3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2015; 6(11):1114-1120. doi:10.7150/jca.11959 This issue Cite
Research Paper
CDKN2 Gene Deletion as Poor Prognosis Predictor Involved in the Progression of Adult B-Lineage Acute Lymphoblastic Leukemia Patients
1. Department of Hematology, Nanfang Hospital, Southern Medical University, Guangzhou Dadao North Street, 1838, Guangzhou 510515, China
2. Department of Management and Development, Nanfang Hospital, Southern Medical University, Guangzhou Dadao North Street, 1838, Guangzhou 510515, China
* Na Xu and Yuling Li contributed equally to this study and should be considered as co-first authors
Received 2015-2-22; Accepted 2015-7-20; Published 2015-9-15
Abstract
Deletion of cyclin-dependent kinase inhibitor 2A/B (CDKN2A/B) is well known in many hematologic malignancies, but only few reports have investigated this deletion effect on clinical prognosis. This study performed analysis of the CDKN2 deletion in 215 adult B- lineage acute lymphoblastic leukemia (B-ALL) patients, and related cytogenetic prognostic factors (BCR/ABL; E2A/PBXl; TEL/AML1; Mixed Lineage Leukemia (MLL) rearrangement; MYC, Immunoglobulin heavy locus (IGH) translocation). The prevalence of CDKN2 deletions in all study populations was 28.4%. There is no difference between patients with CDKN2 deletion and wild-type patients in sex, age, white blood cells (WBC) count, BM blast percentage, extra infiltration and induction complete remission (CR) rate. Analysis in relapse patients revealed that the distribution of CDKN2 deletion is higher in relapse patients (44.6%) than all patients (28.4%, P=0.006). Deletion of CDKN2 was significantly associated with poor outcomes including decreased overall survival (OS) (P<0.001), lower disease free-survival (DFS) (P<0.001), and increased cumulative incidence of relapse (P=0.002); Also, CDKN2 deletion was strongly associated with IGH translocation (P=0.021); and had an adverse effect on patients with BCR-ABL fusion gene or with MLL rearrangement. Patients with CDKN2 gene deletion benefited from allogenic hematopoietic stem cell transplantation (Allo-HSCT). Deletion of CDKN2 gene was commonly observed through leukemia progression and was poor prognostic marker in long-term outcomes.
Keywords: CDKN2, Acute lymphoblastic leukemia, Stem cell, Deletion.
Introduction
Identification of specific abnormal genes involved in the process of leukemogenesis often suggests possible prognostic markers that may be applied into risk stratification and treatment protocol in leukemia. Specifically, alterations of genes in the 9p21.3 locus have been implicated in many types of cancer. Genes CDKN2A (MTS1) and CDKN2B (MTS2), which encode for p16INK4a/p14ARF and p15INK4b respectively from this locus, are thought to be important growth suppressor genes. INK4a/ARF regulates p53 tumor suppressor function by interacting with MDM2. P15/INK4b works as a CDK inhibitor, which inhibits phosphorylation of Rb by inactivating cyclin/ CDK-4/6 complexes and thereby controls cell-cycle G1 progression [1]. Study also found that CDKN2 gene with the role of a gate keeper can sustain the balance between HSC self-renewal and the early differentiation stages which resulted in lineage commitment. [2-4]. It has been reported that inactivation of the CDKN2 occurred in the form of deletion, methylation or mutation in various hematologic malignancies, and genomic deletion was more frequent than mutation or methylation [5, 6]. CDKN2 may be haploinsufficient in human cancer and its inactivation endows differentiated cells with the capacity to inappropriately self-renew which plays an important role in tumor formation [7]. Deletions of CDKN2 have been shown to frequently occur in acute lymphoblastic leukemia (ALL), the incidence ranged from 18% to 45% [1]. Concurrently, the association between the deletion of CDKN2 and the prognosis in child ALL was widely investigated, most of these results found that patients with CDKN2 deletion had an increased poor outcome than wild-type patients [8, 9]. However, only few reports were about prognosis and CDKN2 deletion in adult ALL leukemia. Furthermore, all these researches draw controversial conclusions [10, 11], the identification of the role of CDKN2 deletion in leukemia development remains to be seen. The current study was aim to investigate the prevalence, feature, and specific prognostic relevance of CDKN2 deletion in adult B-ALL patients for the first time in the Chinese Han population.
