3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2015; 6(9):812-818. doi:10.7150/jca.11975 This issue Cite
Research Paper
The Prognostic Significance of Combining VEGFA, FLT1 and KDR mRNA Expressions in Brain Tumors
1. Faculty of Health Sciences, University of Macau, Avenida de Universidade, Macau SAR
2. Center for Cancer Research & Cell Biology and School of Pharmacy, Queen's University Belfast, BT9 7BL, Belfast, United Kingdom
3. Cancer Research UK Cambridge Institute, University of Cambridge, CB2 0RE, Cambridge, United Kingdom
*These authors contributed equally to this work
Received 2015-2-24; Accepted 2015-6-12; Published 2015-7-15
Abstract
Tumor cells require angiogenesis to deliver nutrients and oxygen to support their fast growth and metabolism. The vascular endothelial growth factor (VEGF) pathway plays an important role in promoting angiogenesis, including tumor-induced angiogenesis. Recent clinical trials have demonstrated the benefit of targeting VEGF in the treatment of glioblastoma. However, the prognostic significance of the expression of VEGFA and its receptors VEGFR1 (FLT1) and VEGFR2 (KDR) are still largely elusive. In the present study, we aimed to investigate the prognostic significance of these three factors, alone or in combination, in glioma patients. Gene mRNA expression was extracted from three independent brain tumor cohorts totaling 242 patients and the association between gene expression and survival was tested. We found that when VEGFA, FLT1 and KDR expressions were considered alone, only VEGFA demonstrated a significant association with patient survival. Patients with high expression of both VEGFA and either receptor had significantly worse survival than patients expressing both factors at a low level. Importantly, we found that those patients whose tumors overexpressed all three genes had a significantly shorter survival compared to those patients with a low level expression of these genes. Our results suggest that a high level expression of VEGFA and its receptors, both FLT1 and KDR, may be required for brain tumor progression, and that these three factors should be considered together as a prognostic indicator for brain tumor patients.
Keywords: Brain tumor, VEGFA, FLT1, KDR, Angiogenesis, Survival
Introduction
Gliomas are the most common type of primary brain tumors, of which grade 3 astrocytoma and grade IV glioblastoma are highly malignant and are associated with a very poor prognosis [1]. As with other malignancies, the growth and development of gliomas is reliant on the angiogenic process. VEGF is an important factor promoting tumor angiogenesis, and the activation of VEGF pathway requires the binding of the ligand, such as VEGFA, to one of its receptors, such as FLT1 or KDR, thus generating downstream signals to stimulate the proliferation and structural reorganization of endothelial cells [2]. The VEGF family is divided into four members, including VEGFA, VEGFB, VEGFC and VEGFD, and VEGF receptors consist of three subtypes, including VEGFR1 (FLT1), VEGFR2 (KDR), and VEGFR3 [3]. VEGFA is a key regulator of developmental vasculogenesis and angiogenesis and it mainly interacts with FLT1 and KDR to generate downstream signaling [3]. FLT1 is a tyrosine kinase receptor that binds VEGFA with about 10-fold higher affinity than KDR, and although it is poorly activated by VEGF ligands, it has been shown to promote tumor growth and metastasis [3]. On the other hand, VEGF ligand binding to KDR, also a tyrosine kinase receptor, generates potent downstream signals that leads to endothelial cell growth and migration [3]. The development of targeted therapy for malignant glioma has had limited success mainly due to the parallel and converging complex interactions between several important pathways, including pathways that regulate tumor angiogenesis [4].
VEGF, FLT1 and KDR, were all shown to be up-regulated in brain tumor vasculature [5], signifying the importance of angiogenesis in glioma. Suppressing expression of VEGF in tumoral cells by anti-sense RNA technology was shown to inhibit the growth of glioma cells in vivo in nude mice [6]. In addition, VEGF also plays an important role in angiogenesis induced by stem cell-like glioma cells [7]. Inhibition of angiogenesis acts synergistically with chemotherapy in reducing the tumor sphere forming ability of brain tumor stem-like cells in glioma xenograft model [8]. Together, these results suggest that the VEGF pathway plays a central role in initiation and progression of glioma, especially in the glioma stem cells.
