3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2015; 6(12):1230-1235. doi:10.7150/jca.12546 This issue Cite
Research Paper
MicroRNA-222 Controls Human Pancreatic Cancer Cell Line Capan-2 Proliferation by P57 Targeting
1. Division of Gastroenterology and Hepatology, Digestive Disease Institute, Shanghai Tongji Hospital, Tongji University School of Medicine, Shanghai 200065, China.
2. Department of Gastroenterology, Changhai Hospital, Second Military Medical University, Shanghai 200433, China.
3. Department of Gastroenterology, No. 411 Hospital of PLA, Shanghai 200434, China.
4. Division of Gastroenterology, Zhejiang Hospital, Hangzhou 310000, China.
* These authors contributed equally to this work.
Received 2015-4-29; Accepted 2015-8-25; Published 2015-10-16
Abstract
Pancreatic cancer (PC) is one of the most common cancers and has a poor prognosis due to late diagnosis and ineffective therapeutic multimodality. MicroRNAs (miRNAs, miRs) are a group of non-coding, small RNAs with active biological activities. In our investigation, human pancreatic cancer cell line Capan-2 were transfected with miR-222 mimics, inhibitors or their negative controls. Cell proliferation was assessed by Cell Counting Kit-8 (CCK-8), EdU incorporation assay and cell cycle determination by flow cytometry. MiR-222 and putative target gene expression levels including p27, p57 and PTEN were determined using quantitative reverse transcription polymerase chain reactions and Western blotting. Our results showed that miR-222 could lead to increased vitality and proliferative rate of Capan-2 cells, and also higher S-phase and lower G1-phase of cell cycle. Further, we found p57 at protein level, but not p27 nor PTEN, was regulated by miR-222 in Capan-2 cells. Finally, we co-transfected miR-222 inhibitor and p57 si-RNA into Capan-2 cells, and found that proliferation-suppressing effects of miR-222 inhibitor on Capan-2 cells could be partially reversed by silencing p57. Our results indicate that miR-222 controls Capan-2 cell proliferation by targeting p57. This study provides a novel idea for developing effective therapeutic strategy for PC patients through inhibiting miR-222.
Keywords: MicroRNA, MiR-222, Pancreatic cancer, Capan-2, Proliferation, P57.
Introduction
Pancreatic cancer (PC) is among cancers with worst prognosis and is also a leading cause of death caused by cancer in the United States. In addition, in other developed countries, PC is responsible for over 20% of deaths due to gastrointestinal malignancies [1]. Although recent diagnostic and therapeutic advances have greatly improved the survival of patients with PC, the overall prognosis of these patients is still extremely poor [1]. Overall, the molecular mechanisms responsible for the growth of pancreatic cancer are largely unclear [1].
MicroRNAs (miRNAs, miRs) are a group of non-coding, small RNAs with about 20-22 nucleotides in length, and act as gene regulators at posttranscriptional level through degradation of transcripts or inhibition of translation mainly by binding to 3'-untranslated region (3'-UTR) [2]. miRNAs have been implicated in numerous essential biological processes including development [3], proliferation [4, 5], differentiation [6], metabolism, apoptosis, necrosis, and autophagy [7]. The aberrant expressions of local as well as circulating miRNAs have been reported to be closely associated with a variety of diseases [8-10], which could thus be regarded as potential biomarkers for disease assessment [11-13]. In fact, multiple miRNAs such as miR-21, -29, -34a, -155, -196a, -215, -221, and -301a have been identified as potential regulators for pancreatic cancer [14-21]. Moreover, an upregulation of miR-222 has been reported in human pancreatic tumor tissues compared to normal tissues, which was associated with increased Ki-67 expression and poor prognosis in PC patients [22]. However, the functional and cellular mechanisms of miR-222 overexpression in the tumorigenesis of PC are poorly understood. Besides that, p27, p57 and phosphatase and tensin homolog (PTEN) are three potential target genes of miR-222 reported in other types of cancers [23, 24], however their functional relationship with miR-222 in PC remains unclear.