Materials and methods
Study Population
A total of 278 newly diagnosed Chinese Han adult B-ALL patients were identified retrospectively from January 2008 to December 2013 at South Medical University, South hospital center. Approval was obtained from the Ethics committee of the South hospital Affiliated to South Medical University. Written informed consent was obtained from all patients. Specimens were operated follow national ethical and legal standards.. B-ALL was diagnosed based on standard criteria which include bone marrow morphologic, cytochemical, immunophenotypic and cytogenetic analysis. Among them, 215 patients received all the examinations and systematic treatments. 63 patients who terminate the treatment due to personal reason was excluded from this study. The treatment procedure was: all patients received a 4-week induction therapy (vincristine, daunorubicin or idarubicin, L-asparagines, and prednisone) at first and 53 patients with Philadelphia-positive (Ph+) ALL received extra imatinib 400 mg qd (daily) or dasatinib 70 mg (bid) twice a day. After complete remission, all patients received consolidation treatment with-Hyper CVAD A scheme (cyclophosphamide, vincristine, Adriamycin, dexamethasone) and Hyper CVAD B scheme (high-dose methotrexate and cytarabine) alternately. Then, 100 patients received allogeneic hematopoietic stem cell transplantation (Allo-HSCT) after two to five courses of consolidation chemotherapy. All treatment regimens for ALL patients include central nervous system (CNS) prophylaxis. All 215 patients were de novo ALL cases at the time of diagnosis. 92 relapse cases analyzed only at the time of treatment failure. Both diagnosed and relapsed samples were collected for further testing. Cytogenetic Risk stratification was defined as follows: Standard risk: Hyperdiploidy (51-65 chromosomes and /DNA index >1.16; cases with trisomy of chromosomes 4,10and 17 appear to have the most favorable outcome); t(11;21)(p13;q22): TEL-AML1; high risk: Hypodiploidy (<44 chromosomes and/or DNA index<0.81); t(v;11q23):MLL rearranged; t(9;22)(q34;q11.2):BCR-ABL; Complex karyotype (5 or more chromosomal abnormalities). The definition and assessment for adult ALL was determined from NCCN Guideline Version 1.2014 Acute Lymphoblastic leukemia [12].
FISH and probes
Bone marrow cells of all patients were available for detection of CDKN2 (covers a 193kd region of 9q21.3,extending from 105kd telomere of p16 gene to 46kd centromere of CDKN2B), BCR/ABL, E2A/PBXl, TEL/AML1, MLL rearrangement, MYC (8q24.21)translocation, IGH(14q32) translocation and hyperdiploid (regions of chromosomes 4,10 and 17) based by interphase fluorescence in situ hybridization (I-FISH). The results were analyzed follow the ISCN (2005) criteria [13]. Briefly, probe and slide preparation as well as hybridization and washing steps were performed with two green-labeled chromosome 9 probe and two red-labeled CDKN2 probe kit (Cat. No. LH009, Cytocell, Cambridge, UK) according to the manufacturers' protocols (Figure 1). The sample was first put in to pretreatment solution and incubated at 110°C for 1 minute. Then, samples was digested with pepsin (Sigma, St Louis, MO, USA) at 37°C for 10 minutes. Before hybridization The probes were denatured for 5 minutes at 95°C. The slides were then hybridized for 48 hours at 37°C and stained with DAPI (Life Science, USA).
Statistical analysis
Pearson Chi-square analysis and Fisher exact test were carried out to compare the difference of categorical variables between patients groups. For TEL/AML1 and Hyperdiploidy, we used Fisher's exact test, others used Pearson Chi-square test. For WBC count, we used Wilcoxon test. We already added this detail into statistical section in our revised paper. Overall survival (OS) was analyzed according to the Kaplan-Meier method and Log rank tests were employed to compare survival curves between groups. For all analyses, statistical analysis was performed using SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). And a p value of less than 0.05 was considered statistically significant.
Fluorescence in situ hybridization showing A) Normal cells with two green and two red signals; B) hemizygous which loss of one red signal; C) homozygous which loss of both red signals (p16) and only two green signals represent chromosome 9.
