3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2015; 6(9):893-900. doi:10.7150/jca.11865 This issue Cite
Research Paper
GOLPH3L is a Novel Prognostic Biomarker for Epithelial Ovarian Cancer
1. Department of Gynecologic Oncology, Sun Yat-sen University Cancer Center; State Key Laboratory of Oncology in South China; Collaborative Innovation Center for Cancer Medicine, Guangzhou 510080, China.
2. Department of Molecular Oncology, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL 33612, USA
3. Department of Biology, University of Illinois at Chicago, Chicago, IL 60607, USA
4. Department of Forensic Medicine, Zhongshan Medical School, Sun Yat-Sen University, Guangzhou, Guangdong 510080, P.R. China
# These authors contributed equally to this work and share first authorship.
Received 2015-2-11; Accepted 2015-6-11; Published 2015-7-27
Abstract
Objective: Golgi phosphoprotein 3 (GOLPH3) is a highly conserved membrane protein that is involved in a variety of cancers such as colorectal cancer, gastric cancer, ovarian cancer, and breast cancer. GOLPH3L is a paralog of GOLPH3. Although these proteins share a similar amino acid sequence, much less is known regarding the subcellular functions or effects of GOLPH3L on cancer compared with GOLPH3. The role of GOLPH3L in epithelial ovarian cancer (EOC) has not yet been investigated.
Methods: Using western blot, PCR and immunohistochemical analyses, we studied the clinical significance of GOLPH3L expression in EOC. The correlations between GOLPH3L expression and the clinicopathological variables of patients with EOC were assessed using Pearson's χ2 test. Kaplan-Meier analysis was used to compare the postoperative survival between groups of patients with EOC with varying levels of GOLPH3L expression.
Results: High expression of GOLPH3L was more frequently observed in EOC tissues than in corresponding adjacent non-tumor tissues. The expression of GOLPH3L correlated closely with pre-operative CA125 level (P=0.031). Univariate analysis showed that age, FIGO stage, pre-operative cancer antigen (CA) 125, pre-operative albumin concentration (AC), optimal cytoreductive surgery (CRS) and GOLPH3L expression correlate significantly with overall survival (OS). Multivariate analysis revealed that GOLPH3L expression was an independent prognostic factor for OS of patients with EOC (102 months versus 72 months; P=0.013). What's more, knocked down of GOLPH3L with small interfering RNA (siRNA) technology of OVCAR3 and SKOV3 cell lines reduced cell viability obviously, compared to the negative control and blank control groups.
Conclusions: Our data show that increased expression of GOLPH3L is associated with poor prognosis of patients with EOC and may act as a novel, useful and independent prognostic indicator. Therefore, further studies are warranted.
Keywords: GOLPH3L, epithelial ovarian cancer, prognosis
Introduction
Ovarian cancer (OC) is the most lethal gynecological malignancy because of its stealthy development. Epithelial ovarian cancer (EOC) accounts for over 90% of primary OC, and only 15% of OCs are diagnosed at an early stage [1]. Currently, the primary methods to screen for OC are bimanual pelvic examination, cancer antigen (CA) 125 detection, and transvaginal ultrasound, which have limited specificity and sensitivity [2]. Despite current advances in surgery and adjuvant chemotherapy, the clinical outcome of patients with OC remains uncertain. In 2012, approximately 2,280 new cases of OC were diagnosed in the United States, and the 5-year survival rate was approximately 44% [1]. Because the development of OC is associated with a series of genetic mutations and molecular signals, to identifying an effective molecular biomarker for early diagnosis to improve the prognosis of OC is necessary.
GOLPH3L (also referred to in the literature as GPP34R, GMx33β or FLJ10687) is a paralog of Golgi phosphoprotein 3 (GOLPH3). GOLPH3, which is located at 5p13, has been demonstrated to be a novel oncogene in many cancer types, such as colorectal cancer, gastric cancer, OC, breast cancer, colon cancer, prostate cancer and esophageal cancer [3-7]. GOLPH3 is a highly conserved membrane protein that localizes to the Golgi complex in almost all human cells. This protein moves between the Golgi apparatus and the endosomal system so that it can participate in Golgi trafficking and morphology maintenance [8]. Although this protein shares a similar amino acid sequence, much less is known regarding the subcellular functions or effects of GOLPH3L on cancers compared with GOLPH3 [9]. Only Osamu Kunigou reported that both GOLPH3 and GOLPH3L act as oncogenes in rhabdomyosarcoma [10].
