3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2015; 6(10):953-961. doi:10.7150/jca.12067 This issue Cite
Research Paper
Loss of IRF8 Inhibits the Growth of Diffuse Large B-cell Lymphoma
1. Department of Pathology, Ningbo University School of Medicine, Ningbo, Zhejiang, China
2. The Pathology Service Center, Ningbo, Zhejiang, China
3. The National Institute of Allergy and Infectious Diseases, the National Institutes of Health, Rockville, MD, USA
4. Department of Hematology, Ningbo University Affiliated First Hospital, Ningbo, Zhejiang, China
* These authors contributed equally to this work.
Received 2015-3-8; Accepted 2015-6-12; Published 2015-8-7
Abstract
IRF8 is a transcription factor with a critical role in B lymphocyte development and functions. Its role in human diffuse large B-cell lymphoma (DLBCL), however, remained elusive. In this study, using shRNA-mediated knockdown of IRF8 expression, we found that the loss of IRF8 significantly reduced the proliferation of DLBCL cells (P<0.05). Mechanistically, decreasing the levels of IRF8 led to a suppression of the phosphorylation of p38 and ERK, molecules critical for B cell proliferation. Furthermore, using a xenograft lymphoma mouse model, we found that the loss of IRF8 significantly inhibited the growth of lymphomas in vivo (P<0.05). Immunohistochemical analysis of human DLBCL tissues revealed that the levels of IRF8 were significantly greater in non-germinal center B-cell-like (non-GCB) subtype than that in GCB subtype (P<0.05). Analysis of public available data also suggested that the expression levels of IRF8 mRNA in human DLBCL tissues were inversely correlated with patients' overall survival time. Taken together, this study suggested that IRF8 may play an oncogenic role in human DLBCL by promoting cell proliferation.
Keywords: lymphoma, IRF8, DLBCL
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common type of aggressive non-Hodgkin lymphoma in adults. DLBCL is a highly heterogeneous malignancy with respect to their morphology, pathology, clinical manifestation, and prognosis [1-4]. Gene expression profiling revealed the existence of two major molecular subtypes of DLBCL, namely germinal center B-cell-like (GCB) and activated B-cell-like (ABC) subtypes [5]. These two subgroups of DLBCL have distinct responsiveness to current standard chemotherapy regimen (cyclophosphamide, doxorubicin, vincristine, and prednisone), and have significant different clinical outcomes [6]. While for GCB-subtype DLBCL the 5-year survival rate could reach 59%, for ABC-subtype DLBCL it is usually about 30% [7]. This suggests that the different molecular makeups of DLBCL attributes to the heterogeneity in tumor progression and clinical outcome of this malignance. It is clinically important to further understand the molecular pathology of DLBCL in order for more efficiently managing this disease.
IFN regulatory factor 8 (IRF8), a member of the IFN regulatory factor family, is a transcription factor playing critical roles in hematopoietic cell development, survival, and functions [8]. IRF8 was essential for the development of myeloid lineage cells including basophiles, mast cells, and macrophages [9]. Mice deficient in Irf8 exhibited abnormality in granulocytes and developed a disorder similar to human chronic myelogenous leukemia (CML) [10]. In addition, studies found that IRF8 played a critical role in the development of dendritic cells [11, 12]. For lymphoid lineage, IRF8 was critical in controlling early stage B cell development as well as marginal zone and follicular B-cell development [13, 14]. IRF8 positively regulated BCL6, a transcription factor critical for the development and function of germinal center B cells, through direct and indirect actions [15]. Mice deficient in Irf8 had impaired germinal center (GC) formation [16]. Irf8 was also found to directly up-regulate Mdm2 transcription while suppressing the levels of p21 and p53, thus helping GC B cells tolerate double-strand DNA breaks occurring physiologically during somatic hypermutations and Ig class switch that otherwise would induce apoptosis [17]. In B-cell lymphomas, IRF8 was highly expressed in those of GC origin with lower levels present in mantle cell lymphomas, chronic lymphocytic leukemia, and marginal zone lymphomas [18]. Bouamar et al. recently reported the occurrence of genes fusion between IRF8 and IGH genes in DLBCL that led to the overexpression of IRF8 in tumor tissues [19]. Nevertheless, the role of IRF8 in DLBCL remained elusive.
In a preliminary analysis of publicly available datasets, we found that the levels of IRF8 mRNA were inversely correlated with the overall survival time of patients with DLBCL. In the present study, we aimed to investigate the effect of IRF8 on DLBCL, and found that decreased levels of IRF8 inhibited DLBCL cell proliferation in vitro and suppressed tumor growth in vivo.