Bevacizumab is a monoclonal antibody that binds to VEGFA and neutralizes VEGFA from binding to FLT1 and KDR, thereby inhibiting the downstream growth signal that could induce angiongenesis [9]. Indeed, bevacizumab has been shown to be effective in combination with irinotecan for recurrent high grade glioma with a radiographic response noted in around 60% of patients, and a 6-month overall survival of 77% in patients treated with this regimen [10, 11]. Other phase II trials have shown and confirmed that bevacizumab as a single agent or in combination with irinotecan are active and tolerable in patients with recurrent glioblastoma [12, 13]. The Food and Drug Administration in United States approved bevacizumab as a single agent in patients with gliobastoma multiforme (GBM) whose disease had progressed following prior therapy[14]. Recently, the results from a phase II study reported that bevacizumab in combination with lomustine has clinical activity to recurrent GBM [15]. Two studies have assessed the potential of bevacizumab for the treatment of newly diagnosed glioblastoma. Combination of bevacizumab with temozolomide and radiation therapy increased progression-free survival compared to a retrospective control cohort [16], while combination of bevacizumab with other chemotherapeutic agents may improve efficacy in newly diagnosed glioblastoma [17]. The results from two Phase III studies have confirmed that addition of bevaicumab to radiotherapy and chemotherapy led to improvement in progression-free survival but not overall survival [18, 19].
VEGFA, FLT1 and KDR mRNAs are expressed at an elevated level in malignant tumor endothelial compared to normal brain vasculature [20]. Overexpression of VEGFA, but not FLT1, was seen in high-grade astrocytoma compared to low-grade astrocytoma [21], while increased expression of FLT1 was observed in grade 4 diffusely infiltrating astrocytomas [22]. Very recently, a SNP located in the 5' UTR of the VEGFA gene, was demonstrated to be associated with higher plasma VEGFA concentration and a longer progression-free survival in GBM patients treated with bevacizumab [23].
Despite the unquestionable role of angiogenesis in progression of malignancies of the brain, and despite recognition of VEGF pathway-targeting treatment strategies, expression profiling of the VEGF pathway role-players for brain tumor prognostication has not been done. Recently, we demonstrated the prognostic value of combined expression profiling of VEGFA, FLT1 and KDR in lung and colon cancers, in which anti-VEGF therapy has been shown to be effective. In the present study, we aimed to investigate the prognostic significance of these three factors in combination in glioma, whose most malignant subtype, glioblastoma, is susceptible to bevacizumab.
Materials and Methods
Extraction of clinical and microarray gene expression data from glioma patient datasets
Three glioma patient datasets, GSE4271 [24], GSE4412 [25] and GSE7696 [26] were identified in the Gene Expression Omnibus (GEO) Database; datasets using the HG-U133 microarray platform, and comprising >70 patients for whom survival data were available were included in this study. Microarray gene expression data were retrieved from the data matrixes deposited to the GEO database by the original authors. R scripting was used to extract the expression values from probesets of VEGFA, FLT1 and KDR, and the clinical data from the data matrixes downloaded from GEO as previously described [27].
Patient stratification
Expression levels were divided into four groups using quartile as the cut-off points for association analysis. The median expression level was used as the cut-off point for survival analysis. Patients were further divided into three groups based on the expression levels of VEGFA and FLT1; the VEGFA-FLT1-low group consisted of patients who expressed both of these two genes at low levels; the VEGFA-FLT1-high group consisted of patients who expressed both of these two genes at high levels; the VEGFA-FLT1-intermediate group consisted of the rest of patients; patients were similarly stratified into VEGFA-KDR subgroups. Patients were also divided into four groups based on the number of genes, among VEGFA, FLT1 and KDR, that were expressed at a high level.
