3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2014; 5(8):646-654. doi:10.7150/jca.9955 This issue Cite
Research Paper
Gene Expression Analysis of the 26S Proteasome Subunit PSMB4 Reveals Significant Upregulation, Different Expression and Association with Proliferation in Human Pulmonary Neuroendocrine Tumours
1. Institute of Pathology, University Hospital Essen, University of Duisburg-Essen, Germany.
2. Ruhrlandklinik, West German Lung Centre, University Hospital Essen, University of Duisburg-Essen, Germany.
3. Institute of Pathology, University Hospital Cologne, University of Cologne, Germany.
4. Department of Medical Oncology, University Hospital Essen, University of Duisburg-Essen, Germany.
5. Department of Thoracic Surgery, Ruhrlandklinik, West German Lung Centre, University Hospital Essen, University of Duisburg-Essen, Germany.
6. Department of Pathology, Helios Klinikum Emil von Behring, Berlin, Germany.
7. Pulmonary-Oncology, ``G. Papanikolaou`` General Hospital, Aristotle University of Thessaloniki, Thessaloniki, Greece.
8. Department of Respiratory Diseases, Changhai Hospital/First Affiliated Hospital of the Second Military Medical University, Shanghai, People's Republic of China, China.
9. Thoracic Surgery Department, ``Saint Luke`` Private Clinic, Thessaloniki, Panorama, Greece.
* Authors equally contributed.
Received 2014-5-17; Accepted 2014-6-20; Published 2014-8-1
Abstract
Background: Proteasomal subunit PSMB4 was suggested to be a survival gene in an animal model of hepatocellular carcinoma and in glioblastoma cell lines. In pulmonary adenocarcinoma, a high expression of these genes was found to be associated with poor differentiation and survival. This study investigates the gene expression levels of 26S proteasome subunits in human pulmonary neuroendocrine tumours including typical (TC) and atypical (AC) carcinoid tumours as well as small cell (SCLC) and large cell (LCNEC) neuroendocrine carcinomas.
Material and methods: Gene expression levels of proteasomal subunits (PSMA1, PSMA5, PSMB4, PSMB5 and PSMD1) were investigated in 80 neuroendocrine pulmonary tumours (each 20 TC, AC, LCNLC and SCLC) and compared to controls. mRNA levels were determined by using TaqMan assays. Immunohistochemistry on tissue microarrays (TMA) was performed to determine the expression of ki67, cleaved caspase 3 and PSMB4.
Results: All proteasomal subunit gene expressions were significantly upregulated in TC, AC, SCLC and LCNEC compared to controls. PSMB4 mRNA is differently expressed between all neuroendocrine tumour subtypes demonstrating the highest expression and greatest range in LCNEC (p=0.043), and is significantly associated with proliferative activity (p=0.039).
Conclusion: In line with other 26S proteasomal subunits PSMB4 is significantly increased, but differently expressed between pulmonary neuroendocrine tumours and is associated with the proliferative activity. Unlike in pulmonary adenocarcinomas, no association with biological behaviour was observed, suggesting that increased proteasomal subunit gene expression is a common and probably early event in the tumorigenesis of pulmonary neuroendocrine tumours regardless of their differentiation.
Keywords: 26S Proteasome, TaqMan qPCR, neuroendocrine lung tumours, carcinoid tumours, neuroendocrine lung carcinoma.
Background
Neuroendocrine tumours (NET) of the lung include the low grade typical carcinoid (TC), the intermediate grade atypical carcinoid (AC) and the two high grade tumours large cell neuroendocrine carcinoma (LCNEC) and small cell neuroendocrine carcinoma (SCLC). Whereas TC and AC typically occur in younger patients, are not related to cigarette smoking and have a favourable prognosis, LCNEC and SCLC are mainly observed in older individuals with a history of smoking and are associated with poor survival. Whereas TC and SCLC usually occur centrally in the lung, AC and LCNEC more often show a peripheral localization. Until now, NET grading is based on morphological parameters reflecting the proliferative activity of a given tumour: TC is defined as a neuroendocrine tumour greater than 5 mm which lacks necrosis with not more than one mitosis per 10 HPF. AC is diagnosed when a NET shows between two and ten mitoses per 10 HPF and/or comedo-like necrosis, while SCLC and LCNEC are characterized by a high degree of cellular polymorphism and more than 10 mitoses per HPF [1]. While traditionally the carcinoid tumours have been thought to represent a continuum with the high grade tumours (LCNEC and SCLC), carcinoid tumours differ significantly from the high grade tumours on clinical, immunohistochemical and molecular grounds, thus challenging the notion that these tumours originate from a common precursor cell type [2-4]. Moreover, gene expression profiling studies using genome-wide approaches reported that high grade neuroendocrine carcinomas clustered in different groups than carcinoid tumours [4].
Proteasomes are large multicatalytic proteases located in the cytosol, both free and attached to the endoplasmic reticulum, and in the nucleus that recognize, unfold and digest protein substrates that have been marked for degradation by an ubiquitin moiety. The most common form is the 26S proteasome [5]. This enzyme complex is composed of the 20S proteolytic core particle (CP) capped on one or both ends with a 19S regulatory particle (RP) [6]. The 20S proteasome is the central proteolytic structure consisting of two pairs of rings each containing seven subunits.