Materials and Methods
Cell Culture
DLBCL cells OCI-Ly01 and OCI-Ly10 were purchased from the ATCC (Shanghai, China) and cultured in RPMI-1640 medium (Hyclone, UT, USA) containing 10% fetal bovine serum (FBS) (Ausgenex, Australia), 4 mM L-glutamine (Sigma-Aldrich, MO, USA), and 100 units/mL of penicillin (Hyclone) and 100 units/mL of streptomycin (Hyclone). HEK-293T cells (a gift from Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) were cultured in Dulbecco modified Eagle media (DMEM) (Hyclone) supplemented with 10% FBS. All cell culture were carried out in a humidified chamber at 37°C with an atmosphere of 5% CO2.
Knockdown of IRF8 expression in DLBCL cells
The knockdown of IRF8 expression in DLBCL cells was carried out using human IRF8-specific small hairpin RNAs (shRNA) mediated by lentiviral infection. The IRF8-specific shRNA and scrambled control sequences shRNA were designed using the RNAi Consortium tool (http://www.broadinstitute.org/rnai/trc). The sequences for IRF8 shRNAs were GCATGTATCCAGGACTGATTT (shRNA-1) and GCCTTCTGTGGACGATTACAT (shRNA-2). The sequence for scramble control was CCTAAGGTTAAGTCGCCCTCG. Double-stranded oligonucleotides representing the complementary sequences separated by a hairpin loop were cloned into pLKO.1puro plasmids. The plasmids were transfected into 293T cells together with packaging plasmid pCMV-dR8.2 dvpr and envelope plasmid pCMV-VSVG. The supernatant containing lentiviral particles was harvested, filtered through a 0.45 μm-diameter filter, and used to infect DLBCL cells. Subsequently, cells were undergone for selection for positive clones using puromycin. The levels of IRF8 mRNA and proteins were evaluated using real-time PCR and western blotting, respectively.
RNA preparation, reverse transcription, and real-time PCR
Total RNA was isolated from cells using Trizol reagent (Invitrogen, Shanghai, China) according to the manufacturer's instructions. RNA was reverse-transcribed into cDNA using Thermo Scientific RevertAid First Strand cDNA Synthesis Kit (Thermo, NY, USA) according to the manufacturer's protocol. Quantitative real-time PCR (qRT-PCR) was performed using SYBR Green PCR Master Mix (Roche, Shanghai, China) on a LightCycler 480II system (Roche). The levels of IRF8 expression were normalized to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The primers (5'-3') used for qRT-PCR included IRF8 forward TTCCGAGCCATACAAAGTTTACC and reverse CGACCGCACTCCATCTCTG, GAPDH forward GCGACACCCACTCCTCCACCTTT and reverse TGCTGTAGCCAAATTCGTTGTCATA.
Western blotting analysis
Cells were lysed using sodium dodecyl sulfate (SDS) buffer containing proteinase inhibitors (Roche). Equal amounts of proteins (50 μg) were separated using 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred onto PVDF membranes (Bio-Rad, CA, USA). The membrane was blocked and incubated with specific antibodies overnight at 4°C. Antibodies against GAPDH, p38, phospho-p38 (Thr180/182), ERK, and phospho-ERK (Thr202/204) were purchased from Cell Signaling Technology, Inc. (CA, USA), and antibody against IRF8 (C-19) was purchased from Santa Cruz Biotechnology (CA, USA). Then, membranes were incubated with horseradish peroxidase-labeled secondary antibody (Beyotime Institute of Biotechnology, Jiangsu, China). The protein bands were visualized using enhanced chemiluminescence reagent.
Immunohistochemical analysis
Immunohistochemical (IHC) analysis was performed on formalin-fixed paraffin-embedded tissue sections (5-μm thickness) of DLBCL tumors. The pathological diagnoses of all tumors were confirmed by a hematological pathologist (JF). All cases were immuno-phenotyped and categorized into GCB or non-GCB subtypes based on the Visco-Young algorithm [20]. The monoclonal antibodies used for IHC included CD10 (2A1H5E1, dilution 1:100) (Santa Cruz Biotechnology, CA, USA), FOXP1 (ab32010, dilution 1:1000) (Abcam, Cambridge, UK), BCL6 (D-8, dilution 1:100) (Santa Cruz Biotechnology), and IRF8 (E-9, dilution 1:1000) (Santa Cruz Biotechnology). IHC was performed as described previously [17]. Briefly, the sections were dewaxed in xylene and rehydrated at graded alcohol. For antigen retrieval, sections were pretreated for 15 min in a microwave oven in citrate buffer solution (pH 6.1), followed by incubation in hydrogen peroxide to quench the endogenous peroxidase activity. Subsequently, sections were incubated with primary antibodies, washed, and incubated sequentially with biotin-labeled secondary antibodies and streptavidin-conjugated peroxidase. Proteins were visualized using the DAB IHC Detection Kit (ZSGB-BIO, Beijing, China), and images were acquired using an OLYMPUS microscope. The immuno-reactivity score for IRF8 was based on the proportion of positive cells and the intensity of staining. The number of positive-staining cells in five random microscopic fields was counted and the percentage of positive cells was calculated. The positivity of staining was ranked as 0, 1, 2, 3, and 4 for no positive cells, <25% of positive cells, 25 - 50% of positive cells, 50 - 75% of positive cells, and >75% of positive cells, respectively. The intensity of staining was ranked as 0, 1, 2, and 3 for no staining, weak staining, moderate staining, and strong staining, respectively. A final score was calculated by multiplying the positivity and the intensity scores for each tumor sample. The levels of IRF8 protein in each sample were further classified as low (final score < median level) or high (final score ≥ median level).