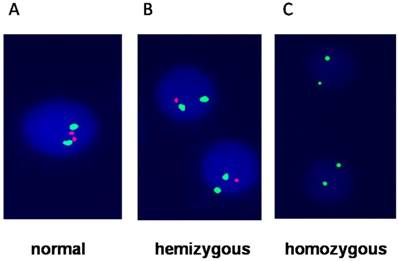
Results
Patient characteristics
215 newly diagnosed B-ALL patients were collected. Aged between 15 and 60 years old, the median age at diagnosis was 31years old, the median white blood cell(WBC) count was 47.×109/L (range: 1.5∼207.8). Of 215 cases, 61(28.4%) patients with CDKN2 deletion, and patients characteristics are summarized in Table 1. There was no significant difference between CDKN2 deletion carrier and non-carrier for age, sex, and WBC counts at presentation, percentage of BM blasts at diagnosis, hepatosplenomegaly, and risk stratification (Table 1).
CDKN2 deletion and its correlation with other molecular alterations
Furthermore, the association of CDKN2 deletion with other known molecular alterations such as E2A/PBXl fusion gene, TEL/AMLl fusion gene, BCR-ABL fusion gene, MLL rearrangement, hyperdiploid, and MYC and IGH translocation were conducted and results are summarized in Table 1. In this cohort study, CDKN2 deletion was strongly associated with IGH translocation (P=0.021), while no association was observed with BCR/ABL fusion gene and MLL rearrangement (p=0.735, p=0.359,). Meanwhile, patients with wild CDKN2 gene had higher MYC translocation or E2A/PBX1 frequencies (5.2%, 5.2%), but no hyperdiploid and TEL-AML fusion gene was detected in CDKN2 deletion patients.
CDKN2 deletion distribution and patients characteristics
| deletion CDKN2 (n=61) | Wild CDKN2 (n=154) | p-value | |
|---|---|---|---|
| Female/ Male | 23/38 | 72/82 | 0.228 |
| 15-35years | 37 | 95 | 0.889 |
| 35-60years | 24 | 59 | 0.889 |
| WBC count(x109/L) | 55.3(15.9∼125.7) | 43.8(1.5∼207.8) | 0.307 |
| hepatosplenomegaly Risk stratification | 24(39.3%) | 50(32.5%) | 0.339 |
| high risk | 39(63.9%) | 104(67.5%) | 0.614 |
| Standard risk | 22(36.1%) | 50(32.5%) | 0.614 |
| Molecular abnormalities | |||
| BCR/ABL | 16(26.2%) | 37(24%) | 0.735 |
| TEL/AML1 | 0 | 1(0.6%) | 1.000 |
| E2A/PBX1 | 1(1.6%) | 8(5.2%) | 0.451 |
| MLL rearrangement | 2(3.3%) | 11(7.1%) | 0.359 |
| MYC | 1(1.6%) | 8(5.2%) | 0.451 |
| Hyperdiploidy | 0 | 5(3.2%) | 0.325 |
| IGH rearrangement* | 5 (8.2%) | 2(1.3%) | 0.021 |
| CR rate | 56(91.8%) | 145(94%) | 0.546 |
| Allo-HSCT | 25(41%) | 75(48.7%) | 0.306 |
| Relapse* | 36(59%) | 56(36.4%) | 0.002 |
Note: WBC: White blood cell; CR: Complete remission; Allo-HSCT: allogeneic hematopoietic stem cell transplantation; *: Comparison between CDKN2 deletion carriers and non carriers.
Analysis of CDKN2 gene deletion in relapse patients
Among 92 (42.7%) relapse patients, 34 patients had CDKN2 deletion both in denovo diagnosis and relapse, 7 patients acquired CDKN2 gene deletion at relapse. This deletion rate was significantly increased at relapse compared to that in all patients. (44.6% vs. 28.4%, p=0.006). Further analysis of the results showed those with CDKN2 deletion had shorter DFS (p=0.004) and OS (p=0.001), but there was no statistical difference in re-induced CR rate (p=0.901) (Table 2).