Three of these subunits are crucial for this activity, the β1 subunit with "caspase-like" activity, the β2 subunit with trypsin-like activity, and the β5 subunit with chymotrypsin-like activity [7]. As proteins play crucial roles in virtually all biological processes in eukaryotic cells, an intact and properly regulated ubiquitin-proteasome-system (UPS) is important for cellular homeostasis [8-13]. The UPS degrades and hereby regulates numerous short-lived and functionally different proteins involved in crucial cellular processes encompassing the modulation of cell surface and nuclear receptors, antigen presentation, signal transduction [14-17], and importantly, cellular proliferation by controlling the cell cycle and transcriptional activity [8, 18-20], as well as apoptosis [21]. In particular, proteasomes are engaged in the proteolysis of many tumour suppressor proteins related to cell division [22, 23], as well as of proteins involved in the NF-κB- [24], the mitogen activated protein kinases (MAPK)- [25], the P53/21- [26, 27] and the retinoblastoma protein (Rb)/E2F [28-30] pathways, resulting in increased proliferation and invasive and metastasizing properties of somatic cells. Consequently, dysfunction or defects of the UPS have been implicated in numerous human diseases including malignancy [25, 31-33]. In this context, overexpression of PSMD2, a subunit of the 19S regulatory complex, was shown to be associated with a metastatic phenotype and poor prognosis in non-small-cell lung cancer (NSCLC) patients. Whereas there are reports on the effects of proteasome inhibition by Bortezomib in SCLC [34] and MG-132 in human pulmonary (NCI-H727) and gastrointestinal (BON) carcinoid cell lines [35], in general little is known about the role of the 26S Proteasome in pulmonary neuroendocrine tumours, in particular about a potential different expression pattern in TC, AC and LCNEC/SCLC with respect to the above mentioned changes in signal transduction, cell cycle regulation or cancer-associated epithelial-mesenchymal transition (EMT) [36]. Against this background, we sought to determine the mRNA expression of specific proteasomal subunits of the 26S proteasome in pulmonary neuroendocrine tumours of varying differentiation.
Methods
Patient data
80 paraffin-embedded tissue samples of pulmonary neuroendocrine tumours with histolopathologically confirmed diagnosis were retrieved from the archives of the Institute of Pathology and Neuropathology of the University Hospital Essen (Germany), including 20 cases of each typical and atypical carcinoid tumours (TC/AC) as well as small and large cell neuroendocrine carcinomas (SCLC/LCNEC) resected between 2005 and 2012 at the Department of Thoracic Surgery (Ruhrlandklinik Essen, Germany). The tumours were classified according to the WHO diagnostic criteria outlined in the 2004 classification of tumours of the lung and pleura. Further requirements encompassed sufficient amount of viable tumour tissue and a low content of both necrosis and non-neoplastic (inflammatory or desmoplastic) cells contaminating the sample. All cases were reviewed by two experienced pathologists (JW, TH) to re-evaluate the initial diagnosis. Additionally, 20 non-tumorous lung tissues obtained from resections in patients with pneumothorax were used as benign tissue controls. These 20 samples were recruited from the Department of Pathology, Helios Klinikum Emil von Behring, Berlin (Berlin, Germany), and were evaluated by an experienced pathologist (PD Dr. T. Mairinger). Inclusion criterion was no noticeable inflammatory reaction. Of these patients, no further clinical and personal data could be evaluated due to anonymisation for ethical aspects.
The patients' mean age of all investigated cohorts was 58.98 years (median 59.58, maximum 84.07, minimum 19.51). In the carcinoid cohort, the mean age was 54 years (median 54, maximum 82, minimum 19), and in the carcinoma cohort 62 years (median 63, maximum 84, minimum 55). 37 patients were males and 43 were females (overall cohort: 43 f, 37 m; carcinoid cohort: 14 f, 26 m; carcinoma cohort: 29 f, 11 m). The investigations conform to the principles outlined in the declaration of Helsinki and were approved by the ethical committee of the University Hospital of Essen (ID: 13-5382-BO).
Real-time TaqMan qPCR
In this study, the subunits α1 and α5, β4 and β5 as well as δ1, also known as RPN2, which is part of the proteasome capping structure, were investigated. To evaluate the expression levels of the subunits of the 26S proteasome in the pulmonary neuroendocrine tumours, TaqMan real-time qPCR was applied. For RNA isolation formalin fixed paraffin embedded (FFPE) tissue was used. Tumour tissue was retrieved from the paraffin blocks with a hollow needle. 3 to 5 4µm thick sections were cut from the blocks using a microtome (Leica, SM 2000 R, Wetzlar, Germany). Tumour tissue was stored at -20°C until used for RNA-isolation. RNA-Isolation was performed with the “miRNeasy® FFPE kit” from Qiagen® according to the manufacturers' protocol, using adapted times and volumes (e.g. overnight digestion). The extracted RNA was stored at -80°C until used. For cDNA synthesis the “iScript® Select cDNA Synthesis Kit” from BioRad® was used. cDNA synthesis was done with the whole RNA-samples. cDNA not immediately used was stored at -20°C. Ct-values were normalized to the mean of two different endogenous reference genes, namely β-actin (ACTB) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Relative cDNA quantification of PSMA1, PSMA5, PSMB4, PSMB5 and PSMD1 and two reference genes (GAPDH and ACTB) as internal reference genes for normalization were measured. For evaluation, commercial TaqMan® Gene Expression- assays (Applied Biosystems®, AoD, Assay-ID: HS 03023943-g1, HS 01027362-g1, HS 00932059-m1, HS 01123843-g1, 01002826-g1, HS 00160631-m1) with optimized primer and probe concentrations were used. 10µl of the sample was sufficient as a reaction volume. For gene expression analysis in formalin fixed paraffin embedded (FFPE) tissue, a set of primers with low amplicon size was used to avoid the limitations of RNA degradation. qPCR and data analysis was performed on a Roche® LightCycler® 480 II. qPCR analysis was performed in concordance to the MIQE-guidelines [37].