Cell proliferation assay
Cell proliferation was examined using CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega, WI, USA). Briefly, OCI-Ly01 or OCI-Ly10 cells were plated in a 96-well tissue culture plate. Following incubation for 0, 24, 48, 72, and 96 h, CellTiter 96® AQueous One Solution was added to each well and the cells were incubated for another 4 h at 37˚C. The absorbance at 490 nm was measured using a spectrophotometer.
Cell division and cell cycle analyses
Cell division was analyzed by labeling cells with carboxyfluorescein diacetate succinimidyl ester (CFSE) (Sigma-Aldrich) as described [21]. Briefly, cells were incubated with CFSE for 10 min at 37 °C in the dark, washed, and continued incubation for additional four days. Cell division was examined at day 0, 1, 2, 3, and 4 using flow cytometry. For cell cycle analysis, cells were washed with PBS and fixed in ethanol for 30 min at 4 °C, and incubated with propidium iodide (PI) and RNase (Sigma-Aldrich) for 30 min at 37 °C in the dark and subjected FACS analysis.
Xenograft mouse model
Five-week-old male nude mice (BALB/C) (Shanghai laboratory animal center, China) were used for xenograft transplantation of DLBCL with a protocol approved by the Institutional Animal Ethics Committee of Ningbo University. OCI-Ly01 cells with or without shRNA-mediated IRF8 knockdown were injected subcutaneously into the right flank of mouse (n=5 for each group). The size of tumors was measured every two days, up to 30 days following tumor cells injection (animal weight losses <15%). Tumor volume (V) was calculated according to the formula V=L × W2/2 (L represents length and W represents width). Every effort was made to minimize animal suffering. Thirty days after tumor cells inoculation, the mice were sacrificed, and the tumors were weighted, and lysed for protein and mRNA preparations.
Statistical Analysis
Data were expressed as mean ± standard deviation, and were analyzed for the statistical significances using Student's t test or Fisher's exact test as appropriate. A P value of less than 0.05 was deemed to be statistically significant.
Results
Reducing levels of IRF8 decreased the proliferation of DLBCL cells in vitro
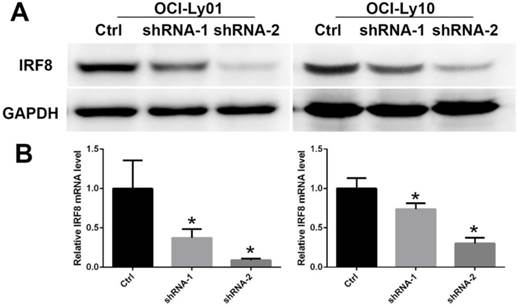
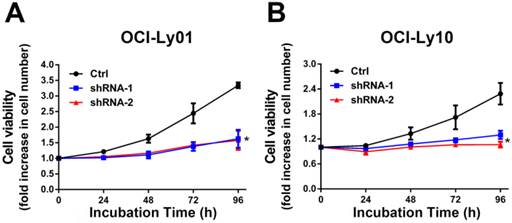
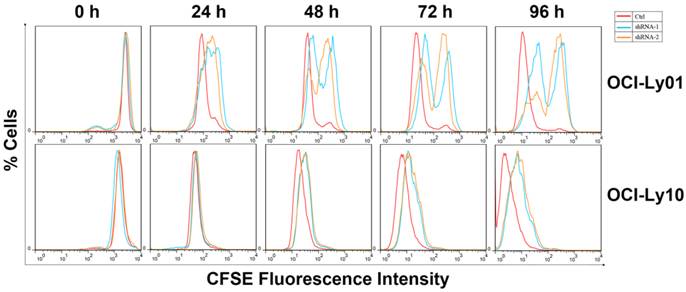
To detect the function of IRF8 in DLBCL, we used lentiviral-mediated shRNA to knock down the levels of IRF8 in DLBCL cells. Two shRNA constructs (shRNA-1 and shRNA-2) were chosen based on their effectiveness in decreasing the levels of IRF8. The results showed that both shRNA constructs decreased the levels of IRF8 mRNA and protein significantly in OCI-Ly01 and OCI-Ly10 cells (P<0.05) (Figure 1). Subsequently, we examined the effect of decreasing levels of IRF8 on DLBCL cells proliferation. Comparing to the control cells, the decrease in levels of IRF8 significantly suppressed the proliferation of DLBCL cells (Figure 2). We further asked if cell cycle arrest attributed to the suppression of proliferation. To our surprise, loss of IRF8 did not change cell cycle distribution (Figure S1). To explore the possible mechanisms responsible for the suppression of cell proliferation induced by IRF8 knockdown, we used fluorescent dye CFSE to label the cells and analyzed the rate of cell division. The fluorescence intensity was partitioned equally among daughter cells during each division. The results showed that the cell division was reduced in OCI-Ly01 and OCI-Ly10 cells with IRF8 knockdown comparing to the control cells (Figure 3).