Therefore, in the present study, we aimed at investigating the role of miR-222 in regulating the proliferation of a human pancreatic cancer cell line Capan-2. Our data demonstrate that miR-222 is sufficient to promote the proliferation of Capan-2 cells while inhibition of miR-222 has inverse effect. P57 is a target gene responsible for the effects of miR-222 in Capan-2 cells.
Methods
Cell culture
Human pancreatic cancer cell line Capan-2 was purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). Capan-2 cells were maintained in RPMI-1640 medium (Hyclone, USA) supplemented with 10% fetal bovine serum (FBS, Hyclone, USA), 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco, USA). Cells were incubated at 37°C in a humidified atmosphere of 95% O2 and 5% CO2.
Cell transfection
The miR-222 mimics, inhibitors and their negative controls (NC) were purchased from RiboBio (China). Capan-2 cells were transfected with miR-222 mimics (50 nM), inhibitors (100 nM) or their negative controls for 48 h using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer's instructions. P57 siRNA was purchased from RiboBio (China). P57 siRNA (100 nM) was transfected into Capan-2 cells with similar method described above.
Cell counting kit-8 assays
The effects of miR-222 on the viability of Capan-2 cells were determined using a Cell counting kit-8 assay (CCK-8; Dojindo, Japan). Cells (1.5×105 cells per milliliter) were seeded in 96-well plates and were allowed to adhere overnight. After 48 h of transfection of miR-222 mimics or inhibitors, CCK-8 solution was added to each well and was incubated for 1 h at 37°C and the absorbance was measured at 450 nm using a spectrophotometer (Bio-Rad, USA).
EdU incorporation assays
Capan-2 cells were plated into 24-well plates at a density of 1.5×105 cells per milliliter and were allowed to adhere overnight in complete medium. Then cells were starved for 6 h with serum-free medium. After transfection, cells were incubated with 5-ethynyl-2'-deoxyuridine (EdU, RiboBio, China) for 6 h before staining. Cell proliferation was detected using Cell-Light™ EdU Cell Proliferation Detection Kit (RiboBio, China) following the manufacturer's instructions. Percentage of proliferative cells was determined by blinded quantitation of EdU-positive cells under fluorescent microscope (Leica, Germany) with 200X magnification.
Cell cycle analysis
Capan-2 cells were plated into 6-well plates at a density of 1.5×105 cells per milliliter and were allowed to adhere overnight in complete medium. Then cells were starved for 6 h with serum-free medium. Cells were harvested at 48 h post transfection of miR-222 mimics or inhibitors, and then were washed twice with PBS. After that, cells were fixed in 70% ethanol overnight at -20°C. Fixed cells were washed once in 4°C PBS, and then were stained with propidium iodide (PI) staining solution (50 µg/mL PI, 100 µg/mL RNase in PBS). PI-stained cells were analyzed for their DNA content by using a Beckman flow cytometry.
Quantitative reverse transcription polymerase chain reactions
Total RNA was isolated from Capan-2 cells using TRIZOL reagent (Invitrogen, USA). For quantitative miRNA analysis, the Bulge-LoopTM miRNA qPCR Primer Set (RiboBio) was used to determine the expression levels of miR-222 by qRT-PCRs with Takara SYBR Premix Ex TaqTM (TliRNaseH Plus) in CFX96TM Real-Time PCR Detection System. 5S ribosomal RNA (5S rRNA) was used as an internal control for miRNA template normalization. The relative expression level for each miRNA was calculated using the 2-ΔΔCt method.
Western blotting
Cells were lysed in RIPA buffer (Beyotime Institute of Biotechnology, China) containing protease inhibitor cocktails (Sigma). Equal amounts of protein (20 to 40 ug) were subjected to SDS-polyacrylamide gel and transferred onto polyvinylidene difluoride membranes. Standard western blot analysis was conducted using p57 primary antibody (Bioworld, dilution of 1:1000), p27 primary antibody (Bioworld, dilution of 1:1000) and PTEN primary antibody (Bioworld, dilution of 1:1000). Beta-actin (Bioworld, dilution of 1:5000) was used as a loading control. After incubation with the appropriate secondary antibodies, signals were visualized using the ECL plus Western blotting detection reagents (Bio-Rad) and the ChemiDoc XRS Plus luminescent image analyzer (Bio-Rad). Densitometric analysis of band intensity was performed using Imagelab software (Bio-Rad).