Immunohistochemistry
For immunohistochemical characterization staining of whole tissue slides with antibodies directed against Pan-Cytokeratin (DAKO, Clone MNF116, Hamburg, Germany), CD56 (Zytomed, clone RCD56, Berlin, Germany), Chromogranin A (Novocastra, clone 5H7, Wetzlar, Germany) and TTF-1 (DAKO, Clone 8G7G3/1, Hamburg, Germany) was performed. The proliferation index was determined by staining with antibodies directed against the proliferation associated marker ki67 (Zytomed, clone K-2, Berlin, Germany) using a fully automated stainer (Dako AutostainerPlus). Apoptosis was determined by immunohistochemistry against cleaved caspase 3 (Cell Signalling, Beverly, MA, USA). The immunoexpression of ki67 was quantified by applying a four stage score (0= 0%, 1= <10%, 2=11 -50%, 3=> 50%), whereas cleaved caspase was scored by a different system (0 = 0%, 1= <5%, 2= 5-10%, 3= > 10%). Because PSMB4 was differentially expressed in any pulmonary NET subtype, the protein expression was assessed by immunohistochemistry (Sigma, polyclonal antibody, Taufkirchen, Germany).
Statistical analysis
For assay validation, standard curves were calculated using six different concentrations of a pool out of all isolated RNAs. Calculation was performed automatically with the Roche® LightCycler® 480 II data analysis program.
PCR was analysed by the 2-ΔCt method. Correlation analysis was performed with a custom-programmed algorithm for the R software. The exact Wilcoxon Mann-Witney Rank Sum test was used to test associations between gene or protein expression and dichotomous variables (gender). To rule out a possible association between gene or protein expression and clinical variables (age, age of blocks, etc.), a Spearman's rank correlation test was performed. The Spearman's rank correlation test was done to test associations between gene expression and IHC staining score. For correlation analysis between the expression statuses of the different investigated genes/proteins Spearman's rank correlation test was applied. Calculation of the overall survival (OS) and progression-free survival (PFS) was performed by Kaplan-Meier curves. Survival analysis for overall and progression-free survival was done by Cox-regression (COXPH-model), statistical significance was determined using Likelihood ratio test and Score (logrank) test. A p-value below 0.05 was considered statistically significant.
Results
TaqMan real-time qPCR
Five genes of the 26S proteasome complex coding for proteins of the catalytic domain and two additional reference genes as endogenous controls were tested using the TaqMan AoD for their mRNA expression levels. Efficacies calculated from the standard curves lay between 1.89 and 2.01. The standard deviation was lower than 0.4 Ct in any cases and assays. None of the non-template controls (NTCs) generated a detectable signal. 2^-delta Ct values for the 26S proteasome subunits were as follows: PSMA1 (3.5E-07 to 53.69; median: 1.35; mean: 2.70), PSMA5 (7.7E-06 to 901.20; median: 2.27; mean: 136.12), PSMB4 (1.3E-05 to 77.36; median: 1.61; mean: 3.54) and PSMD1 (9.9E-07 to 40.38; median: 1.32; mean: 3.78). GAPDH was between 0.58 and 1.96 (median: 0.97; mean: 1.31) and ACTB was used for normalization (min: 1; median: 1, mean: 1, max: 1).
Overall cohort of pulmonary neuroendocrine tumours
The mRNA expression of the 26S proteasome subunits demonstrated significant correlations to each other (p=0.003 and lower; rho between 0.338 and 0.598), except PSMA5/PSMB5 (p=0.69; rho=0.047); indicating that any 26S proteasome subunits are equally expressed in these tumours and mRNA of each subunit was reliably detected.
26S Proteasome subunit mRNA expressions are significantly upregulated in pulmonary neuroendocrine tumours
Compared to non-tumorous lung tissue, the gene expression of all 26S Proteasome subunits was significantly increased in any subtypes of pulmonary neuroendocrine tumours including TC, AC, SCLC and LCNEC (PSMA1: p> 0.0001; PSMA5: p= 0.0002; PSMB4: p> 0.0001; PSMB5: p> 0.0001; PSMD1: p> 0.0001). Except for PSMB4, no significant differences between gene expressions of PSMA1, PSMA5, PSMB5 and PSMD1 could be observed among TC, AC, LCNEC and SCLC (Figure 1).
PSMB4 mRNA gene expression is significantly upregulated and differently expressed between the subtypes of pulmonary neuroendocrine tumours
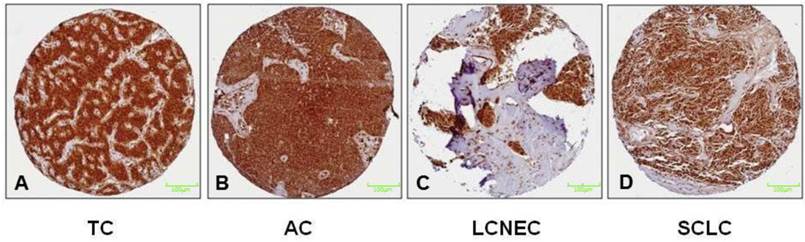
PSMB4 gene expression is significantly different between the different neuroendocrine tumour subtypes (p=0.043), showing both the highest mRNA levels and the greatest range in LCNEC (Figure 1). Immunohistochemically, strong reactivity with antibodies directed against PSMB4 was observed in any of the subtypes of pulmonary NET (Figure 2).
mRNA level of PSMB4 is significantly associated with proliferation in pulmonary neuroendocrine tumours
The proliferation index of pulmonary neuroendocrine tumours measured by nuclear expression of Ki67 showed a significant positive correlation to the mRNA level of PSMB4 (p=0.0039, rho= 0.301). None of the other proteasomal subunits were associated with the proliferation (ki67) or apoptosis (cleaved caspase 3), respectively.