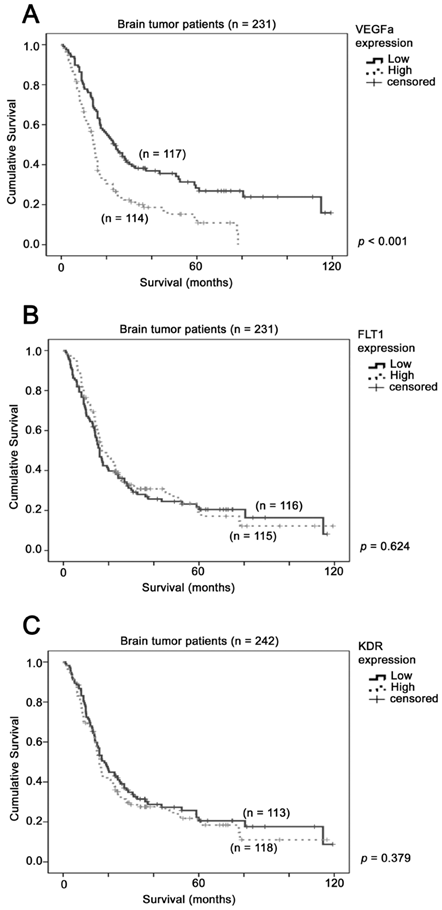
The association between VEGFA mRNA expression and brain tumor patient survival. Kaplan-Meier analyses for mRNA expression of (A) VEGFA (B) FLT1 and (C) KDR in the brain tumor patient cohort consisting of 231 patients. P-value for log-rank test is presented alongside each Kaplan-Meier plot.
Correlations of gene expression levels and clinical data
Since the sample size was small, the three patient cohorts were combined and analyzed together. The combined brain tumor dataset comprised 258 patients, 231 of whose survival data were available. Of these 231 patients, 175 were deceased at data collection. Of the 231 samples, 186 were glioblastoma, and the remaining 45 were of other subtypes. All statistical analyses were performed using SPSS19.0. The associations between expression levels of genes were tested by Chi-Square test. For Kaplan-Meier survival analysis, results were compared by log-rank test. Cox regression analysis was used to correlate the gene expression levels and patient survival.
Results
The prognostic significance of VEGFA, FLT1 and KDR in glioma patients
The prognostic significance of VEGFA, FLT1 and KDR was tested in a combined dataset comprising a total of 231 patients from GSE4271, GSE4412 and GSE7696. Using median expression level as a cut-off point, we found that patients whose tumors expressed a high level of VEGFA had a mean survival of 23 months (95% CI = 18 - 28 months), while those whose tumors expressed a low level of VEGFA had a mean survival of 46 months (95% CI = 37 - 54 months), with a Hazard ratio of 1.86 (95% CI = 1.38 - 2.52, p < 0.001; Figure 1A). This result suggests that a high level of VEGFA in glioma specimens predicted a shorter survival time of glioma patients. On the other hand, the survival time was not significantly different between patients whose tumors expressed different levels of FLT1 (p = 0.624; Figure 1B) or KDR (p = 0.379; Figure 1C). Together, our results suggest that VEGFA is a better prognostic factor for glioma patients.
Association between VEGFA, FLT1 and KDR expression
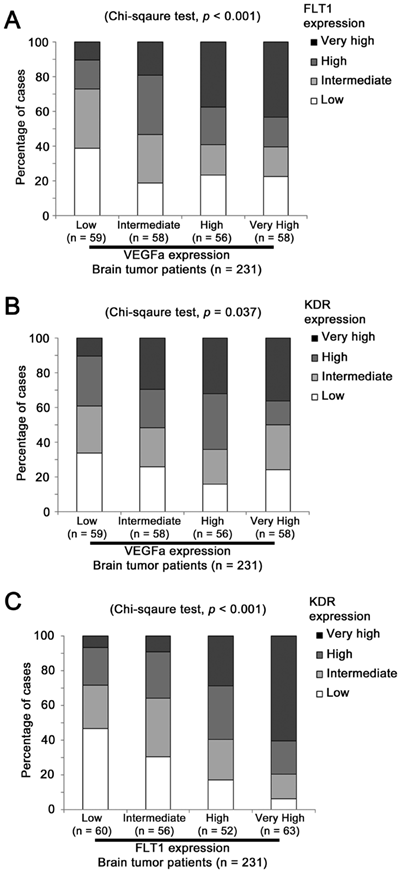
FLT1 expression level was significantly different among patients expressing various levels of VEGFA (Chi Square test, p < 0.001; Figure 2A). Around 11% (25 out of 231) of patients expressed both VEGFA and FLT1 at a very high level and around 10% (23 out of 231) of patients expressed both VEGFA and FLT1 at a low level, while only 3% (6 out of 231) of patients who expressed VEGFA at a low level expressed FLT1 at a very high level and only 6% (13 out of 231) of patients who expressed VEGFA at a very high level expressed FLT1 at a low level. Similar results were observed when the association between VEGFA and KDR expression levels was tested (Chi Sqaure test, p = 0.037; Figure 2B). Merely 3% (6 out of 231) of patients who expressed VEGFA at a low level expressed KDR at a very high level, but around 18% patients expressed these two genes both at a very high level or both at a low level. The expression levels between the two receptors, FLT1 and KDR, were significantly associated (Chi Square test, p < 0.001; Figure 2C). As shown in Figure 2C, only 4% patients expressed FLT1 at a low level and KDR at a very high level, and vice versa, but 29% of patients expressed FLT1 and KDR at a very high level, and at a low level, respectively. Our results suggest that these three factors were co-expressed at a high level in glioma specimens leading us to further investigate the prognostic significance of the co-expression of these factors.