Association of CDKN2 gene distribution and prognosis in relapse patients
| deletion CDKN2 (n=41) | Wild CDKN2 (n=51) | p-value | |
|---|---|---|---|
| Re-induce CR rate (%) | 35(85.3) | 44(86.3) | 0.901 |
| DFS*(Median time) | 5(0∼38) | 16(0∼57) | 0.004 |
| OS*(Median time) | 8(1∼39) | 18(2∼58) | 0.001 |
CR: Complete remission; DFS: Disease Free Survival; OS: Overall Survival;
*: Comparison between CDKN2 deletion carriers and non carriers.
Effect of CDKN2 deletion on prognosis with Allo-HSCT treatment
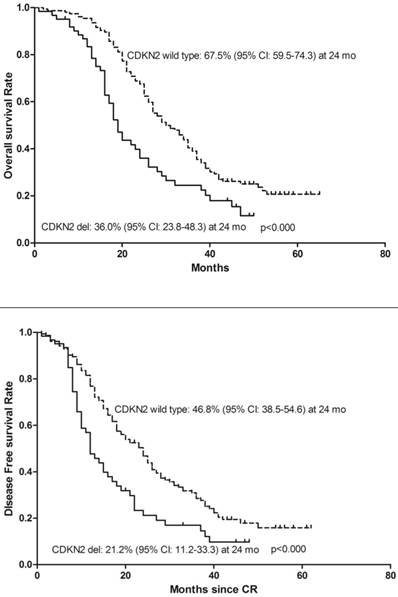
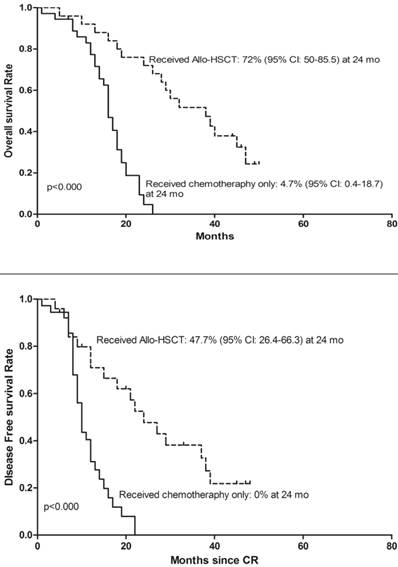
To determine whether deletion of CDKN2 gene has an effect on treatment outcome in B-ALL patients, a follow-up study was taken with median 27 months (range: 1-65). 143 patients were classified as the high-risk group and 72 patients were classified as the standard-risk group. There was no significant difference in the CDKN2 deletion distribution in the two risk groups. 25 (41%) of the 61 patients with CDKN2 deletion and 75(48.7%) patients with wild type CDKN2 gene received Allo-HSCT and there was no difference in treatment protocol and induction CR rate between these two groups (p=0.306; p=0.546). However, patients with deletion of CDKN2 had a higher relapse rate compared to wild-type patients (59% VS 36.4%, p=0.002) (Table 1). Thereafter, a univariate analysis of the CDKN2 genomic status and its association with outcomes was conducted. A shorter OS and DFS was found in patients with CDKN2 deletion compared with wild-type patients (OS: 19 versus 30 months, respectively, P<0.000; DFS: 12 versus 24 months, respectively, P<0.000)(Figure 2). 25 of the 61 patients with the deletion received Allo-HSCT, the subgroup which received Allo-HSCT treatment had a better OS and DFS compared with the subgroup which received chemotherapy only (OS: p<0.000; DFS: p<0.000)(Figure 3).
Kaplan-Meier survival curves for Overall Survival (OS) and Disease Free Survival (DFS) in enrolled B-ALL patients. CR= Complete remission. Significant differences were shown between CDKN2 deletion and non deletion patients at 24 months. P<0.05.
Kaplan-Meier survival curves for Overall Survival (OS) and Disease Free Survival (DFS) in the clinical treated CDKN2 deletion patients. CR= Complete remission. Significant differences were shown between Allo-HSCT treatment and chemotherapy treatment only at 24 months. P<0.05.
Effect of CDKN2 deletion on IGH rearrangement
In our study, seven patients had IGH rearrangement, two patients with IGH rearrangement only remained in continuous first CR for 45 and 38 months respectively, three of five patients concurrent with IGH rearrangement and CDKN2 deletion died within 1year, the other two patients were still alive during the study time frame. The relationship of CDKN2 deletion and IGH rearrangement and whether CDKN2 gene deletion has adverse effects on IGH rearranged leukemia need further study.