The phosphorylation of p38 and ERK MAPK was inhibited in DLBCL cells with IRF8 knockdown
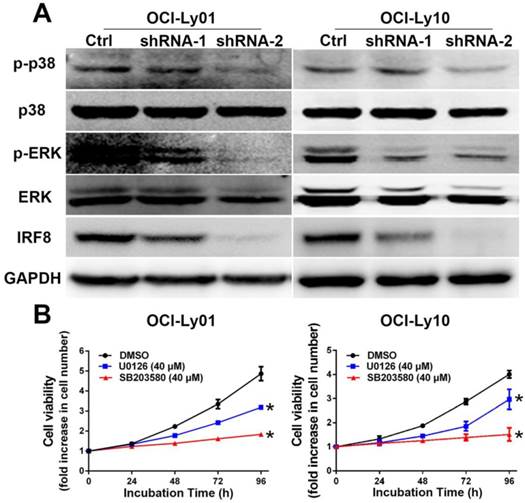
To understand the mechanisms underlying the effect of IRF8 on cell proliferation, we examined the signaling pathways critical to B cell proliferation. The activation of MAPK pathway has been well characterized as essential for B cell proliferation [22-24]. For both OCI-Ly01 and OCI-Ly10 cells, knockdown of IRF8 expression decreased the phosphorylation of p38 and ERK, two key molecules in the MAPK pathway (Figure 4A). To confirm that the inhibition of p38 and ERK was able to suppress the cell proliferation, we treated DLBCL cells with U0126 and SB203580 that were the inhibitors of p38 and ERK, respectively. The results showed that both inhibitors suppressed the proliferation of OCI-Ly01 and OCI-Ly10 cells significantly (P<0.05) (Figure 4B).
The decreased levels of IRF8 in DLBCL cells treated with shRNAs specific for IRF8. Two shRNA clones (shRNA-1 and shRNA-2) were used in the study based on their efficiencies of knocking down the levels of IRF8 in OCI-Ly01 and OCI-Ly10 cells. The levels of both IRF8 proteins (A) and transcripts (B) decreased in specific shRNA-treated DLBCL cells. * depicts statistically significant difference between the IRF8 knockdown cells and control cells in transcripts (P<0.05).
IRF8 knockdown inhibited the proliferation of DLBCL cells. The cell proliferation rate was measured in DLBCL cells with or without IRF8 knockdown at various incubation time points. Both shRNA clones significantly inhibited the proliferation of OCI-Ly01 and OCI-Ly10 cells at 96 h of incubation. Data shown are from four independent experiments (mean ± SD). * depicts statistically significant difference between the IRF8 knockdown cells and control cells after 96 h of incubation (P<0.05).
Cell division was inhibited in DLBCL cells with IRF8 knockdown. OCI-Ly01 and OCI-Ly10 cells were labeled with CFSE and cultured for various lengths of time. The fluorescence intensity of cells decreased by half during successive cell division. Cells with IRF8 knockdown had increase in fluorescence intensity at a much slower rate than cells without IRF8 knockdown did.
The phosphorylation of p38 and ERK MAPK was reduced in DLBCL cells with IRF8 knockdown. A: Western blotting analyses of proteins from OCI-Ly01 and OCI-Ly10 cells with or without IRF8 knockdown using specific antibodies for IRF8, p38, phospho-p38 (p-p38), ERK, phospho-ERK (p-ERK), and GAPDH. GAPDH was used as a loading control. B: The cell proliferation was attenuated in OCI-Ly01 and OCI-Ly10 cells treated with p38 inhibitors SB203580 (40 μM) or ERK inhibitor U0126 (40 μM). Data shown are from four independent experiments (mean ± SD). * depicts statistically significant difference between the cells treated by inhibitors and control cells after 96 h of incubation (P<0.05).