The association between VEGFA, FLT1 and KDR mRNA expression in brain tumor patients. Histograms showing the percentage of patients whose tumor expressed different levels of (A) VEGFA and FLT1, (B) VEGFA and KDR and (C) FLT1 and KDR.
High level expression of both VEGFA and FLT1 mRNA in the same patients predicts shorter survival outcome
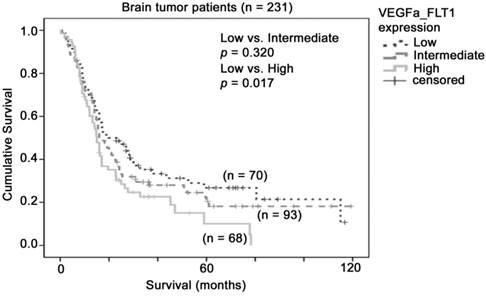
Although FLT1 alone did not predict patient survival in our combined glioma dataset, the molecular mechanism for VEGFA-FLT1 signaling led us to hypothesize that glioma patients whose tumors expressed high levels of both VEGFA and FLT1 may have the worst prognosis. To investigate this hypothesis, we stratified the patients into three groups based on the levels of expression of VEGFA and FLT1. As shown in Figure 3, patients whose tumors expressed both VEGFA and FLT1 at low levels had a mean survival of 43 months (32 - 53 months), patients whose tumors expressed one of these two genes at high level had a mean survival of 37 months (28 - 46 months), and patients whose tumors expressed both genes at high level had a mean survival of 24 months (18 - 31 months), which was significantly shorter than that of the VEGFA-FLT1-low group of patients (p = 0.017). Patients whose tumors expressed both genes at high level had a significantly higher risk of death than those whose tumors expressed both genes at low level with a hazard ratio of 1.57 (95% CI = 1.06 - 2.31, p = 0.023). Our results suggest that glioma patients whose tumors co-expressed VEGFA and FLT1 at a high level had a poorer prognosis.
High level expression of both VEGFA and KDR mRNA in the same patients predicts shorter survival outcome
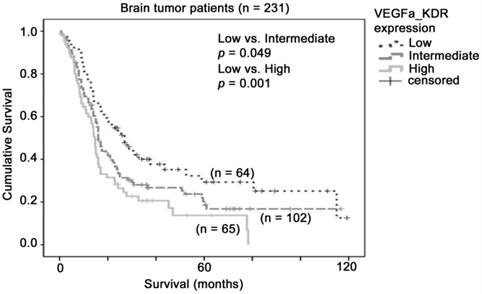
Similarly, although KDR alone did not predict patient survival in our combined glioma dataset, the co-overexpression of VEGFA and KDR may be important for activation of such signals. We stratified the patients into three groups based on the levels of expression of VEGFA and KDR. As shown in Figure 4, patients whose tumors expressed both VEGFA and KDR at low levels had a mean survival of 48 months (36 - 59 months), patients whose tumors expressed one of these two genes at high level had a mean survival of 35 months (27 - 44 months), and patients whose tumors expressed both genes at high level had a mean survival of 24 months (17 - 30 months), which was significantly shorter than the VEGFA-KDR-low group of patients (p = 0.001). Patients whose tumors expressed both genes at high level had a significantly higher risk of death than those whose tumors expressed both genes at low level with a hazard ratio of 1.94 (95% CI = 1.30 - 2.92, p = 0.001). Our results suggest that glioma patients whose tumor co-expressed VEGFA and KDR at a high level had a poorer prognosis.