Association of CDKN2 deletion with BCR-ABL fusion gene and MLL gene rearrangement in ALL patients
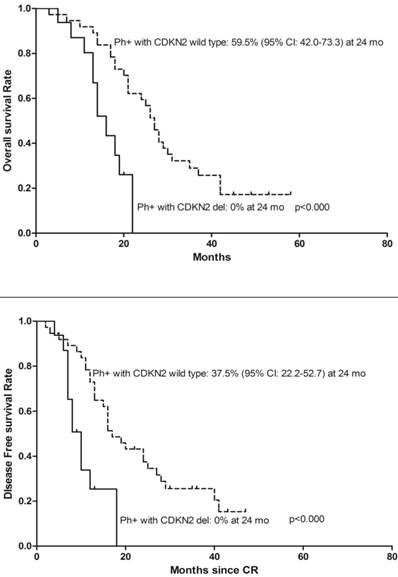
We analyzed the effect of CDKN2 deletion in ALL patients with BCR-ABL fusion gene and found 16 (26.2%) BCR/ABL fusion gene positive patients also had CDKN2 deletion. Moreover, the patients with BCR-ABL fusion gene and CDKN2 gene deletion had a poorer OS, DFS (Figure 4) and were even more likely to relapse compared to patients with BCR-ABL fusion gene only (Relapse: 68.8% vs. 37.8%, p=0.038). Two of thirteen MLL gene rearrangement patients had concurrent CDKN2 gene loss, one patient died during the period of induction chemo-therapy, another relapsed 3 months after diagnosis and died 1 month following the relapse. Meanwhile, four of eleven patients with MLL rearrangement only were still alive on the date of the follow-up censored.
Kaplan-Meier survival curves for Overall Survival (OS) and Disease Free Survival (DFS) in denovo Ph/BCR/ABL positive ALL patients. CR= Complete remission. Ph+= Philadelphia chromosome positive. Significant differences were shown between CDKN2 deletion and non deletion patients at 24 months. P<0.05.
Discussion
Several studies have reported high frequencies of p15/p16 (CDKN2) deletion in both adult and child acute lymphoblastic leukemia and proved FISH as a reliable and accurate method for this deletion detection [1, 10]. However, the majority of these studies were based on a small study cohort and were limited in further analysis of the correlation between CDKN2 deletion and those established prognostic karyotype aberrations in B-ALL including BCR/ABL fusion gene and MLL gene rearrangement. Therefore, we recruited a larger patient cohort, containing 215 newly diagnosis B-ALL patients for this study. Results showed the frequency of CDKN2 deletion was 28.4%, which was higher than Kawamata report (21%) but lower than Bertin (31%) and Mullighan report (34%) [14-16]. Some investigators reported a more closer relationship between CDKN deletion carriers and abnormalities of 9p21 as well as other clinical lab data including high WBC, blast counts, prominent lymphadenopathy, splenomegaly, expression of predominantly T-cell surface markers, and shorter survival timer compared to other types of ALL patients [8, 9, 17], but this association did not found in other studies [11, 18, 19]. Data from our study demonstrated that the frequency of CDKN2 deletion had no significant association with age, sex, WBC counts, percentage of BM blasts and hepatosplenomegaly.
Our research showed that CDKN2 deletion carriers have decreased OS, DFS and a higher rate of recurrence compared with wild-type patients. Those CDKN2 deletion patients who received Allo-HSCT exhibited an improved OS and DFS compared to patients that only received chemotherapy. These results are consistent with previous studies which reported loss of p16 as a major independent negative prognostic indicator in pediatric ALL [20]. Abdulmohsen Alhejaily found that inactive CDKN2A is relatively common in pre-treatment follicular lymphoma associated with more aggressive forms and poor clinical outcome [21]. We also investigated if CDKN2 deletion would have an enhanced predicted effect on survival when took established poor prognostic factors into consideration such as BCR/ABL or MLL rearrangement or hypodiploidy. In our study, no relationship was found between CDKN2 deletion and BCR-ABL1 fusion gene or MLL rearrangement, but CDKN2 deletion always occurred together with IGH translocation. However, this additional IGH translocation changes had no impact on prognosis in CDKN2 deletion carriers in our analysis. This negative results may occurred due to relatively small number of study subjects in the subgroup, further larger study with more clinical and genetic information could benefit this correlation study..