This study is the first to investigate the clinical significance of GOLPH3L in EOC by immunohistochemistry (IHC). We also compared the expression of GOLPH3L in OC and in adjacent non-tumor tissues by western blot (WB) analysis. Small interfering RNA (siRNA) technology was used to assess the effect of GOLPH3L inhibition on the growth of OC cells.
Material and methods
Cell culture
The human OC cells lines OVCAR3 and SKOV3 were obtained from Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China. The cells were cultured in RPMI 1640 medium supplemented with 10% (v/v) FBS in a 37°C humidified incubator with 5% CO2.
Transient interference assay
SiRNA technology was used to silence the expression of GOLPH3L in cell lines. Approximately 2×105 cells/well were seeded in 6-well plates cells and cultured in serum-free RPMI 1640 medium to 40% confluence by the time of transfection. The siRNA duplex-Lipofectamine™ RNAiMAX (Invitrogen, Life Technologies, Carlsbad, CA, USA) complexes (a final volume of 500 μl and a final RNA concentration of 10 nM) were added to each well according to the manufacturer's recommendations. After 4 hours of incubation, the cells were cultured in RPMI 1640 medium with 10% FBS. The GOLPH3L-targeting siRNA and the negative control siRNA were designed and purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). The siRNA sequences used in this study were as follows:
GOLPH3L siRNA, forward, 5'-GAUGGAAAGUGAGGAAGACAGUAAU-3'; antisense sequences, 5'-AUUACUGUCUUCCUCACUUUCCAUC-3'.
Negative control RNA(ncRNA), forward, 5'-UUCUCCGAACGUGUCACGUTT-3'; antisense sequences, 5'-ACGUGACACGUUCGGAGAATT-3'.
All the transfections were repeated twice independently.
Cell viability
At 24 hours after transfection, the cells were seeded at 1×104 cells/well in 24-well plates. PrestoBlue Cell Viability Reagent (Invitrogen, Life Technologies, Carlsbad, CA, USA) was used according to the manufacturer's instructions to test cell viability. The fluorescence was obtained at an excitation wavelength of 560 and an emission wavelength of 590 nm using a microplate reader (SpectraMax M5, Molecular Devices, USA). Then, results are calculated and plotted. Higher fluorescence represents greater total metabolic activity.
Clinical samples
This study was approved by the medical ethics committee of Sun Yat-sen University Cancer Center. The samples in this study were anonymous and were collected from patients for routine pathology use. In total, 92 EOC tissues and adjacent non-tumor tissues were sectioned and confirmed by pathological review by IHC, including samples from 49 serous borderline ovarian tumors, 8 mucinous ovarian epithelial carcinomas, 1 seromucinous carcinoma, 4 endometrioid carcinomas, 28 adenocarcinomas, and 2 others. Pre-operative blood status (albumin concentration (AC), hemoglobin concentration (HC), CA125 levels) was determined by standard methodologies. The blood samples were obtained 2-7 days before surgery.
Reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted using TRIzol Reagent (Invitrogen, USA) according to the manufacturer's protocol. RNAse-free DNase I was used to eliminate DNA contamination. Total RNA concentration and quantity were assessed at 260 nm by measuring the absorbance of samples using a NanoDrop spectrophotometer (ND-1000, Thermo Scientific, USA); the purity of the samples was estimated by the OD ratios (A260/A280, range: 1.8-2.2) and was visualized on an agarose gel to verify the quality. Reverse transcription was performed in a 25-µl reaction volume with 2 µg of total RNA, 0.5 µl of oligo(dT), 25 U RNase inhibitor, 200 U M-MLV reverse transcriptase and 2.5 mM dNTPs to synthesize first-strand cDNA (Promega, USA) according to the manufacturer's recommendations. The reaction system was incubated at 70°C for 5 minutes and then at 42°C for 1 hour. Then, the cDNA was stored at -20°C until use. The resulting cDNA was subjected to RT-PCR to evaluate the relative expression levels of β-actin (as an internal control) and GOLPH3L. The sequences of the sense and anti-sense primers were as follows: 5'-ATGACCCTCAGCGTATGGAC-3' (F) and 5'-TTTGTCCCTTCCACTTCAGG-3' (R). Gene-specific amplification was performed using an Applied Biosystems (ABI 7900HT) RT-PCR machine that measures the binding of SYBR Green I to double-stranded DNA. Each sample was tested with a no template control for each pair of oligonucleotide primers to limit contamination and primer-dimer formation; moreover, each experiment was repeated at least twice with the cDNA samples.