High level expression of VEGFA, FLT1 and KDR mRNA in the same patients predicts shorter survival outcome
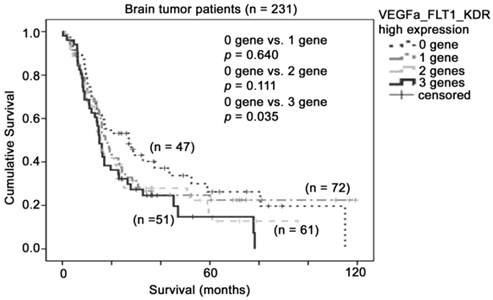
Since both FLT1 and KDR could be activated upon binding by its ligand, VEGFA, we then investigated whether the co-expression of these three factors have value in patient prognosis. Interestingly, we found that patient survival time shortened with increasing number of VEGF pathway genes overexpressed. As shown in Figure 5, patients whose tumors expressed none of these three genes at a high level had a mean survival time of 43 months (95% CI = 30 - 57 months), patients whose tumors expressed one of these three genes at a high level had a mean survival time of 39 months (95% CI = 29 - 50 months), patients whose tumors expressed two of these three genes at a high level had a mean survival time of 30 months (95% CI = 22 - 38 months), and patients whose tumors expressed all three genes at a high level had a mean survival time of 26 months (95% CI = 19 - 34 months), which was significantly shorter than group of patients who expressed all three genes to a low degree. Importantly, patients whose tumors expressed all three genes at a high level had a higher risk of death compared to those whose tumors expressed none of these three genes at a high level (Hazard Ratio = 1.56, 95% CI = 0.98 - 2.47, p = 0.06). Together, our results suggest that co-overexpression of VEGFA and its two receptors, FLT1 and KDR, in glioma biopsies is a prognostic indicator for glioma patients.
The association between combination of VEGFA-FLT1 mRNA expression and brain tumor patient survival. Kaplan-Meier analyses for VEGFA and FLT1 mRNA expression in combination in the brain tumor patient cohort
The association between combination of VEGFA-KDR mRNA expression and brain tumor patient survival. Kaplan-Meier analyses for VEGFA and KDR mRNA expression in combination in the brain tumor patient cohort
The association between combination of VEGFA-FLT1-KDR mRNA expression and brain tumor patient survival. Kaplan-Meier analyses for VEGFA, FLT1 and KDR mRNA expression in combination in the brain tumor patient cohort.
Discussion
Anti-angiogenesis has been shown to be an effective strategy in treatment of patients with glioblastoma, the most aggressive type of glioma. Previously, we have shown that the co-overexpression of VEGFA and its receptors, FLT1 and KDR, is associated with poor prognosis in both lung and colon cancers [28, 29], malignancies that benefit from anti-angiogenesis therapy. Interestingly, in the present study, we have shown that co-expression of these genes also confers prognostic significance in glioma, suggesting that VEGFA-FLT1-KDR three-gene signature may be a potential prognostic indicator for glioma patients. We previously reported that expression profiling of this three-gene signature could also predict response to bevacizumab in colon cancer patients [29]. Bevacizumab is an anti-angiogenic agent that has also been shown to be effective in glioblastoma patients; as no dataset comprising glioblastoma patients who have received bevacizumab exist in the public domain, and as there is also a lack of success in identification of biomarker for anti-angiogenic agents [30], the value of this three-gene signature for prediction of response to bevacizumab in glioblastoma patients warrants further investigation.