Loss of IRF8 suppressed the growth of B-cell lymphoma in vivo
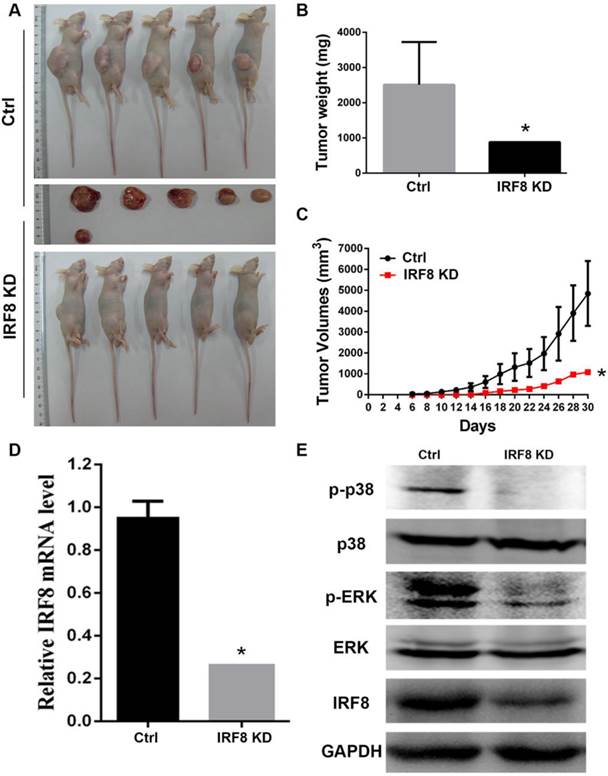
As we identified the effect of IRF8 on DLBCL cells in vitro, we were interested to examine if IRF8 had similar effect in an in vivo situation. We transplanted DLBCL cells with or without IRF8 knockdown into nude mice, and observed the tumor growth in vivo. After 30 days of tumor cells transplantation, in the group of mice with IRF8 knockdown (n=5), there was only one mouse developed tumor; while in the group of control mice (n=5), all animals developed tumors (Figure 5A). The tumor weights and volumes in the mouse received IRF8-knockdown DLBCL cells were both lesser than that in the mice received control DLBCL cells (P<0.05) (Figure 5B, 5C). qRT-PCR and Western blot analysis confirmed that the levels of IRF8 mRNA and protein in the tumor from mouse received IRF8-knockdown DLBCL cells were significantly lower than that in the tumors from the control mice (P<0.05) (Figure 5D, 5E). Finally, we analyzed the phosphorylation levels of p38 and ERK in the tumors, and found that as occurred in the in vitro condition, the tumor derived from the mouse received IRF8-knockdown DLBCL cells showed significant decrease in the phosphorylation of both p38 and ERK in comparison to the control group (Figure 5E).
Loss of IRF8 suppressed the tumor growth in vivo. A: Photograph shows the tumors developed in mice received OCI-Ly01 cells with or without IRF8 for 30 days. B: The increase in tumor weights over 30 days of period showed that the tumor derived from cells with IRF8 knockdown grew significant lowerer than those derived from control cells. C: The increase in tumor volumes over 30 days of period showed that the tumor derived from cells with IRF8 knockdown grew significant slower than those derived from control cells (P<0.05). D: The levels of IRF8 transcripts in tumors derived from OCI-Ly01 cells with IRF8 knockdown were significant lesser than that derived from control cells (P<0.05). E: Western blotting analyses of proteins from tumor tissues showed that the levels of p-p38 and p-ERK were decreased tumors derived from OCI-Ly01 cells with IRF8 knockdown than that derived from control cells. IRF8 KD: IRF8 knockdown. * depicts statistically significant difference between the IRF8 knockdown and control tumors (P<0.05).
The levels of IRF8 in DLBCL were associated with molecular subtypes
To study the clinical significance of IRF8 in DLBCL, we analyzed the association of IRF8 levels with molecular subtypes of DLBCL. The DLBCL samples were subtyped using IHC-based Visco-Young algorithm, a well-characterized method for classifying DLBCL into GCB and non-GCB subtypes based on the positivity of CD10, FOXP1, and BCL6 staining of the tissue [20]. In 73 DLBCL samples used in this study, there were 39 GCB cases and 34 non-GCB cases (Table 1) (Figure S2). The results showed that the levels of IRF8 was significantly associated with the molecular subtypes of DLBCL, in which low levels of IRF8 were associated with GCB subtype whereas high levels of IRF8 were associated with non-GCB subtype (P<0.05) (Table 1).