The reactions were performed in a volume of 15 µl that contained 0.5 µl of cDNA, 7.5 µl of 2× SYBR Green Master Mix (Invitrogen USA), and 200 nM each pair of oligonucleotide primers. The amplification was performed as follows: an initial step at 95°C for 10 minutes, followed by 45 cycles of 95°C for 30 seconds and 60°C for 60 seconds. Regression curves were calculated for each sample, and the threshold cycles were calculated with the instrument's software (SDS 2.0). The resultant RT-PCR amplicons were analyzed by gel electrophoresis to confirm the specificity of the generated products. The relative expression levels of the target genes were normalized to the geometric mean of the internal control gene β-actin. The averaged generated data are expressed in relative units of normalized expression. The data were analyzed using the comparative threshold cycle (2-∆∆CT) method.
WB analysis
The frozen samples of OVCAR3 and SKOV3 cells after RNA interference were lysed in mammalian cell lysis buffer containing protease and phosphatase inhibitor cocktails (1:1000) for 30 minutes on ice. The raw homogenate was centrifuged (13,000×g) at 4°C for 15 minutes. Additionally, tumor and non-tumor tissue samples from patients with EOC were homogenized in RIPA lysis buffer, and the lysates were cleared by centrifugation (13,000×g) at 4°C for 15 minutes.
Approximately 40 µg of each protein sample was run on a 10% SDS-PAGE gel and then transferred to PVDF membranes. After blocking the non-specific binding sites for 60 minutes with 5% non-fat milk, the membranes were incubated overnight at 4°C with primary monoclonal antibodies directed against GOLPH3L (at a 1:1000 dilution) and β-actin (at a 1:3000 dilution). Then, the membranes were washed 3 times with Tris-Buffered Saline-Tween 20 for 10 minutes each and probed with HRP-conjugated secondary antibody (at a 1:2000 dilution) for 1 hour at room temperature. Next, the membranes were washed three times with Tris-Buffered Saline-Tween 20 and developed using an enhanced chemiluminescence system (ECL, Pierce).
GOLPH3L IHC
The tissue sections were deparaffinized with dimethylbenzene and rehydrated through 100%, 95%, 90%, 80% and 70% ethanol. After the tissue sections were washed 3 times in phosphate-buffered saline, the slides were boiled in antigen retrieval buffer containing 0.01 M sodium citrate-hydrochloric acid (pH 6) for 15 minutes in a microwave oven. Then, the slides were treated with 3% hydrogen peroxide in methanol to quench the endogenous peroxidase activity. Next, 1% bovine serum albumin was used to block nonspecific binding. Then, the sections were incubated with rabbit anti-human polyclonal antibody directed against GOLPH3L (1:1000 dilution; ab121999, Abcam, USA) at 4°C overnight in a moist chamber. Blocking solution without the primary antibody was used as a negative control. After incubation with horseradish peroxidase for 30 min at 37°C, the slides were incubated with 3,3'-diaminobenzidine solution for visualization. Negative control adjacent sections were processed as described above. However, these sections were incubated overnight at 4°C in blocking solution without the primary antibody.