Tumor angiogenesis is a highly dynamic process that is controlled by the interaction between tumor and endothelial cells as well as the tumor microenvironment and is required for tumor growth, invasion, and metastasis [31]. Activation of a cell surface receptor requires the presence of its ligand, however, overexpression of the ligand alone may not be sufficient to drive sufficient signaling to promote cancer progression. Overexpression of a cell surface receptor, such as HER2, has been shown to be sufficient to promote breast cancer progression and predict tumor response to anti-HER2 therapy [32]. In addition, mutation that led to constitutive activation of a cell surface receptor, EGFR, was found to be sufficient to promote lung cancer progression and predict tumor response to EGFR tyrosine kinase inhibitors [33]. In the present report, we have demonstrated the importance of considering the co-overexpression of both the ligand, VEGFA, and its receptors, FLT1 and KDR, in prognosis of glioma patients.
Glioblastoma is a very aggressive subtype of glioma with very short life expectancy and limited treatment options. Although anti-angiogenesis has been shown to be effective in delaying tumor progression in glioblastoma patients, mixed results on other clinical endpoints in phase III trials studying bevacizumab in newly diagnosed glioblastoma patients suggest that not all patients will be benefit from anti-angiogenic therapy [18, 19]. Similarly, mixed results have also been observed in phase III trials in lung [34, 35] and colon cancers [36, 37]. Together with our previous findings in lung and colon cancer patient cohorts [28, 29], our results suggest that this three-gene signature may have clinical application in terms of predicting response towards anti-angiogenic agents in various types of cancer, and that this warrants further clinical evaluation, especially in a prospective trial setting.
Since glioblastoma patients have only limited treatment options, bevacizumab, which has been shown to relieve symptoms related to brain tumor, could be a valuable option to them. However, the clinical use with bevacizumab is still hindered by the lack of a suitable biomarker for identifying a sub-population of patients that could definitely benefit from treatment of bevacizumab. Although we have been unable to determine the anti-angiogenesis response-prediction capacity of this three-gene signature in glioblastoma patients, we have validated its use for predicting patient morbidity, and encourage others who may have access to data on glioblastoma patient response to bevacizumab to test the value of the three-gene signature for predicting response to the therapy.
Acknowledgements
We would like to thank Omic Science and Technology Ltd. for performing the initial gene analysis in this study. This study was supported by the University of Macau Faculty of Health Sciences Fund, the Start-Up Research Grant (SRG2014-00006-FHS) and the Multi-Year Research Grant (MYRG2015-00065-FHS) to Hang Fai Kwok's research group.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Khasraw M, Ameratunga MS, Grant R, Wheeler H, Pavlakis N. Antiangiogenic therapy for high-grade glioma. Cochrane Database Syst Rev. 2014;9:CD008218. doi:10.1002/14651858.CD008218.pub3
2. Ferrara N. VEGF and the quest for tumour angiogenesis factors. Nat Rev Cancer. 2002;2:795-803 doi:10.1038/nrc909
3. Takahashi S. Vascular endothelial growth factor (VEGF), VEGF receptors and their inhibitors for antiangiogenic tumor therapy. Biol Pharm Bull. 2011;34:1785-8 doi:JST.JSTAGE/bpb/34.1785 [pii]
4. Omuro AM, Faivre S, Raymond E. Lessons learned in the development of targeted therapy for malignant gliomas. Mol Cancer Ther. 2007;6:1909-19 doi:10.1158/1535-7163.MCT-07-0047