Target gene validation
P27, p57 and PTEN, three potential target genes of miR-222 reported in other types of cancer cells, were chosen as candidates in Capan-2 cells. Firstly, Capan-2 cells were transfected with miR-222 mimics, inhibitors or their negative controls. Forty-eight hours after the transfection, the protein levels of p27, p57 and PTEN were determined by Western blot. Secondly, as p27 and PTEN were found to be unchanged while p57 was endogenous regulated in Capan-2 cells by miR-222. Finally, the miR-222 inhibitor (100 nM) and p57 siRNA (75 nM) co-transfection was used to check if miR-222 take effects through p57 in Capan-2 cells.
Statistical analysis
All quantitative data were presented as the mean ± SEM. An independent Student T test or one-way ANOVA was conducted to evaluate the one-way layout data. If a significant difference was observed, Bonferroni's post-hoc test was conducted to identify groups with significant differences. P-values < 0.05 were considered significant. All statistical analyses were performed with the software package SPSS statistics19.0 (IBM, USA) for Windows.
Results
miR-222 controls the proliferation and cell cycle process in Capan-2 cells
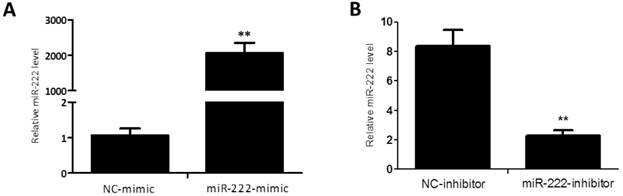
Capan-2 cells were transfected with miR-222 mimics, inhibitors or their negative controls for 48 h. qRT-PCRs were used to determine the effects of miR-222 mimics and inhibitors. As shown in Figure 1, miR-222 expression levels were significantly increased by miR-222 mimics while decreased by miR-222 inhibitors, confirming that miR-222 mimics and inhibitors successfully regulated miR-222 expression levels in Capan-2 cells.
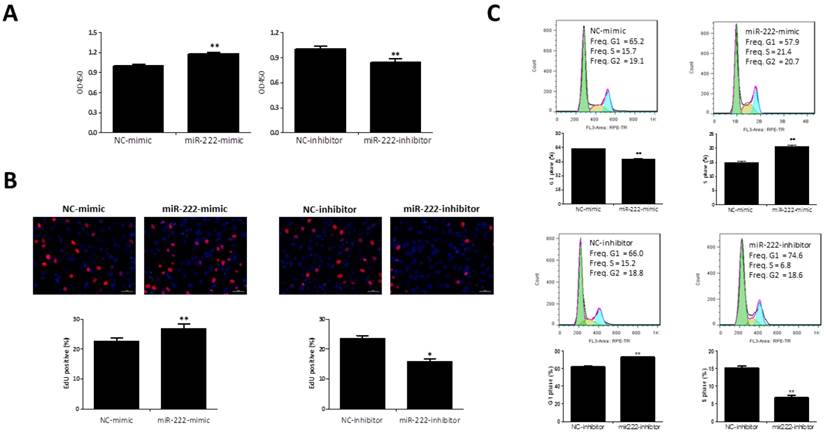
CCK-8 was added to the medium for 1 h and the presence of metabolically active viable cells was used as an index of cell proliferation. As showed by results based on CCK-8, miR-222 mimics could increase Capan-2 cell vitality while miR-222 inhibitors decreased that (Figure 2A).
To determine if this viability advantage was due to higher proliferation rate, we studied the effects of miR-222 in EdU incorporation and cell cycle regulation. The EdU assay confirmed that miR-222 mimics-transfected cells had a significant proliferative rate when compared with control cells, and miR-222 inhibitors reduced that (Figure 2B). Cell cycle analysis also further confirmed that miR-222 could accelerate cell cycle of Capan-2 cells by increasing S phase cells rate and decreasing G1 phase cells rate (Figure 2C).