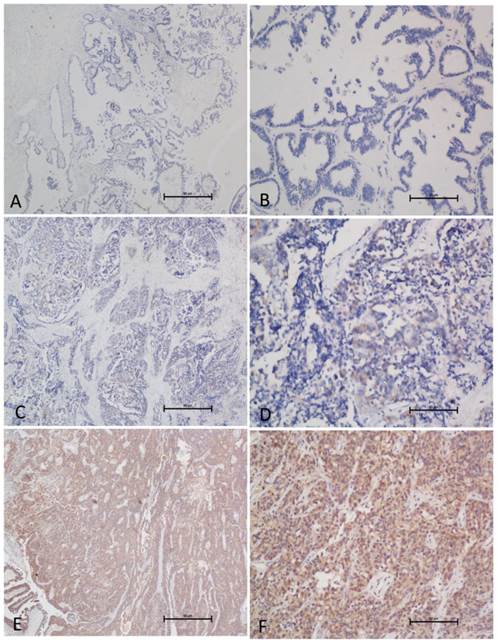
Two independent pathologists who were blinded with respect to the clinicopathological information performed the immunoreactivity scoring (IRS) for GOLPH3L expression (Fig. 3). The staining results were scored based on the following criteria: (a) percentage of positive tumor cells in the tumor tissue: zero (0%), 1 (1-10%), 2 (11-50%), 3 (51-75%), and 4 (76-100%); (b) signal intensity: zero (no signal), 1 (weak), 2 (moderate), and 3 (strong). The IRS was calculated by multiplying the score for the percentage of positive cells by the intensity score (range: 0-12). The specimens were rescored if the difference between the scores determined by the two pathologists was greater than 3. The cutoff value for low and high expression was determined by the receiver operating characteristic (ROC, MedCalc for Windows statistics) method.
Statistical analysis
Statistical analysis was performed using SPSS software (standard version 17.0, SPSS, Chicago, IL, USA). The correlation between GOLPH3L expression and the clinicopathological variables of the patients was assessed using Pearson's χ2 test. Overall survival (OS) was assessed using the Kaplan-Meier method and compared by the log-rank test. The Cox regression model was employed for multivariate survival analysis. A two-sided probability value of less than 0.05 was considered statistically significant.
Results
Cell viability
The viability of OVCAR3 cells was detected by PrestoBlue Cell Viability Reagent, which was used to compare GOLPH3L siRNA-transfected cells, ncRNA-transfected cells and untreated controls. As shown in Fig. 1, GOLPH3L siRNA-transfected OVCAR3 cells demonstrated reduced cell viability compared with the ncRNA-transfected OVCAR3 cells and the blank control. The same result was observed in SKOV3 cells.
Cell viability was detected by PrestoBlue Cell Viability Reagent. siRNA-transfected OVCAR3 cells (A) demonstrated reduced cell viability compared with the ncRNA OVCAR3 and the blank control. A similar result was observed in SKOV3 cells (B). Solid lines represent GOLPH3L siRNA groups. Dashed lines represent the blank control groups. Dashed dotted lines represent the ncRNA groups.
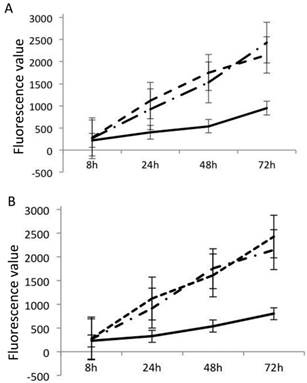
GOLPH3L up-regulation was detected in EOC tissues
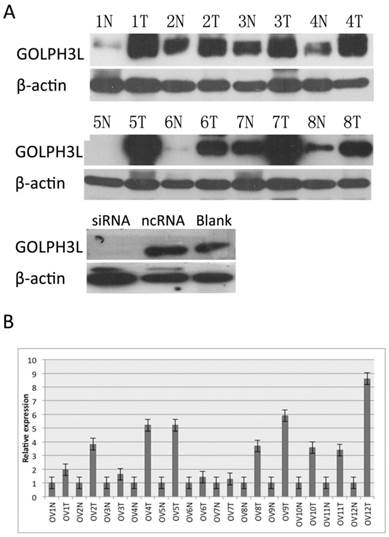
The protein level of GOLPH3L was determined in 12 pairs of resected specimens (tumor and matched adjacent non-tumor tissue samples) from patients with OC by RT-PCR and WB analyses. The results showed that GOLPH3L protein levels were significantly increased in tumor tissues compared with the levels in the adjacent non-tumor tissues. In addition, when GOLPH3L was knocked down by siRNA, the GOLPH3L protein levels decreased significantly compared with the levels of GOLPH3L in SKOV3 cells that were treated with ncRNA and with the blank control.