We identified CDKN2 deletion in 30.2% of BCR/ABL positive ALL patients, similar to previous reports [22] in which the frequency was 26.2% at diagnosis. Moreover, our research suggested that CDKN2 gene deletion resulted in poor prognosis in BCR/ABL positive ALL patients and agreed with Ilaria Iacobucci [21]. Other studies found that p16 gene deletion emerged in some of CML-BC and Ph-positive acute lymphoblastic leukemia ALL patients, which had not been detected in CML-CP patients [23, 24]. Transducted p16(INK4a) or p14(ARF) virus into primary blast cells from chronic myeloid leukemia in blast crisis (CML-BC) and Ph-positive acute lymphoblastic leukemia (ALL) showed, inhibited growth of cells apoptosis promoted. Furthermore, these cells had increased sensitivity to imatinib [25]. The results illustrated that p16 gene deletion is a poor prognostic predictor.
MLL rearrangement is another adverse factor resulting in poor prognosis, the frequency of CDKN2 deletion in MLL rearranged patients was 15.4% in our study, the incidence rate was lower than Ohnishi, H's report [26]. In our research, patients with MLL rearrangement and CDKN2 deletion had a short survival time, especially one patient with both gene alterations died in the early period of induction chemotherapy, and another patient died within 4 months. Study found down-regulation of p16 gene could restore the cell colony forming capacity in MLL/ENL transduced cells and exacerbates leukemia. On the contrary, the up-regulation of p16 had a curative effect on the MLL/ENL leukemia cells by diminishing their leukemia initiating capacity [27]. However, the relationship between p16 gene and MLL rearrangement and whether CDKN2 gene deletion has an adverse effect in MLL rearranged leukemia needs further study.
Both TEL/AML1 and hypodiploidy (regions of chromosomes 4, 10 and 17) were a common cytogenetic alteration and associated with an improved prognosis in child ALL, but rare in adult ALL. This cytogenetic discrepancy may partially explain the different clinical outcomes between childhood and adult ALL. In our research, there were only a few patients with TEL/AML1 and hypodiploidy and even fewer in CDKN2 deletion carriers. Kim found that ALL cell lines with TEL/AML1 fusion gene and deletion of 9p21 had a strong ability to proliferate compared with the cell lines with TEL/AML1 fusion gene only [28], and deletion of 9p21 may weakened or even reverse the favorable impact of the TEL/AML1 fusion proliferation.
In relapse patients, CDKN2 loss rate (44.6%) was dramatically higher compared with that in all patients at diagnosis, and together with impaired OS, DFS. These results suggest that loss of this genomic region may be involved in disease progression. It is well known that loss of CDKN2A/B could result in removal of tumor suppressors and activation of tumor growth factors such as MDM2 and CDK4/6 which promote tumor cell proliferation and thus be considered as attractive drugs points. Mllighan's study showed that CDKN2A/B deletion may be a reason for relapse by escaping drug-induced lethal effects and clone reproduction [29]. Sranks G's study showed that three NHL cell lines with Biallelic deletions of CDKN2 had undergone transformation from low- to high-grade disease [30].
In summary, the present study found more frequent CDKN2 deletion in relapse adult B-ALL, and are frequently acquired in leukemia progression. The study also provides evidence of CDKN2 deletion as a negative prognostic factor for OS and DFS. These results imply that more focused attention should be shown to the patients who carry the CDKN2 deletion. A new therapeutic strategy could be developed in the future based on fully understand for genetic events in ALL.
Acknowledgements
We thank all researchers, clinical providers and especially the patients and their families who participated in this study.
Funding
This study was financially supported by National Natural Science Foundation of China (No. 81170521) and Presidential Foundation of Nan fang Hospital (No.2012Z013)
Conflict of Interests
We have read and understood policy on conflicts of interest disclosure and declare no competing interests.