No significant correlation between mRNA levels of 26S proteasome subunits with a metastatic phenotype in pulmonary neuroendocrine tumours
No significant correlation between the mRNA levels of the proteasome subunits and the occurrence of metastases to either lymph nodes or distant locations could be observed. However, there was a statistical trend for PSMB4 (p=0.076, rho=0.241) and PSMB5 (p=0.054; rho=0.262) mRNA expression and tumour size.
No significant correlations between the mRNA levels of the Proteasome subunits and the immunoexpression of Pan-Cytokeratin (MNF116), TTF-1, CD56, Synaptophysin or Chromogranin A immunoexpression was noted. Neither the expression of the immunohistochemical markers mentioned above, nor the proliferation index or the mRNA levels of the proteasome subunits demonstrated any significant association with the patients' gender and age at time of diagnosis. Also the sample age, measured as the time period between surgical excision of the tissue until the isolation RNA and immunohistochemistry did not exert any effect on the analyses performed.
Pulmonary neuroendocrine carcinomas (SCLC/LCNEC)
As observed in the overall cohort any measured mRNA expression levels of the 26S-proteasome were significantly correlated to each other (p=0.010 and lower) except PSMA5/PSMB5 (p=0.332; rho=0.159).
Box plots illustrating significantly upregulated mRNA expression of PSMA1 (A), PSMA5 (B), PSMB4 (C), PSMB5 (D) and PSMD1 (E) in the different subtypes of pulmonary neuroendocrine tumours (NET; TC = typical carcinoid tumour, AC = atypical carcinoid tumour, LCNEC = large cell neuroendocrine carcinoma and SCLC = small cell neuroendocrine carcinoma). The gene expression is shown using log(2^-dCt) of each target. Horizontal bars indicate significant differences. Note that any proteasomal subunit mRNA expression is significantly upregulated compared to controls obtained from non-tumourous lung tissue of patients with pneumothorax. Especially PSMB4 and PSMD1 show the strongest upregulation compared to benign lung. LCNEC show the broadest range of proteasomal expression pattern, maybe due to the heterogeneity of this entity. Remarkable is the relatively low PSMA5 expression level when compared to the control.
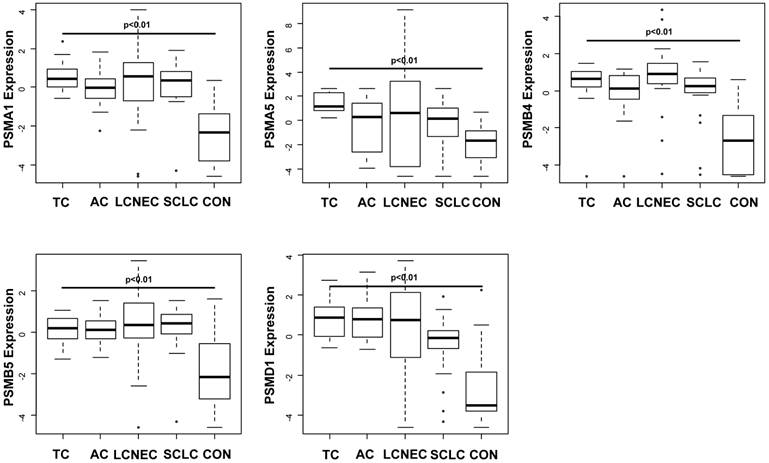
Figure 2 illustrates the results of the immunohistochemical staining of the proteasomal subunit PSMB4 in both, low-grade (A+B) and high-grade (C+D) pulmonary NET. Immunohistochemically, PSMB4 reactivity is strong in any subtypes of pulmonary NET. All NET are silhouetted against the encircling stroma. In particular, the PSMB4 mRNA expression is upregulated in pulmonary NET compared to controls and differs significantly among the subtypes of pulmonary NET. PSMB4 mRNA expression, which has been suggested to be a survival gene in cancer, is higher in LCNEC/SCLC than in TC/AC. Nevertheless, immunohistochemically no differences between the four tumor entities could be detected.
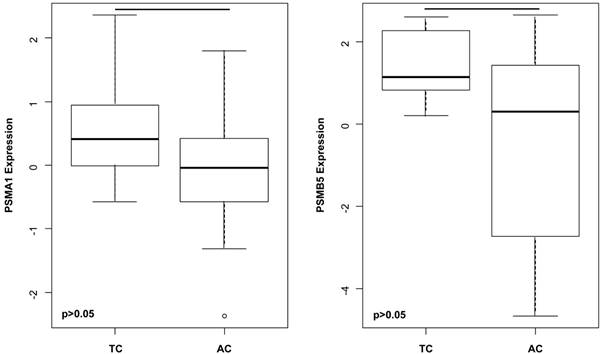
Among others, the gene expressions of A) PSMA1 (p= 0.030) and B) PSMB5 (p= 0.035) are significantly higher in TC compared to AC. This suggests that upregulated gene expressions of proteasomal subunits do not simply reflect the grade or the malignant potential, but might influence a wide spectrum of crucial cellular processes including signal transduction, cell cycle, survival and differentiation.
Significantly increased PSMB4 mRNA expression in LCNEC compared to SCLC
PSMB4 gene expression level is significantly higher in LCNEC compared to SCLC (p=0.036).
Pulmonary carcinoids tumours (TC/AC)
mRNA levels of several Proteasome subunits significantly differ between TC and AC
PSMA1 (p=0.030), PSMB5 (p=0.035) (Figure 3) as well as PSMA5 (p=0.029) and PSMB4 (p=0.050) were significantly increased in TC compared to AC.
