cell Mol Physiol. 2004;287(6):L1116-26
30. Volckaert T, Dill E, Campbell A, Tiozzo C, Majka S, Bellusci S. et al. Parabronchial smooth muscle constitutes an airway epithelial stem cell niche in the mouse lung after injury. J Clin Invest. 2011;121(11):4409-19
31. Bhushan A, Itoh N, Kato S, Thiery J, Czernichow P, Bellusci S. et al. Fgf10 is essential for maintaining the proliferative capacity of epithelial progenitor cells during early pancreatic organogenesis. Development. 2001;128(24):5109-17
32. Hart A, Papadopoulou S, Edlund H. Fgf10 maintains notch activation, stimulates proliferation, and blocks differentiation of pancreatic epithelial cells. Dev Dyn. 2003;228(2):185-93
33. Meyer M, Müller A, Yang J, Moik D, Ponzio G, Ornitz D. et al. FGF receptors 1 and 2 are key regulators of keratinocyte migration in vitro and in wounded skin. J Cell Sci. 2012;125(pt23):5690-701
34. Neganova I, Vilella F, Atkinson S, Lloret M, Passos J, von Zglinicki T. et al. An important role for CDK2 in G1 to S checkpoint activation and DNA damage response in human embryonic stem cells. Stem Cells. 2011;29(4):651-9
35. Calegari F. CyclinD2 at the edge: splitting up cell fate. EMBO J. 2012;31(8):1850-2
36. Dedov V, Dedova I, Nicholson G. Equilibrium between cell division and apoptosis in immortal cells as an alternative to the G1 restriction mechanism in mammalian cells. Cell Cycle. 2004;3(4):491-5
37. Fulda S. Evasion of apoptosis as a cellular stress response in cancer. Int J Cell Biol. 2010 370835
38. Masola V, Gambaro G, Tibaldi E, Brunati A, Gastaldello A, D'Angelo A. et al. Heparanase and syndecan-1 interplay orchestrates fibroblast growth factor-2-induced epithelial-mesenchymal transition in renal tubular cells. J Biol Chem. 2012;287(2):1478-88
39. Hardy K, Yatskievych T, Konieczka J, Bobbs A, Antin P. FGF signalling through RAS/MAPK and PI3K pathways regulates cell movement and gene expression in the chicken primitive streak without affecting E-cadherin expression. BMC Dev Biol. 2011;11(20):11-20
40. Katoh M, Katoh M. Cross-talk of WNT and FGF signaling pathways at GSK3beta to regulate beta-catenin and SNAIL signaling cascades. Cancer Biol Ther. 2006;5(9):1059-64
41. Puntervoll P, Linding R, Gemünd C, Chabanis-Davidson S, Mattingsdal M, Cameron S. et al. ELM server: A new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31(13):3625-30
42. Guo X, Ramirez A, Waddell D, Li Z, Liu X, Wang X. Axin and GSK3- control Smad3 protein stability and modulate TGF- signaling. Genes Dev. 2008;22(1):106-20
Author contact
![]() Corresponding author: Reza Shirkoohi, Group of Genetics, Cancer Research Center, Cancer Institute of Iran, Keshavarz Blvd, Imam Khomeini Hospital Complex, Tehran, Iran. Postal Code: 1419733141 Fax: +98 21 66581526 Tel: +98 21 66914545 E-mail: rshirkoohiac.ir
Corresponding author: Reza Shirkoohi, Group of Genetics, Cancer Research Center, Cancer Institute of Iran, Keshavarz Blvd, Imam Khomeini Hospital Complex, Tehran, Iran. Postal Code: 1419733141 Fax: +98 21 66581526 Tel: +98 21 66914545 E-mail: rshirkoohiac.ir

 Global reach, higher impact
Global reach, higher impact