The levels of IRF8 in GCB and non-GCB human DLBCL tumor tissues.
| IRF8 Levels | GCB | Non-GCB | Totals |
|---|---|---|---|
| Low | 27 | 14 | 41 |
| High | 12 | 20 | 32 |
| Totals | 39 | 34 | 73 |
Note: The levels of IRF8 in tumor tissues were classified as “low” or “high” as described in the Materials and Methods section. The non-GCB DLBCL had greater numbers of samples with high levels of IRF8 than the GCB DLBCL did (P<0.05).
Discussion
As a malignant disease with high degree of heterogeneity, DLBCL has a complex molecular pathology that underlies highly variable clinical outcomes. As an effort to decipher the molecular mechanisms underlying the complexity of DLBCL, in current study we presented data that loss of IRF8 reduced DLBCL cell proliferation in vitro and suppressed lymphoma growth in vivo. Mechanistically, decreasing levels of IRF8 in DLBCL cells diminished the phosphorylation of p38 and ERK MAPK, two essential components in the proliferation of B lymphocytes.
A number of studies have shown that IRF8 is one of the key players in regulating B-cell development and functions [14, 15, 25, 26]. However, IRF8's role in the development of B-cell malignancies remained elusive. Li et al. recently reported that there were about 6% of follicular lymphomas had IRF8 gene mutation [27]. In DLBCL, using capture-sequencing technology, Bouamar et al. found the occurrence of gene fusion between IRF8 and IGH in tumor tissues that resulted in an increased expression of IRF8 [19]. By ectopically over-expressing IRF8 in B lymphoma cells, Bouamar et al. further found that the increased levels of IRF8 conferred resistance to apoptosis of B lymphoma cells [19]. Previously, we also observed that IRF8 exerted an anti-apoptosis role by regulating MDM2 and TP53 in germinal center B cells [17]. These data together were in full agreement with our current findings and strongly support an oncogenic role of IRF8 in DLBCL.
We initiated this study based on the analysis results of public available datasets showing an inverse correlation between the levels of IRF8 transcripts and the overall survival time of patients with DLBCL. Unfortunately, our own cohort of Chinese DLBCL patients used in the current study did not have sufficient data for survival time analysis. Although this precluded us to perform analysis for the association between IRF8 levels and survival time of patients with DLBCL, we did find that tumor tissues from non-GCB subtype of DLBCL patients had significantly greater levels of IRF8 than that from GCB patients. It has been well documented that patients with GCB subtype of DLBCL generally have superior survival time than those with non-GCB subtype of DLBCL [5]. This suggests that high levels of IRF8 in tumor tissues may be associated with an inferior survival of patients with DLBCL. As a matter of fact, our in vivo data from tumor xenograft animal studies supported this note by showing that loss of IRF8 inhibited DLBCL tumor growth in vivo.
An interesting feature for IRF8 is that its biological functions are cell type-dependent. In particular, as we discussed above, IRF8 exerted an anti-apoptosis effect in B cells and may play an oncogenic role in B-cell lymphoma. In T cells, IRF8 was shown to inhibit inflammation by suppressing the development of TH17 cells [28]. However, in sarcoma tumor cells, Yang et al. reported that Irf8 exerted a pro-apoptosis effect through its regulation on Fas-mediated apoptosis [29]. In colon cancer cells, the expression of IRF8 was reported to be silenced due to DNA methylation, and the levels of IRF8 in colon tumors were inversely associated with cancer metastasis [30]. Furthermore, it has been reported that IRF8 deficiency in mice led to the production of a myeloid cell population which was highly homologous to tumor-induced myeloid-derived suppressor cells (MDSCs), a group of immature myeloid cells able to suppress antitumor immunity [31]. Recently, it was reported that in renal cell carcinoma, IRF8 expression was frequently inhibited by DNA methylation, and ectopic expression of IRF8 induced cell cycle G2/M arrest and apoptosis [32]. These data clearly demonstrated the complexity of IRF8 functions, and that the role of IRF8 in B lymphocytes and B-cell lymphomas would be different from other types of cells and their corresponding malignancies.
It has been well characterized that p38 MAPK is essential for the proliferation of B lymphocytes [22]. In B lymphoma tissues and cell lines, the phosphorylation of p38 was increased [33]. Blocking p38 activation led to the suppression of B-cell proliferation [23]. In current study, we found that decreasing levels of IRF8 suppressed the phosphorylation of p38 in DLBCL cells. These data provided an insight for IRF8's effect on DLBCL cell proliferation. Similarly, ERK activation is also an essential component for B cell proliferation. Pharmacological inhibition of ERK in B lymphocytes resulted in inhibition of cell proliferation and induction of apoptosis [34]. Our current results showed that IRF8 knockdown also inhibited ERK phosphorylation, adding another mechanism for IRF8's effect on DLBCL cell proliferation.