5. Chan AS, Leung SY, Wong MP, Yuen ST, Cheung N, Fan YW. et al. Expression of vascular endothelial growth factor and its receptors in the anaplastic progression of astrocytoma, oligodendroglioma, and ependymoma. Am J Surg Pathol. 1998;22:816-26
6. Im SA, Gomez-Manzano C, Fueyo J, Liu TJ, Ke LD, Kim JS. et al. Antiangiogenesis treatment for gliomas: transfer of antisense-vascular endothelial growth factor inhibits tumor growth in vivo. Cancer Res. 1999;59:895-900
7. Bao S, Wu Q, Sathornsumetee S, Hao Y, Li Z, Hjelmeland AB. et al. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 2006;66:7843-8 doi:10.1158/0008-5472.CAN-06-1010
8. Folkins C, Man S, Xu P, Shaked Y, Hicklin DJ, Kerbel RS. Anticancer therapies combining antiangiogenic and tumor cell cytotoxic effects reduce the tumor stem-like cell fraction in glioma xenograft tumors. Cancer Res. 2007;67:3560-4 doi:10.1158/0008-5472.CAN-06-4238
9. Trevisan E, Bertero L, Bosa C, Magistrello M, Pellerino A, Ruda R. et al. Antiangiogenic therapy of brain tumors: the role of bevacizumab. Neurol Sci. 2014;35:507-14 doi:10.1007/s10072-014-1627-6
10. Vredenburgh JJ, Desjardins A, Herndon JE 2nd, Dowell JM, Reardon DA, Quinn JA. et al. Phase II trial of bevacizumab and irinotecan in recurrent malignant glioma. Clin Cancer Res. 2007;13:1253-9 doi:10.1158/1078-0432.CCR-06-2309
11. Vredenburgh JJ, Desjardins A, Herndon JE 2nd, Marcello J, Reardon DA, Quinn JA. et al. Bevacizumab plus irinotecan in recurrent glioblastoma multiforme. J Clin Oncol. 2007;25:4722-9 doi:10.1200/JCO.2007.12.2440
12. Kreisl TN, Kim L, Moore K, Duic P, Royce C, Stroud I. et al. Phase II trial of single-agent bevacizumab followed by bevacizumab plus irinotecan at tumor progression in recurrent glioblastoma. J Clin Oncol. 2009;27:740-5 doi:10.1200/JCO.2008.16.3055
13. Friedman HS, Prados MD, Wen PY, Mikkelsen T, Schiff D, Abrey LE. et al. Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma. J Clin Oncol. 2009;27:4733-40 doi:10.1200/JCO.2008.19.8721
14. Cohen MH, Shen YL, Keegan P, Pazdur R. FDA drug approval summary: bevacizumab (Avastin) as treatment of recurrent glioblastoma multiforme. Oncologist. 2009;14:1131-8 doi:10.1634/theoncologist.2009-0121
15. Taal W, Oosterkamp HM, Walenkamp AM, Dubbink HJ, Beerepoot LV, Hanse MC. et al. Single-agent bevacizumab or lomustine versus a combination of bevacizumab plus lomustine in patients with recurrent glioblastoma (BELOB trial): a randomised controlled phase 2 trial. Lancet Oncol. 2014;15:943-53 doi:10.1016/S1470-2045(14)70314-6
16. Lai A, Tran A, Nghiemphu PL, Pope WB, Solis OE, Selch M. et al. Phase II study of bevacizumab plus temozolomide during and after radiation therapy for patients with newly diagnosed glioblastoma multiforme. J Clin Oncol. 2011;29:142-8 doi:10.1200/JCO.2010.30.2729
17. Vredenburgh JJ, Desjardins A, Reardon DA, Peters KB, Herndon JE 2nd, Marcello J. et al. The addition of bevacizumab to standard radiation therapy and temozolomide followed by bevacizumab, temozolomide, and irinotecan for newly diagnosed glioblastoma. Clin Cancer Res. 2011;17:4119-24 doi:10.1158/1078-0432.CCR-11-0120
18. Chinot OL, Wick W, Mason W, Henriksson R, Saran F, Nishikawa R. et al. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med. 2014;370:709-22 doi:10.1056/NEJMoa1308345
19. Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, Vogelbaum MA. et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370:699-708 doi:10.1056/NEJMoa1308573
20. Hatva E, Kaipainen A, Mentula P, Jaaskelainen J, Paetau A, Haltia M. et al. Expression of endothelial cell-specific receptor tyrosine kinases and growth factors in human brain tumors. Am J Pathol. 1995;146:368-78
21. Hlobilkova A, Ehrmann J, Knizetova P, Krejci V, Kalita O, Kolar Z. Analysis of VEGF, Flt-1, Flk-1, nestin and MMP-9 in relation to astrocytoma pathogenesis and progression. Neoplasma. 2009;56:284-90