Discussion
NET are a heterogeneous group of tumours and show considerable differences on immunohistochemical and molecular, and in particular on clinical grounds with decreasing survival and increasing metastatic phenotype from TC over AC to SCLC/LCNEC. Gene expression profiling studies using genome-wide approaches revealed significant differences between neuroendocrine carcinomas and carcinoid tumours [4]. Thus, it is doubtful, whether the pulmonary neuroendocrine tumours originate from an identical precursor cell or represent biologically different entities [2, 3, 29, 30]. In general, there is increasing evidence that dysfunction of the UPS is associated with the development of malignant tumours in humans.
The 20S-proteasome subunit forms a cylinder containing the inner β-units and the outer α-subunits. Only the β-subunits show catalytical activity, namely β1 (caspase-like activity), β2 (trypsin-like activity) and β5 (chymotrypsin-like activity). The 19S-proteasome forms the cap and is built of Rpn- and Rpt-proteins. An increased proteasome expression was observed in leukaemia [38], renal carcinoma [39] and human breast cancer cells [40]. Upregulation of the proteasomal subunits PSMA6, PSMB4, PSMC2 and PSMD12 was demonstrated in hepatocellular carcinomas of p21-HBx transgenic mice by using MALDI-TOF analysis [41]. By application of small interfering (si) RNA screening in human glioblastoma cells, Thaker and co-workers were able to identify 55 survival genes encoding proteases, kinases and transferases out of which 12 were subunits of the 20S Proteasome, including PSMB4 [42], suggesting a role of different gene expression of proteasomal subunits in the development of various malignant tumours. Matsuyama and co-workers were able to show that pulmonary adenocarcinoma patients can be clearly divided in two groups; those with and without a general high expression of genes involved in proteasome-mediated protein degradation: a high expression of these genes was associated with higher tumour grade, non-terminal respiratory unit (non-TRU) histology, history of smoking and decreased survival. Of note, treatment of pulmonary adenocarcinoma cell lines with increased PSMD2 expression by siRNA was associated with decreased proteasomal activity, induction of apoptosis and increased numbers of cells in the G1 phase. Moreover, a decrease in phosphorylated AKT and increase of phosphorylated p38 and induction p21 appeared to be consistent with the treatment effects. This study also demonstrated that genes encoding components of the 26S proteasome and those involved in proteasome assembly are co-regulated in lung adenocarcinomas and a high expression of these genes is associated with shortened survival. The authors conclude that such co-regulated upregulation may confer greater advantage to cancer cell growth during cancer development [43].
In contrast to the investigation by Matsuyama and co-workers on pulmonary adenocarcinomas, this study demonstrates that gene expression of proteasomal subunits is generally increased in all subtypes of human pulmonary neuroendocrine tumours regardless of the biological behaviour and tumour grading compared to non-tumorous controls. The strong staining intensity of tumour cell in comparison to benign tissue can be helpful in the case of diagnosing this tumour entity also in more ambitious cases or e.g. biopsic material. However, PSMB4 expression shows a significant association with the degree of proliferation in these neoplasias. This could be explained by the effects of the proteasome engaged in the proteolysis of many tumour suppressor proteins related to cell division [22, 23], as well as of proteins involved in the NF-κB- [24], the mitogen activated protein kinases (MAPK)- [25], the P53/21- [26, 27] and the retinoblastoma protein (Rb)/E2F [28-30] pathways, resulting in increased proliferation of tumour cells. Of note, Wang and co-workers were able to show that under hypoxic conditions, PSMB6 was upregulated and associated with proliferation of smooth muscle cells of the pulmonary artery in an animal model of pulmonary hypertension [44], since hypoxia is also an important factor in carcinogenesis. Despite of a general and nearly equal upregulation of any proteasomal subunits investigated, PSMB4 mRNA expression differs significantly between the different subgroups of pulmonary NET being highest and showing the greatest range in LCNEC. The mRNA expression of PSMB4 is significantly lower in carcinoid tumours and higher in neuroendocrine carcinomas, but is significantly increased in TC compared to AC, and in LCNEC compared to SCLC. These findings suggest (1) in line with the study of Thaker and co-workers that PSMB4 may act as a survival gene in malignant neuroendocrine tumours of varying differentiation [42], and (2) that an increased gene expression and proteasomal enzymatic activity might be a crucial and probably early event in the development of neuroendocrine tumours of the lung, that interferes with numerous cellular processes involved in malignant transformations such as cell cycle regulation, proliferation and apoptosis promoting further malignant transformation. In contrast to the findings of Matsuyama and co-workers, the increased mRNA expression of proteasomal subunits does not necessarily reflect the proliferative activity, biological behaviour or clinical course of pulmonary NET. In particular, the UPS has been shown to play a complex role in the regulation of apoptosis, in which inhibition of the UPS has pro- as well as anti-apoptotic effects [45]. The activation of NFκB by proteasomes through degradation of IκB induces the expression of anti-apoptotic members of the bcl-2 family [46]. Additionally, proteasomes degrade pro-apoptotic proteins including Bax and Bid [47, 48]. Although the 26S Proteasome is involved in the epithelial-mesenchymal transition (EMT) associated with infiltrative growth and metastatic property, no association between the mRNA expressions of any proteasomal subunit investigated (PSMA1, PSMA5, PSMB4, PSMB5 and PSMD1) with apoptosis and the occurrence of a metastatic phenotype could be outlined. This could probably be explained by the relatively small number of cases investigated per group (TC, AC and LCNEC/SCLC). The issue of metastatic phenotype of pulmonary NET and proteasomal gene expression has to be addressed in greater number of cases in the future.