Patient characteristics
After exclusion of the noninformative samples (i.e., unrepresentative samples, samples with too few tumor cells, and lost samples), 92 samples were included in this study. GOLPH3L was highly expressed in 47 patients (51.8%) aged from 25 to 76 years (median 52.0 years). The distribution of International Federation of Gynecology and Obstetrics (FIGO) stages was 25 patients with stage I (27.2%), 19 patients with stage II (20.7%), 43 patients with stage III (46.7%) and 5 patients with stage IV (5.4%). Table 1 summarizes the characteristics of this population.
GOLPH3L IRS cutoff value
The GOLPH3L IRS cutoff value was defined by the ROC curve. The value closest to the point with both maximum sensitivity and specificity (i.e., the point [0.0, 0.1] on the curve) was selected as the cutoff value. The result showed that the cutoff value was 6; thus, expression greater than 6 was considered high, while expression less than 6 was considered low.
Correlation between GOLPH3L expression and the clinicopathological variables of 92 OC cases
| Variables | Cases (n=92) | GOLPH3L expression | P value | |||
|---|---|---|---|---|---|---|
| Low (n=45) | High (n=47) | |||||
| Age (years) | 0.535 | |||||
| ≤52 | 48 | 25 | 23 | |||
| >52 | 44 | 20 | 24 | |||
| FIGO stage | 0.514 | |||||
| stage I | 25 | 12 | 13 | |||
| stage II | 19 | 11 | 8 | |||
| stage III | 43 | 21 | 22 | |||
| stage IV | 5 | 1 | 4 | |||
| Pre-operative CA125 | 0.031 | |||||
| ≤967 | 51 | 30 | 21 | |||
| >967 | 39 | 14 | 25 | |||
| Pre-operative AC | 0.626 | |||||
| ≤35 | 5 | 3 | 2 | |||
| >35 | 82 | 40 | 42 | |||
| Pre-operative HC | 0.489 | |||||
| ≤110 | 21 | 9 | 12 | |||
| >110 | 66 | 34 | 22 | |||
| Optimal CRS | 0.462 | |||||
| Optimal | 48 | 25 | 23 | |||
| Not optimal | 19 | 8 | 11 | |||
* P<0.05, statistically significant.
Optimal CRS is the mean from patients with FIGO stage II and above.
AC: albumin concentration; HC: hemoglobin concentration.
GOLPH3L expression in EOC tissues
The relationships between the clinicopathological features of the patients and the expression of GOLPH3L are summarized in Table 1. The expression of GOLPH3L correlated closely with the pre-operative CA125 level (P=0.031). No statistical correlations were observed between GOLPH3L expression and age, FIGO stage, pre-operative AC, pre-operative HC, or optimal cytoreductive surgery (CRS, P=0.535, P=0.514, P=0.626, P=0.489, and P=0.462, respectively).
GOLPH3L expression and survival
Among the 92 patients with EOC, no patients were lost to follow-up. In total, 35 patients were deceased and 57 were alive at the end of the follow-up period. The 5-year OS for the entire cohort was 58.6%, and the median survival time was 65.2 months.
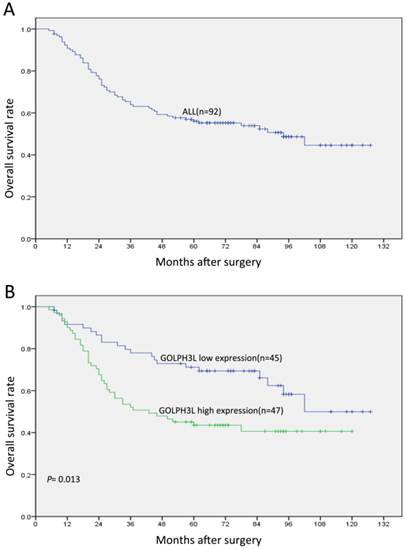
Patients with low GOLPH3L expression demonstrated longer OS compared with those with high GOLPH3L expression (mean, 102 months versus 72 months; P=0.013; Fig. 2, Table 2). The 5-year OS for patients with low GOLPH3L expression was 79.9%, while that for patients with high expression was 51.1%.