P57 is regulated by miR-222 in Capan-2 cells
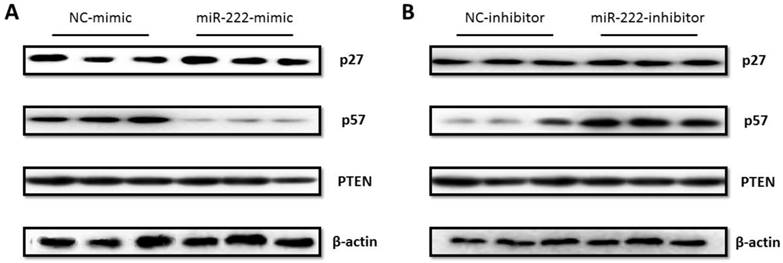
As p27 and p57 have been reported as putative target genes of miR-222 in other types of cancer cells [23, 24], we assessed the effects of miR-222 in endogenous expressions of p27 and p57 in Capan-2 cells by Western blotting, finding that the protein level of p57 but not p27 was endogenously regulated by miR-222 mimic and inhibitor (Figure 3). Furthermore, PTEN, another well-known target of miR-222, was not modified by miR-222 in Capan-2 cells (Figure 3). The results indicate that p57 is a target gene of miR-222 in Capan-2 cells.
Quantitative reverse transcription polymerase chain reactions confirm that miR-222 mimics and inhibitors take effects in Capan-2 cells. The expression level of miR-222 was significantly up-regulated in Capan-2 cells transfected with miR-222 mimics (A), while downregulated with miR-222 inhibitors (B) compared to negative control (NC) (n=6). *p<0.05, ** p<0.01 vs. NC.
Effects of miR-222 in the proliferation of Capan-2 cells. (A) CCK-8 assay (n=10) showed that Capan-2 cells transfected with miR-222 mimic showed an increased cell vitality while Capan-2 cells transfected with miR-222 inhibitor showed a decreased cell vitality than their respective negative controls (NC) (n=10). (B) The EdU positive cells rate was increased by transfection of miR-222 mimic, while miR-222 inhibitor treated Capan-2 cells had a lower EdU positive cells rate (n=6). (C) Decreased G1-phase cells rate and increased S-phase cells rate were observed in miR-222 mimic treated Capan-2 cells by flow cytometry, instead, G1-phase cells rate was increased and S-phase cells rate was decreased in miR-222 inhibitor treated Capan-2 cells (n=6). *p<0.05, ** p<0.01, vs. NC.
P57 was regulated by miR-222 in Capan-2 cells. (A) P57 protein level was decreased in miR-222 mimic transfected Capan-2 cells (A), while was increased in miR-222 inhibitor transfected Capan-2 cells (B). Neither PTEN nor p27 was regulated by miR-222 in Capan-2 cells at protein level (n=3).
P57 is responsible for effects of miR-222 in Capan-2 cells
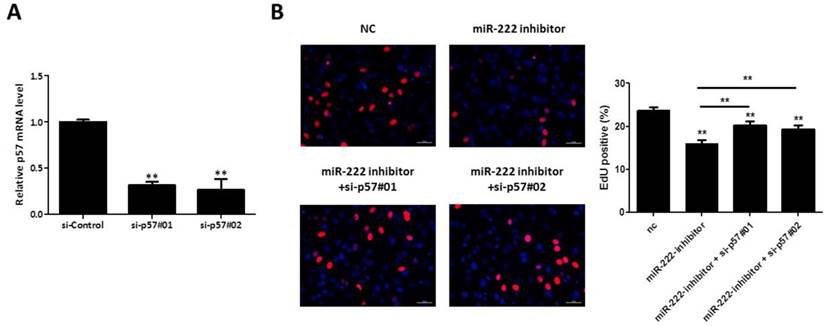
To further investigate if p57 is responsible for the effects of miR-222 in Capan-2 cells, we transfected p57 siRNA into Capan-2 cells, and the effects of p57 knockdown was synchronously confirmed by qRT-PCR (Figure 4A). We then co-transfected p57 siRNA (75nM) and miR-222 inhibitor, to determine whether or not miR-222 increases Capan-2 cells vitality by targeting p57. The EdU results showed that the proliferation-suppressing effects of miR-222 inhibitor in Capan-2 cells could be partially reversed by silencing p57 (Figure 4B), indicating that miR-222 promoted Capan-2 cell proliferation, at least in part, via p57 targeting.