References
1. Usvasalo A, Savola S, Räty R, Vettenranta K, Harila-Saari A, Koistinen P. et al. CDKN2A deletions in acute lymphoblastic leukemia of adolescents and young adults: an array CGH study. Leuk Res. 2008;32:1228-35
2. Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL. et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302-5
3. Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. et al. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164-8
4. Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr Opin Genet Dev. 2003;13:77-83
5. Sulong S, Moorman AV, Irving JA, Strefford JC, Konn ZJ, Case MC. et al. A comprehensive analysis of the CDKN2A gene in childhood acute lymphoblastic leukemia reveals genomic deletion, copy number neutral loss of heterozygosity, and association with specific cytogenetic subgroups. Blood. 2009;113:100-7
6. Ohnishi H, Kawamura M, Ida K, Sheng XM, Hanada R, Nobori T. et al. Homozygous deletions of p16/MTS1 gene are frequent but mutations are infrequent in childhood T-cell acute lymphoblastic leukemia. Blood. 1995;86:1269-75
7. Chapman EJ, Harnden P, Chambers P, Johnston C, Knowles MA. Comprehensive analysis of CDKN2A status in microdissected urothelial cell carcinoma reveals potential haploinsufficiency, a high frequency of homozygous co-deletion and associations with clinical phenotype. Clin Cancer Res. 2005;11:5740-7
8. Carter TL, Watt PM, Kumar R, Burton PR, Reaman GH, Sather HN. et al. Hemizygous p16(INK4A) deletion in pediatric acute lymphoblastic leukemia predicts independent risk of relapse. Blood. 2001;97:572-4
9. Graf Einsiedel H, Taube T, Hartmann R, Wellmann S, Seifert G, Henze G. et al. Deletion analysis of p16(INKa) and p15(INKb) in relapsed childhood acute lymphoblastic leukemia. Blood. 2002;99:4629-31
10. Kim M, Yim SH, Cho NS, Kang SH, Ko DH, Oh B. et al. Homozygous deletion of CDKN2A (p16, p14) and CDKN2B (p15) genes is a poor prognostic factor in adult but not in childhood B-lineage acute lymphoblastic leukemia: a comparative deletion and hypermethylation study. Cancer Genet Cytogenet. 2009;195:59-65
11. Faderl S, Kantarjian HM, Manshouri T, Chan CY, Pierce S, Hays KJ. et al. The prognostic significance of p16INK4a/p14ARF and p15INK4b deletions in adult acute lymphoblastic leukemia. Clin Cancer Res. 1999;5:1855-61
12. Acute Lymphoblastic Leukemia, NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines ®) Version 1, 2014. Fort Washington, USA: National Comprehensive Cancer Network. http://www.tri-kobe.org/nccn/guideline/hematologic/english/all.pdf
13. Shaffer LG, Tommerup N. ISCN 2005: an international system for human cytogenetic nomenclature (2005): recommendations of the International Standing Committee on Human Cytogenetic Nomenclature. Basel, Swizerland: Karger Medical and Scientific Publishers. 2005
14. Mullighan CG, Williams RT, Downing JR, Sherr CJ. Failure of CDKN2A/B (INK4A/B-ARF)-mediated tumor suppression and resistance to targeted therapy in acute lymphoblastic leukemia induced by BCR-ABL. Genes Dev. 2008;22:1411-5
15. Kawamata N, Ogawa S, Zimmermann M, Kato M, Sanada M, Hemminki K. et al. Molecular allelokaryotyping of pediatric acute lymphoblastic leukemias by high-resolution single nucleotide polymorphism oligonucleotide genomic microarray. Blood. 2008;111:776-84
16. Bertin R, Acquaviva C, Mirebeau D, Guidal-Giroux C, Vilmer E, Cave H. CDKN2A, CDKN2B, and MTAP gene dosage permits precise characterization of mono- and bi-allelic 9p21 deletions in childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2003;37:44-57
17. Takeuchi S, Bartram CR, Seriu T, Miller CW, Tobler A, Janssen JW. et al. Analysis of a family of cyclin-dependent kinase inhibitors: p15/MTS2/INK4B, p16/MTS1/INK4A, and p18 genes in acute lymphoblastic leukemia of childhood. Blood. 1995;86:755-60