In summary, in the present study we found that the loss of IRF8 suppressed DLBCL cell proliferation in vitro and tumor growth in vivo, and the regulatory effect may be mediated by the inhibition of p38 and ERK activation. Future studies would be performed to further analyze the clinical significance of IRF8 in DLBCL.
Abbreviations
DLBCL: diffuse large B-cell lymphoma; GC: germinal center; IRF: IFN regulatory factor; shRNA: short hairpin RNA; qRT-PCR: quantitative real-time PCR.
Supplementary Material
Figures S1 -S2.
Acknowledgements
This work was supported by the National Science Foundation of China (NSFC) (grant # 81172251 to JXZ and # 81400098 to LJ) and by the K.C. Wong Magna Fund at Ningbo University.
Conflict of interest
The authors declare no financial or commercial conflict of interest.
References
1. Schneider C, Pasqualucci L, Dalla-Favera R. Molecular pathogenesis of diffuse large B-cell lymphoma. Semin Diagn Pathol. 2011;28:167-77
2. Huang JZ, Sanger WG, Greiner TC, Staudt LM, Weisenburger DD, Pickering DL. et al. The t(14;18) defines a unique subset of diffuse large B-cell lymphoma with a germinal center B-cell gene expression profile. Blood. 2002;99:2285-90
3. Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RC. et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8:68-74
4. Colomo L, Lopez-Guillermo A, Perales M, Rives S, Martinez A, Bosch F. et al. Clinical impact of the differentiation profile assessed by immunophenotyping in patients with diffuse large B-cell lymphoma. Blood. 2003;101:78-84
5. Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A. et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503-11
6. Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI. et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937-47
7. De Paepe P, De Wolf-Peeters C. Diffuse large B-cell lymphoma: a heterogeneous group of non-Hodgkin lymphomas comprising several distinct clinicopathological entities. Leukemia. 2007;21:37-43
8. Shukla V, Lu R. IRF4 and IRF8: Governing the virtues of B Lymphocytes. Front Biol (Beijing). 2014;9:269-82
9. Sasaki H, Kurotaki D, Osato N, Sato H, Sasaki I, Koizumi S. et al. Transcription factor IRF8 plays a critical role in the development of murine basophils and mast cells. Blood. 2015;125:358-69
10. Holtschke T, Lohler J, Kanno Y, Fehr T, Giese N, Rosenbauer F. et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87:307-17
11. Gabriele L, Ozato K. The role of the interferon regulatory factor (IRF) family in dendritic cell development and function. Cytokine Growth Factor Rev. 2007;18:503-10
12. Tsujimura H, Tamura T, Ozato K. Cutting edge: IFN consensus sequence binding protein/IFN regulatory factor 8 drives the development of type I IFN-producing plasmacytoid dendritic cells. J Immunol. 2003;170:1131-5
13. Wang H, Morse HC 3rd. IRF8 regulates myeloid and B lymphoid lineage diversification. Immunol Res. 2009;43:109-17
14. Feng J, Wang H, Shin DM, Masiuk M, Qi CF, Morse HC 3rd. IFN regulatory factor 8 restricts the size of the marginal zone and follicular B cell pools. J Immunol. 2011;186:1458-66
15. Yoon J, Feng X, Kim YS, Shin DM, Hatzi K, Wang H. et al. Interferon Regulatory Factor 8 (IRF8) Interacts with the B Cell Lymphoma 6 (BCL6) Corepressor BCOR. J Biol Chem. 2014;289:34250-7
16. Lee CH, Melchers M, Wang H, Torrey TA, Slota R, Qi CF. et al. Regulation of the germinal center gene program by interferon (IFN) regulatory factor 8/IFN consensus sequence-binding protein. J Exp Med. 2006;203:63-72
17. Zhou JX, Lee CH, Qi CF, Wang H, Naghashfar Z, Abbasi S. et al. IFN regulatory factor 8 regulates MDM2 in germinal center B cells. J Immunol. 2009;183:3188-94
18. Martinez A, Pittaluga S, Rudelius M, Davies-Hill T, Sebasigari D, Fountaine TJ. et al. Expression of the interferon regulatory factor 8/ICSBP-1 in human reactive lymphoid tissues and B-cell lymphomas: a novel germinal center marker. Am J Surg Pathol. 2008;32:1190-200