Kaplan-Meier survival analysis (log-rank test) according to GOLPH3L expression in patients with OC
| Variable | Case (n=92) | OS | |||
|---|---|---|---|---|---|
| Mean | Median | P value | |||
| Age | 0.001* | ||||
| ≤52 | 48 | 103.34 | NA | ||
| >52 | 44 | 68.36 | 57 | ||
| FIGO stage | <0.001* | ||||
| stage I | 25 | 116.16 | NA | ||
| stage II | 19 | 100.47 | NA | ||
| stage III | 43 | 62.11 | 44 | ||
| stage IV | 5 | 51.40 | 33 | ||
| Pre-operative CA125 | 0.002* | ||||
| ≤967 | 51 | 98.26 | NA | ||
| >967 | 39 | 74.63 | 78 | ||
| Pre-operative AC | <0.001* | ||||
| ≤35 | 5 | 24.40 | 20 | ||
| >35 | 82 | 92.08 | NA | ||
| Pre-operative HC | 0.643 | ||||
| ≤110 | 21 | 92.71 | NA | ||
| >110 | 66 | 85.28 | NA | ||
| Optimal CRS | <0.001* | ||||
| Optimal | 48 | 89.95 | NA | ||
| Not optimal | 19 | 34.26 | 26 | ||
| GOLPH3L expression | 0.013* | ||||
| Low | 45 | 101.84 | NA | ||
| High | 47 | 71.47 | 78 | ||
* P<0.05, statistically significant.
AC: albumin concentration; HC: hemoglobin concentration.
WB and RT-PCR analyses of the expression pattern of GOLPH3L in EOC tissues and cells. GOLPH3L protein (A) levels were significantly increased in tumor tissues of patients with EOC compared with levels in the adjacent non-tumor tissues. A similar result was detected with respect to the mRNA (B) levels. The GOLPH3L protein (A) level in OC cells decreased after RNA interference. For convenience, tumor is abbreviated to T and normal is abbreviated to N. As a loading control, β-actin level is detected to ensure that equal amounts of protein is loaded in each lane.
A univariate analysis using Cox's proportional hazard model showed that the following parameters correlated significantly with OS: age, FIGO stage, pre-operative CA125 level, pre-operative AC, optimal CRS and GOLPH3L expression (Table 2). When the above parameters were included in a multivariate analysis, the results suggested that age, optimal CRS and GOLPH3L expression were independent factors that affected OS (Table 3).
Cox multivariate analysis of the clinicopathological parameters for OS
| Variables | Hazard ratio | 95% confidence interval | P value |
|---|---|---|---|
| Age | 3.166 | 1.32~7.56 | 0.01* |
| FIGO stage | 0.290 | 0.72~3.02 | 0.290 |
| Pre-operative CA125 | 0.770 | 0.34~1.75 | 0.533 |
| Pre-operative AC | 0.477 | 0.14~1.59 | 0.227 |
| Optimal CRS | 0.398 | 0.16~0.99 | 0.047* |
| GOLPH3L expression | 2.105 | 1.07~4.67 | 0.033* |
* P<0.05, statistically significant.
AC: albumin concentration.
GOLPH3L expression as determined by IHC. (A, B) Normal ovarian tissue demonstrated no or low expression of GOLPH3L protein in the cytoplasm of ovarian cells (magnification: A, ×40; B, ×200). (C, D) An EOC case that demonstrates a low expression level of GOLPH3L (magnification: C, ×40; D, ×200). (E, F) High expression levels of GOLPH3L were detected in EOC tissues (magnification: E, ×40; F, ×200).
Discussion
The GOLPH3 family is an emerging area of cancer research. Recently, many researchers have reported the involvement of GOLPH3 in the maintenance of Golgi morphology and trans-Golgi transportation. In addition, GOLPH3 has been shown to function as an oncogene in multiple types of cancer via the regulation of mTOR molecular signals [11]. GOLPH3 up-regulation in cancers where patients have a decreased OS has attracted the interest of scientists; therefore, further studies are required to verify its potential as a molecular anti-cancer drug target [12]. However, compared with GOLPH3, relatively little attention has been given to its isoform GOLPH3L. Neither its physical functions nor its involvement in cancer has been accurately characterized.