18. van Zutven LJ, van Drunen E, de Bont JM, Wattel MM, Den Boer ML, Pieters R. et al. CDKN2 deletions have no prognostic value in childhood precursor-B acute lymphoblastic leukemia. Leukemia. 2005;19:1281-4
19. Mirebeau D, Acquaviva C, Suciu S, Bertin R, Dastugue N, Robert A. et al. The prognostic significance of CDKN2A, CDKN2B and MTAP inactivation in B-lineage acute lymphoblastic leukemia of childhood. Results of the EORTC studies 58881 and 58951. Haematologica. 2006;91:881-5
20. Carter TL, Terry P, Gottardo N, Baker DL, Kees UR, Watt PM. Deletion of one copy of the p16INK4A tumor suppressor gene is implicated as a predisposing factor in pediatric leukemia. Biochem Biophys Res Commun. 2004;318:852-5
21. Alhejaily A, Day AG, Feilotter HE, Baetz T, Lebrun DP. Inactivation of the CDKN2A tumor-suppressor gene by deletion or methylation is common at diagnosis in follicular lymphoma and associated with poor clinical outcome. Clin Cancer Res. 2014;20:1676-86
22. Iacobucci I, Ferrari A, Lonetti A, Papayannidis C, Paoloni F, Trino S. et al. CDKN2A/B alterations impair prognosis in adult BCR-ABL1-positive acute lymphoblastic leukemia patients. Clin Cancer Res. 2011;17:7413-23
23. Lee DS, Lee JH, Min HC, Kim TY, Oh BR, Kim HY. et al. Application of high throughput cell array technology to FISH: investigation of the role of deletion of p16 gene in leukemias. J Biotechnol. 2007;127:355-60
24. Drexler HG. Review of alterations of the cyclin-dependent kinase inhibitor INK4 family genes p15, p16, p18 and p19 in human leukemia-lymphoma cells. Leukemia. 1998;12:845-59
25. Bai Y, Lu Z, Lin Y, Sun B, Wang S, Wang G. Restoration of INK4a/ARF gene inhibits cell growth and cooperates with imatinib mesylate in Philadelphia chromosome-positive leukemias. Oncol Res. 2013;21:23-31
26. Ohnishi H, Guo SX, Ida K, Taki T, Naritaka S, Bessho F. et al. Alterations of p16 and p15 genes in acute leukemia with MLL gene rearrangements and their correlation with clinical features. Leukemia. 1997;11:2120-4
27. Ueda K, Yoshimi A, Kagoya Y, Nishikawa S, Marquez VE, Nakagawa M. et al. Inhibition of histone methyltransferase EZH2 depletes leukemia stem cell of mixed lineage leukemia fusion leukemia through upregulation of p16. Cancer Sci. 2014;105:512-9
28. Kim DH, Moldwin RL, Vignon C, Bohlander SK, Suto Y, Giordano L. et al. TEL-AML1 translocations with TEL and CDKN2 inactivation in acute lymphoblastic leukemia cell lines. Blood. 1996;88:785-94
29. Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA. et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377-80
30. Stranks G, Height SE, Mitchell P, Jadayel D, Yuille MA, De Lord C. et al. Deletions and rearrangement of CDKN2 in lymphoid malignancy. Blood. 1995;85:893-901
Author contact
![]() Corresponding authors: Xiaoli Liu, Department of Hematology, Nanfang Hospital, Southern Medical University, Guangzhou, 510515, China. Phone: +86 (020) 6164-1616; Fax: +86 (020) 6164-1616; E-mail: lxl2405com or Qingfeng Du, Department of Management and Development, Nanfang Hospital, Southern Medical University, Guangzhou 510515, China. Phone: +86 (020) 6164-1616; Fax: +86 (020) 6164-1616; E-mail: njs41195com
Corresponding authors: Xiaoli Liu, Department of Hematology, Nanfang Hospital, Southern Medical University, Guangzhou, 510515, China. Phone: +86 (020) 6164-1616; Fax: +86 (020) 6164-1616; E-mail: lxl2405com or Qingfeng Du, Department of Management and Development, Nanfang Hospital, Southern Medical University, Guangzhou 510515, China. Phone: +86 (020) 6164-1616; Fax: +86 (020) 6164-1616; E-mail: njs41195com

 Global reach, higher impact
Global reach, higher impact