19. Bouamar H, Abbas S, Lin AP, Wang L, Jiang D, Holder KN. et al. A capture-sequencing strategy identifies IRF8, EBF1, and APRIL as novel IGH fusion partners in B-cell lymphoma. Blood. 2013;122:726-33
20. Visco C, Li Y, Xu-Monette ZY, Miranda RN, Green TM, Li Y. et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Leukemia. 2012;26:2103-13
21. Fontan L, Yang C, Kabaleeswaran V, Volpon L, Osborne MJ, Beltran E. et al. MALT1 small molecule inhibitors specifically suppress ABC-DLBCL in vitro and in vivo. Cancer Cell. 2012;22:812-24
22. Khiem D, Cyster JG, Schwarz JJ, Black BL. A p38 MAPK-MEF2C pathway regulates B-cell proliferation. Proc Natl Acad Sci U S A. 2008;105:17067-72
23. Craxton A, Shu G, Graves JD, Saklatvala J, Krebs EG, Clark EA. p38 MAPK is required for CD40-induced gene expression and proliferation in B lymphocytes. J Immunol. 1998;161:3225-36
24. Yasuda T, Sanjo H, Pages G, Kawano Y, Karasuyama H, Pouyssegur J. et al. Erk kinases link pre-B cell receptor signaling to transcriptional events required for early B cell expansion. Immunity. 2008;28:499-508
25. Shin DM, Lee CH, Morse HC 3rd. IRF8 governs expression of genes involved in innate and adaptive immunity in human and mouse germinal center B cells. PLoS One. 2011;6:e27384
26. Wang H, Lee CH, Qi C, Tailor P, Feng J, Abbasi S. et al. IRF8 regulates B-cell lineage specification, commitment, and differentiation. Blood. 2008;112:4028-38
27. Li H, Kaminski MS, Li Y, Yildiz M, Ouillette P, Jones S. et al. Mutations in linker histone genes HIST1H1 B, C, D, and E; OCT2 (POU2F2); IRF8; and ARID1A underlying the pathogenesis of follicular lymphoma. Blood. 2014;123:1487-98
28. Ouyang X, Zhang R, Yang J, Li Q, Qin L, Zhu C. et al. Transcription factor IRF8 directs a silencing programme for TH17 cell differentiation. Nat Commun. 2011;2:314
29. Yang D, Thangaraju M, Browning DD, Dong Z, Korchin B, Lev DC. et al. IFN regulatory factor 8 mediates apoptosis in nonhemopoietic tumor cells via regulation of Fas expression. J Immunol. 2007;179:4775-82
30. McGough JM, Yang D, Huang S, Georgi D, Hewitt SM, Rocken C. et al. DNA methylation represses IFN-gamma-induced and signal transducer and activator of transcription 1-mediated IFN regulatory factor 8 activation in colon carcinoma cells. Mol Cancer Res. 2008;6:1841-51
31. Waight JD, Netherby C, Hensen ML, Miller A, Hu Q, Liu S. et al. Myeloid-derived suppressor cell development is regulated by a STAT/IRF-8 axis. J Clin Invest. 2013;123:4464-78
32. Zhang Q, Zhang L, Li L, Wang Z, Ying J, Fan Y. et al. Interferon regulatory factor 8 functions as a tumor suppressor in renal cell carcinoma and its promoter methylation is associated with patient poor prognosis. Cancer Lett. 2014;354:227-34
33. Ding H, Gabali AM, Jenson SD, Lim MS, Elenitoba-Johnson KS. P38 mitogen activated protein kinase expression and regulation by interleukin-4 in human B cell non-Hodgkin lymphomas. J Hematop. 2009;2:195-204
34. O'Reilly LA, Kruse EA, Puthalakath H, Kelly PN, Kaufmann T, Huang DC. et al. MEK/ERK-mediated phosphorylation of Bim is required to ensure survival of T and B lymphocytes during mitogenic stimulation. J Immunol. 2009;183:261-9
Author contact
![]() Corresponding author: Jeff X. Zhou, Ningbo University School of Medicine, Ningbo, Zhejiang 315211, China; email: zhouxiwuedu.cn; Tel.: 86-574-87609595; Fax: 86-574-87608638; or Guifang Ouyang, Ningbo University Affiliated First Hospital, Ningbo, Zhejiang 315000, China; email: ouyangguifangcom.cn; Tel: 86-574-87085591; Fax: 86-574-87291583
Corresponding author: Jeff X. Zhou, Ningbo University School of Medicine, Ningbo, Zhejiang 315211, China; email: zhouxiwuedu.cn; Tel.: 86-574-87609595; Fax: 86-574-87608638; or Guifang Ouyang, Ningbo University Affiliated First Hospital, Ningbo, Zhejiang 315000, China; email: ouyangguifangcom.cn; Tel: 86-574-87085591; Fax: 86-574-87291583

 Global reach, higher impact
Global reach, higher impact