This study is the first to investigate GOLPH3L expression in a large number of EOC samples. By IHC, we found that high GOLPH3L expression in EOC tissues was more frequently observed than in corresponding adjacent non-malignant tissues. We also used WB analysis to confirm the result of IHC and obtained a similar conclusion. In addition, a further analysis demonstrated that GOLPH3L overexpression loosely correlated with the clinicopathological parameter of CA125 concentration. Furthermore, the cumulative 5-year OS rates of patients with high GOLPH3L expression were lower than were those with low GOLPH3L expression. Thus, patients with high GOLPH3L expression have a poorer prognosis than those with low or absent GOLPH3L expression. Kaplan-Meier analysis and Cox multivariate analysis revealed that GOLPH3L expression is an independent prognostic factor for EOC. Osamu Kunigou compared the expression of GOLPH3 and GOLPH3L in rhabdomyosarcoma and came to the similar conclusion that both GOLPH3 and GOLPH3L are oncogenes [10]. However, the study by Michelle M. Ng showed that GOLPH3L antagonizes GOLPH3 in the determination of Golgi morphology [13]. This contradiction could be explained by two observations. First, the experiments in the study by Michelle M. Ng were performed in only the HEK293 cell line. Rhabdomyosarcoma and OC have completely different characteristics. Specifically, EOC is a heterogeneous disease that contains several histological subtypes, including serous, mucinous, endometrioid, clear cell, transitional cell, mixed, and undifferentiated. Second, the author focused on the effects of GOLPH3L on the regulation of Golgi morphology. The author mentioned that the tissues that most highly express GOLPH3L are those that are highly secretory and speculated as to the existence of some unknown downstream pathways of GOLPH3L. Moreover, we used siRNA to interfere with GOLPH3L expression temporarily and detected a difference in proliferation between SKOV3 and OVCAR3 cells. Compared with the cells that were treated with ncRNA or with the blank control, the growth of the cells that were treated with GOLPH3L siRNA was significantly reduced. As mentioned in existing studies, scientists found that both of GOLPH3 and GOLPH3L protein can affect the process of cell secretion. It is possible that elevated expression of GOLPH3L protein can promote the autocrine signaling loop of some growth- promoting cytokines among cancer cells and thereby promotes cell growth. Because our experiments are mainly limited in clinical indications, we are not eligible to say too much about its underlying mechanism. Therefore, additional evidence and studies are required to clarify the effects of GOLPH3L on cancer development.
The poor diagnostic methods of OC can be attributed primarily to several factors, the first of which is the histopathological complexity of EOC itself. As above-mentioned, EOC is a heterogeneous disease that comprises several histological subtypes [14]. These types of differences in cell origin and differentiation have led to great difficulties in understanding EOC. Second, medical imaging technology is not sufficiently effective. Currently, transvaginal ultrasound is considered the most effective noninvasive diagnostic method for high-risk women [15, 16]. However, currently, no recommended OC screening is available for average-risk women. Finally, too few effective biomarkers are available for OC diagnosis. CA125 is one of the most widely used parameters in clinical practice [17]. However, CA125 may also be elevated in women with benign ovarian disease or in healthy women, which limits its specificity [18]. Thus, the discovery of effective novel biomarkers that may assist in the diagnosis of OC and that may help to predict patient survival is of great value. We sincerely hope that our study leads to the establishment of a comprehensive network of OC diagnostic and treatment evaluation systems. However, we are aware that the present study also has some limitations. The samples were obtained from a single institution, and the sample numbers were insufficient for subgroup analyses.
In conclusion, our findings demonstrate that patients with EOC with high GOLPH3L expression have worse OS compared with those with low GOLPH3L expression. GOLPH3L may be involved in the proliferation of OC cells, and GOLPH3L expression detected by IHC can serve as an independent predictor of OC in patients with EOC. Therefore, further studies are required to reveal the role of GOLPH3L in OC.
OS curves for patients with EOC according to their GOLPH3L expression status. (A) OS curves: all patients with levels of GOLPH3L expression. (B) OS curves: patients with low and high GOLPH3L expression levels.
Conflicts of interest
The authors declare that no conflicts of interest exist.