The strong expression of all subunits of the 26S-proteasome, especially in comparison to benign lung tissue, suggests the conclusion that neuroendocrine tumours of the lung strongly depend on their proteasomal expression level. This hypothesis leads the door wide open for potential therapeutic approaches regarding a potential anti-proteasome therapeutic concept. In particular, a therapy with bortezomib, a proteasome inhibitor approved by the FDA for the treatment of relapsed multiple myeloma and mantle-cell lymphoma [49], chances to be a promising approach for the treatment of generally less chemosensitive carcinoid tumours of the lung with syndrome or even high-grade pulmonary NETs. Even when the most trials using bortezomib up to now focused on lymphoma and leukaemia [50-55], also in different entities of solid tumours an inhibitory and apoptotic effect could be shown [55-63]. Also in SCLC [34] and carcinoid cell lines [35], a beneficial effect of bortezomib in a therapeutic concept was found. The question, whether proteasomal inhibition in this class of tumours should find ones way into standard therapy, must be addressed in further studies.
Conclusion
In summary, these data show that proteasomal subunits are upregulated in pulmonary neuroendocrine neoplasms.
Although it is very likely from clinical and molecular grounds, that pulmonary carcinoid tumours and SCLC/LCNEC are different entities that probably arise from different cells of origin; all subtypes show an almost equal upregulation of proteasomal subunit mRNA expression. This suggests that proteasomal overexpression is a common and sustained phenomenon occurring in pulmonary NET regardless of their differentiation and biological behaviour, which might provide the biochemical milieu for proliferative growth for these neoplasms. In contrast to pulmonary adenocarcinomas, the gene expression of proteasomal subunits does not seem to be able to discriminate between different biological behaviours of pulmonary NET. The interplay between increased proteasomal activity and different cellular processes in the different subtypes of pulmonary NET such as cell cycle regulation and apoptosis has to be investigated in more detail in the future. Nevertheless, these data might prompt further investigation of proteasome inhibitors in animal models of pulmonary NET.
Acknowledgements
The skilful assistance of Dorothe Möllmann is highly appreciated.
Conflict of Interest
No conflicts of interest declared.
References
1. Travis WD. et al. Pathology and genetics of tumours of the lung, pleura, thymus and heart. Lyon, Oxford: IARC Press. 2004
2. Flieder DB. Neuroendocrine tumors of the lung: recent developments in histopathology. Current opinion in pulmonary medicine. 2002;8:275-80
3. Lim E, Goldstraw P, Nicholson AG, Travis WD, Jett JR, Ferolla P. et al. Proceedings of the IASLC International Workshop on Advances in Pulmonary Neuroendocrine Tumors 2007. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2008;3:1194-201 doi:10.1097/JTO.0b013e3181861d7b
4. Swarts DR, Ramaekers FC, Speel EJ. Molecular and cellular biology of neuroendocrine lung tumors: evidence for separate biological entities. Biochimica et biophysica acta. 2012;1826:255-71 doi:10.1016/j.bbcan.2012.05.001
5. Chitra S, Nalini G, Rajasekhar G. The ubiquitin proteasome system and efficacy of proteasome inhibitors in diseases. International journal of rheumatic diseases. 2012;15:249-60 doi:10.1111/j.1756-185X.2012.01737.x
6. Tomko RJ Jr, Hochstrasser M. Order of the proteasomal ATPases and eukaryotic proteasome assembly. Cell biochemistry and biophysics. 2011;60:13-20 doi:10.1007/s12013-011-9178-4
7. Calise J, Powell SR. The ubiquitin proteasome system and myocardial ischemia. American journal of physiology Heart and circulatory physiology. 2013;304:H337-49 doi:10.1152/ajpheart.00604.2012
8. Hershko A, Ciechanover A. The ubiquitin system. Annual review of biochemistry. 1998;67:425-79 doi:10.1146/annurev.biochem.67.1.425
9. Goldberg AL. Functions of the proteasome: from protein degradation and immune surveillance to cancer therapy. Biochemical Society transactions. 2007;35:12-7 doi:10.1042/BST0350012
10. Gudmundsdottir K, Lord CJ, Ashworth A. The proteasome is involved in determining differential utilization of double-strand break repair pathways. Oncogene. 2007;26:7601-6 doi:10.1038/sj.onc.1210579
11. McBride WH, Iwamoto KS, Syljuasen R, Pervan M, Pajonk F. The role of the ubiquitin/proteasome system in cellular responses to radiation. Oncogene. 2003;22:5755-73 doi:10.1038/sj.onc.1206676
12. van Tijn P, Hol EM, van Leeuwen FW, Fischer DF. The neuronal ubiquitin-proteasome system: murine models and their neurological phenotype. Progress in neurobiology. 2008;85:176-93 doi:10.1016/j.pneurobio.2008.03.001
13. Yi JJ, Ehlers MD. Emerging roles for ubiquitin and protein degradation in neuronal function. Pharmacological reviews. 2007;59:14-39 doi:10.1124/pr.59.1.4
14. Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: biological regulation via destruction. BioEssays: news and reviews in molecular, cellular and developmental biology. 2000;22:442-51 doi:10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q
15. Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiological reviews. 2002;82:373-428 doi:10.1152/physrev.00027.2001