22. Xiang F, Tanaka J, Takahashi J, Fukuda T. Expression of vascular endothelial growth factor (VEGF) and its two receptors in diffusely infiltrating astrocytomas and relationship to proliferative activity of tumor cells. Brain Tumor Pathol. 2001;18:67-71
23. Di Stefano AL, Labussiere M, Lombardi G, Eoli M, Bianchessi D, Pasqualetti F. et al. VEGFA SNP rs2010963 is associated with vascular toxicity in recurrent glioblastomas and longer response to bevacizumab. J Neurooncol. 2015;121:499-504 doi:10.1007/s11060-014-1677-x
24. Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD. et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157-73 doi:10.1016/j.ccr.2006.02.019
25. Freije WA, Castro-Vargas FE, Fang Z, Horvath S, Cloughesy T, Liau LM. et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64:6503-10 doi:10.1158/0008-5472.CAN-04-0452
26. Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF. et al. Stem cell-related "self-renewal" signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26:3015-24 doi:10.1200/JCO.2007.15.7164
27. Yuen HF, Gunasekharan VK, Chan KK, Zhang SD, Platt-Higgins A, Gately K. et al. RanGTPase: a candidate for Myc-mediated cancer progression. J Natl Cancer Inst. 2013;105:475-88 doi:10.1093/jnci/djt028
28. Zhang SD, McCrudden CM, Kwok HF. Prognostic significance of combining VEGFa, FLT1 and KDR mRNA expressions in lung cancer. Oncology Letters. 2015 In press. doi: 10.3892/ol.2015.3415
29. Zhang SD, McCrudden CM, Chen M, Yao L, Kwok HF. The significance of combining VEGFa, FLT1 and KDR expressions in colon cancer patient prognosis and predicting response to bevacizumab. OncoTargets and Therapy. 2015;8:835-843 doi: 10.2147/OTT.S80518
30. Murukesh N, Dive C, Jayson GC. Biomarkers of angiogenesis and their role in the development of VEGF inhibitors. Br J Cancer. 2010;102:8-18 doi:10.1038/sj.bjc.6605483
31. Mittal K, Ebos J, Rini B. Angiogenesis and the tumor microenvironment: vascular endothelial growth factor and beyond. Semin Oncol. 2014;41:235-51 doi:10.1053/j.seminoncol.2014.02.007
32. Ballinger TJ, Sanders ME, Abramson VG. Current HER2 Testing Recommendations and Clinical Relevance as a Predictor of Response to Targeted Therapy. Clin Breast Cancer. 2014 doi:10.1016/j.clbc.2014.11.009
33. Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M. et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786-92 doi:10.1056/NEJMoa044238
34. Sandler A, Gray R, Perry MC, Brahmer J, Schiller JH, Dowlati A. et al. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355:2542-50 doi:10.1056/NEJMoa061884
35. Reck M, von Pawel J, Zatloukal P, Ramlau R, Gorbounova V, Hirsh V. et al. Phase III trial of cisplatin plus gemcitabine with either placebo or bevacizumab as first-line therapy for nonsquamous non-small-cell lung cancer: AVAil. J Clin Oncol. 2009;27:1227-34 doi:10.1200/JCO.2007.14.5466
36. Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W. et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350:2335-42 doi:10.1056/NEJMoa032691
37. Saltz LB, Clarke S, Diaz-Rubio E, Scheithauer W, Figer A, Wong R. et al. Bevacizumab in combination with oxaliplatin-based chemotherapy as first-line therapy in metastatic colorectal cancer: a randomized phase III study. J Clin Oncol. 2008;26:2013-9 doi:10.1200/JCO.2007.14.9930
Author contact
![]() Corresponding author: Hang Fai Kwok, Room 4006, Faculty of Health Sciences (E12), University of Macau, Avenida de Universidade, Macau SAR. hfkwokmo / h.f.kwokac.uk / hfk24ac.uk
Corresponding author: Hang Fai Kwok, Room 4006, Faculty of Health Sciences (E12), University of Macau, Avenida de Universidade, Macau SAR. hfkwokmo / h.f.kwokac.uk / hfk24ac.uk

 Global reach, higher impact
Global reach, higher impact