References
1. Siegel R, Naishadham D, Jemal A. Cancer statistics for Hispanics/Latinos, 2012. CA: a cancer journal for clinicians. 2012;62:283-98
2. Jelovac D, Armstrong DK. Recent progress in the diagnosis and treatment of ovarian cancer. CA: a cancer journal for clinicians. 2011;61:183-203
3. Wang JH, Chen XT, Wen ZS, Zheng M, Deng JM, Wang MZ. et al. High expression of GOLPH3 in esophageal squamous cell carcinoma correlates with poor prognosis. PloS one. 2012;7:e45622
4. Hu BS, Hu H, Zhu CY, Gu YL, Li JP. Overexpression of GOLPH3 is associated with poor clinical outcome in gastric cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2013;34:515-20
5. Ma Y, Ren Y, Zhang X, Lin L, Liu Y, Rong F. et al. High GOLPH3 expression is associated with a more aggressive behavior of epithelial ovarian carcinoma. Virchows Archiv: an international journal of pathology. 2014;464:443-52
6. Zeng Z, Lin H, Zhao X, Liu G, Wang X, Xu R. et al. Overexpression of GOLPH3 promotes proliferation and tumorigenicity in breast cancer via suppression of the FOXO1 transcription factor. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:4059-69
7. Wang Z, Jiang B, Chen L, Di J, Cui M, Liu M. et al. GOLPH3 predicts survival of colorectal cancer patients treated with 5-fluorouracil-based adjuvant chemotherapy. Journal of translational medicine. 2014;12:15
8. Scott KL, Chin L. Signaling from the Golgi: mechanisms and models for Golgi phosphoprotein 3-mediated oncogenesis. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16:2229-34
9. Bell AW, Ward MA, Blackstock WP, Freeman HN, Choudhary JS, Lewis AP. et al. Proteomics characterization of abundant Golgi membrane proteins. The Journal of biological chemistry. 2001;276:5152-65
10. Kunigou O, Nagao H, Kawabata N, Ishidou Y, Nagano S, Maeda S. et al. Role of GOLPH3 and GOLPH3L in the proliferation of human rhabdomyosarcoma. Oncology reports. 2011;26:1337-42
11. Abraham RT. GOLPH3 links the Golgi network to mTOR signaling and human cancer. Pigment cell & melanoma research. 2009;22:378-9
12. Ali MF, Chachadi VB, Petrosyan A, Cheng PW. Golgi phosphoprotein 3 determines cell binding properties under dynamic flow by controlling Golgi localization of core 2 N-acetylglucosaminyltransferase 1. The Journal of biological chemistry. 2012;287:39564-77
13. Ng MM, Dippold HC, Buschman MD, Noakes CJ, Field SJ. GOLPH3L antagonizes GOLPH3 to determine Golgi morphology. Molecular biology of the cell. 2013;24:796-808
14. Silverberg SG. Histopathologic grading of ovarian carcinoma: a review and proposal. International journal of gynecological pathology: official journal of the International Society of Gynecological Pathologists. 2000;19:7-15
15. Fishman DA, Cohen L, Blank SV, Shulman L, Singh D, Bozorgi K. et al. The role of ultrasound evaluation in the detection of early-stage epithelial ovarian cancer. American journal of obstetrics and gynecology. 2005;192:1214-21 discussion 21-2
16. Menon U, Gentry-Maharaj A, Hallett R, Ryan A, Burnell M, Sharma A. et al. Sensitivity and specificity of multimodal and ultrasound screening for ovarian cancer, and stage distribution of detected cancers: results of the prevalence screen of the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS). The lancet oncology. 2009;10:327-40
17. Cannistra SA. Cancer of the ovary. New Engl J Med. 2004;351:2519-29
18. Bast RC, Klug TL, Stjohn E, Jenison E, Niloff JM, Lazarus H. et al. A Radioimmunoassay Using a Monoclonal-Antibody to Monitor the Course of Epithelial Ovarian-Cancer. New Engl J Med. 1983;309:883-7
Author contact
![]() Corresponding author: Jihong Liu, Department of Gynecologic Oncology, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou 510060, P.R. China. Email: liujhorg.cn; Tel: +86- 20-87343105.
Corresponding author: Jihong Liu, Department of Gynecologic Oncology, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou 510060, P.R. China. Email: liujhorg.cn; Tel: +86- 20-87343105.

 Global reach, higher impact
Global reach, higher impact