16. Goldberg AL, Elledge SJ, Harper JW. The cellular chamber of doom. Scientific American. 2001;284:68-73
17. Nawaz Z, O'Malley BW. Urban renewal in the nucleus: is protein turnover by proteasomes absolutely required for nuclear receptor-regulated transcription? Mol Endocrinol. 2004;18:493-9 doi:10.1210/me.2003-0388
18. Adams J, Palombella VJ, Elliott PJ. Proteasome inhibition: a new strategy in cancer treatment. Investigational new drugs. 2000;18:109-21
19. Adams J. Preclinical and clinical evaluation of proteasome inhibitor PS-341 for the treatment of cancer. Current opinion in chemical biology. 2002;6:493-500
20. Varshavsky A. The ubiquitin system. Trends in biochemical sciences. 1997;22:383-7
21. Arlt A, Bauer I, Schafmayer C, Tepel J, Muerkoster SS, Brosch M. et al. Increased proteasome subunit protein expression and proteasome activity in colon cancer relate to an enhanced activation of nuclear factor E2-related factor 2 (Nrf2). Oncogene. 2009;28:3983-96 doi:10.1038/onc.2009.264
22. Diehl JA, Ponugoti B. Ubiquitin-dependent proteolysis in G1/S phase control and its relationship with tumor susceptibility. Genes & cancer. 2010;1:717-24 doi:10.1177/1947601910382902
23. Masamha CP, Benbrook DM. Cyclin D1 degradation is sufficient to induce G1 cell cycle arrest despite constitutive expression of cyclin E2 in ovarian cancer cells. Cancer research. 2009;69:6565-72 doi:10.1158/0008-5472.CAN-09-0913
24. Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78:773-85
25. Orlowski RZ, Small GW, Shi YY. Evidence that inhibition of p44/42 mitogen-activated protein kinase signaling is a factor in proteasome inhibitor-mediated apoptosis. The Journal of biological chemistry. 2002;277:27864-71 doi:10.1074/jbc.M201519200
26. Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296-9 doi:10.1038/387296a0
27. Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237-45
28. Berezutskaya E, Bagchi S. The human papillomavirus E7 oncoprotein functionally interacts with the S4 subunit of the 26 S proteasome. The Journal of biological chemistry. 1997;272:30135-40
29. Wang J, Sampath A, Raychaudhuri P, Bagchi S. Both Rb and E7 are regulated by the ubiquitin proteasome pathway in HPV-containing cervical tumor cells. Oncogene. 2001;20:4740-9 doi:10.1038/sj.onc.1204655
30. Ying H, Xiao ZX. Targeting retinoblastoma protein for degradation by proteasomes. Cell Cycle. 2006;5:506-8
31. Chen W, Hu XT, Shi QL, Zhang FB, He C. Knockdown of the novel proteasome subunit Adrm1 located on the 20q13 amplicon inhibits colorectal cancer cell migration, survival and tumorigenicity. Oncology reports. 2009;21:531-7
32. Dahlmann B. Role of proteasomes in disease. BMC biochemistry. 2007;8(Suppl 1):S3. doi:10.1186/1471-2091-8-S1-S3
33. Orlowski RZ, Dees EC. The role of the ubiquitination-proteasome pathway in breast cancer: applying drugs that affect the ubiquitin-proteasome pathway to the therapy of breast cancer. Breast cancer research: BCR. 2003;5:1-7
34. Lara PN Jr, Bold RJ, Mack PC, Davies AM, Gumerlock PH, Gandara DR. Proteasome inhibition in small-cell lung cancer: preclinical rationale and clinical applications. Clinical lung cancer. 2005;7(Suppl 2):S67-71
35. Chen JY, Cook MR, Pinchot SN, Kunnimalaiyaan M, Chen H. MG-132 inhibits carcinoid growth and alters the neuroendocrine phenotype. The Journal of surgical research. 2010;158:15-9 doi:10.1016/j.jss.2009.05.032
36. Voutsadakis IA. The ubiquitin-proteasome system and signal transduction pathways regulating Epithelial Mesenchymal transition of cancer. Journal of biomedical science. 2012;19:67. doi:10.1186/1423-0127-19-67
37. Huggett JF, Foy CA, Benes V, Emslie K, Garson JA, Haynes R. et al. The digital MIQE guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments. Clinical chemistry. 2013;59:892-902 doi:10.1373/clinchem.2013.206375
38. Kumatori A, Tanaka K, Inamura N, Sone S, Ogura T, Matsumoto T. et al. Abnormally high expression of proteasomes in human leukemic cells. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:7071-5
39. Kanayama H, Tanaka K, Aki M, Kagawa S, Miyaji H, Satoh M. et al. Changes in expressions of proteasome and ubiquitin genes in human renal cancer cells. Cancer research. 1991;51:6677-85
40. Choi YH. Proteasome-mediated degradation of BRCA1 protein in MCF-7 human breast cancer cells. International journal of oncology. 2001;19:687-93
41. Cui F, Wang Y, Wang J, Wei K, Hu J, Liu F. et al. The up-regulation of proteasome subunits and lysosomal proteases in hepatocellular carcinomas of the HBx gene knockin transgenic mice. Proteomics. 2006;6:498-504 doi:10.1002/pmic.200500218
42. Thaker NG, Zhang F, McDonald PR, Shun TY, Lewen MD, Pollack IF. et al. Identification of survival genes in human glioblastoma cells by small interfering RNA screening. Molecular pharmacology. 2009;76:1246-55 doi:10.1124/mol.109.058024
43. Matsuyama Y, Suzuki M, Arima C, Huang QM, Tomida S, Takeuchi T. et al. Proteasomal non-catalytic subunit PSMD2 as a potential therapeutic target in association with various clinicopathologic features in lung adenocarcinomas. Molecular carcinogenesis. 2011;50:301-9 doi:10.1002/mc.20632
44. Wang J, Xu L, Yun X, Yang K, Liao D, Tian L. et al. Proteomic analysis reveals that proteasome subunit beta 6 is involved in hypoxia-induced pulmonary vascular remodeling in rats. PloS one. 2013;8:e67942. doi:10.1371/journal.pone.0067942
45. Naujokat C, Hoffmann S. Role and function of the 26S proteasome in proliferation and apoptosis. Laboratory investigation; a journal of technical methods and pathology. 2002;82:965-80
46. Catz SD, Johnson JL. Transcriptional regulation of bcl-2 by nuclear factor kappa B and its significance in prostate cancer. Oncogene. 2001;20:7342-51 doi:10.1038/sj.onc.1204926
47. Li B, Dou QP. Bax degradation by the ubiquitin/proteasome-dependent pathway: involvement in tumor survival and progression. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3850-5 doi:10.1073/pnas.070047997
48. Breitschopf K, Zeiher AM, Dimmeler S. Ubiquitin-mediated degradation of the proapoptotic active form of bid. A functional consequence on apoptosis induction. The Journal of biological chemistry. 2000;275:21648-52 doi:10.1074/jbc.M001083200
49. Twombly R. First proteasome inhibitor approved for multiple myeloma. Journal of the National Cancer Institute. 2003;95:845
50. Elstrom RL, Ruan J, Christos PJ, Martin P, Lebovic D, Osborne J. et al. Phase 1 study of radiosensitization using bortezomib in patients with relapsed non-Hodgkin lymphoma receiving radioimmunotherapy with I-tositumomab. Leukemia & lymphoma. 2014:1-5 doi:10.3109/10428194.2014.914195
51. Furman RR, Martin P, Ruan J, Cheung YK, Vose JM, LaCasce AS. et al. Phase 1 trial of bortezomib plus R-CHOP in previously untreated patients with aggressive non-Hodgkin lymphoma. Cancer. 2010;116:5432-9 doi:10.1002/cncr.25509
52. Xu W, Wei W, Yu Q, Wu C, Ye C, Wu Y. et al. Arsenic trioxide and bortezomib interact synergistically to induce apoptosis in chronic myelogenous leukemia cells resistant to imatinib mesylate through Bcr/Abldependent mechanisms. Molecular medicine reports. 2014 doi:10.3892/mmr.2014.2333
53. Richardson PG, Baz R, Wang M, Jakubowiak AJ, Laubach JP, Harvey RD. et al. Phase 1 study of twice-weekly dosing of investigational oral proteasome inhibitor ixazomib in patients with relapsed and/or refractory multiple myeloma. Blood. 2014 doi:10.1182/blood-2014-01-548826
54. Kumar SK, Bensinger WI, Zimmerman TM, Reeder CB, Berenson JR, Berg D. et al. Weekly dosing of the investigational oral proteasome inhibitor ixazomib in relapsed/refractory multiple myeloma: results from a phase 1 study. Blood. 2014 doi:10.1182/blood-2014-01-548941
55. Einsele H. Bortezomib. Recent results in cancer research Fortschritte der Krebsforschung Progres dans les recherches sur le cancer. 2014;201:325-45 doi:10.1007/978-3-642-54490-3_20
56. Poklepovic A, Youssefian LE, Winning M, Birdsell CA, Crosby NA, Ramakrishnan V. et al. Phase I trial of bortezomib and dacarbazine in melanoma and soft tissue sarcoma. Investigational new drugs. 2013;31:937-42 doi:10.1007/s10637-012-9913-8
57. Ambati SR, Lopes EC, Kosugi K, Mony U, Zehir A, Shah SK. et al. Pre-clinical efficacy of PU-H71, a novel HSP90 inhibitor, alone and in combination with bortezomib in Ewing sarcoma. Molecular oncology. 2014;8:323-36 doi:10.1016/j.molonc.2013.12.005
58. Shapovalov Y, Benavidez D, Zuch D, Eliseev RA. Proteasome inhibition with bortezomib suppresses growth and induces apoptosis in osteosarcoma. International journal of cancer Journal international du cancer. 2010;127:67-76 doi:10.1002/ijc.25024
59. Bauer S, Parry JA, Muhlenberg T, Brown MF, Seneviratne D, Chatterjee P. et al. Proapoptotic activity of bortezomib in gastrointestinal stromal tumor cells. Cancer research. 2010;70:150-9 doi:10.1158/0008-5472.CAN-09-1449
60. Bersani F, Taulli R, Accornero P, Morotti A, Miretti S, Crepaldi T. et al. Bortezomib-mediated proteasome inhibition as a potential strategy for the treatment of rhabdomyosarcoma. Eur J Cancer. 2008;44:876-84 doi:10.1016/j.ejca.2008.02.022
61. Ciombor KK, Feng Y, Benson AB 3rd, Su Y, Horton L, Short SP. et al. Phase II trial of bortezomib plus doxorubicin in hepatocellular carcinoma (E6202): a trial of the Eastern Cooperative Oncology Group. Investigational new drugs. 2014 doi:10.1007/s10637-014-0111-8
62. Kobrinsky B, Joseph SO, Muggia F, Liebes L, Beric A, Malankar A. et al. A phase I and pharmacokinetic study of oxaliplatin and bortezomib: activity, but dose-limiting neurotoxicity. Cancer chemotherapy and pharmacology. 2013;72:1073-8 doi:10.1007/s00280-013-2295-6
63. Gilbert J, Lee JW, Argiris A, Haigentz M Jr, Feldman LE, Jang M. et al. Phase II 2-arm trial of the proteasome inhibitor, PS-341 (bortezomib) in combination with irinotecan or PS-341 alone followed by the addition of irinotecan at time of progression in patients with locally recurrent or metastatic squamous cell carcinoma of the head and neck (E1304): a trial of the Eastern Cooperative Oncology Group. Head & neck. 2013;35:942-8 doi:10.1002/hed.23046
Author contact
![]() Corresponding author: Paul Zarogoulidis, Pulmonary Department-Oncology Unit, ``G. Papanikolaou`` General Hospital, Aristotle University of Thessaloniki, Thessaloniki, Greece. Tel: +306977271974, Fax: +302310992433, e-mail: pzarogcom.
Corresponding author: Paul Zarogoulidis, Pulmonary Department-Oncology Unit, ``G. Papanikolaou`` General Hospital, Aristotle University of Thessaloniki, Thessaloniki, Greece. Tel: +306977271974, Fax: +302310992433, e-mail: pzarogcom.

 Global reach, higher impact
Global reach, higher impact