Discussion
Although extensive efforts have been made into the elucidation of the molecular pathogenesis of PC, few effective treatment strategies for PC currently exist. Therefore, PC is still one of the most common cancer-related death causes and has the worst prognosis of any major malignancy, with less than 5% of patients alive 5 years after diagnosis. For these reasons, further understanding of aberrant regulatory mechanisms involved in PC progression is fairly critical. Here we provide novel data showing that miR-222 controls Capan-2 cell proliferation by targeting p57, indicating that inhibition of miR-222 might be a therapeutic way for PC.
Several studies have indicated that miR-222 was aberrantly expressed in multiple cancer types, including meningioma, prostate cancer, papillary thyroid cancer and breast cancer [25-29], and these results implied that miR-222 might be an important regulator for cancer. In vitro studies found that miR-222 led to enhanced cancer cell proliferation rate and migration [30-32], and these results indicated miR-222 as a potential onco-microRNA. However, a recent study demonstrated that miR-222 and its homologue miR-221 inhibited non-small cell lung cancer cell growth on 6 lung cancer cell lines [33], indicating the role of miR-222 in cancer might be an interesting duality. Importantly, miR-222 has previously been reported to be upregulated in human pancreatic tumor tissues compared to normal controls, which was associated with increased Ki-67 expression and reduced survival rate in PC patients [22]. Nevertheless, the functional and cellular mechanisms underlying the potential roles of miR-222 on the tumorigenesis of PC are largely unknown. Thus, in the current study, we further clarified the functional roles of miR-222 on the proliferation and cell cycle process of pancreatic cancer cells. Noteworthy, our data show that miR-222 is sufficient to induce the proliferation and the G1-to-S phase transition of human pancreatic cancer cell line Capan-2 in vitro.
P57 is a cyclin-dependent kinase (CDK) inhibitor, which is homologous to p27 and p21, belonging to the Cip/Kip family, and a well-established tumor suppressor [34, 35]. P57 was initially considered to inhibit cell cycle transition in cancer development, as tumor cells lacking p57 showed increased cell growth [36, 37]. As p57 is a well-known target gene of miR-222 [38], here we also further showed that p57, but not p27 and PTEN, was negatively regulated by miR-222 in Capan-2 cells. Importantly, inhibitory effect of miR-222 inhibitors on the proliferation of Capan-2 cells could be partially reversed by silencing p57, indicating that the effect of miR-222 on Capan-2 cell proliferation is, at least in part, mediated by p57.
Last but not least, given that miR-222 is essential for various cellular biological processes in different types of cancer, including cell proliferation, migration, apoptosis and others [3, 39], and that p57 has also been proved to regulate other aspects of cancer cells, for example, DNA methylation and histone modification [40, 41], it will also be of great interest to further study the impact of miR-222/p57 on the other biological processes of pancreatic cancer cells.
In conclusion, our study suggests that miR-222 controls the proliferation of Capan-2 cells, and this effect may be achieved by targeting p57. This study provides a novel idea for developing effective therapeutic strategy for PC through inhibiting miR-222 in PC patients.
Silencing P57 restored the proliferation-suppressing effect of miR-222 inhibitor on Capan-2 cells. (A) The p57 mRNA level was downregulated by p57 siRNA, as demonstrated by quantitative real-time PCR (n=6). (B) Silencing p57 could partially reverse the inhibitory effect of miR-222 inhibitor on the proliferation of Capan-2 cells (n=6). *p<0.05, ** p<0.01.
Acknowledgements
This work was supported by the grants from National Natural Science Foundation of China (81070343 and 81370559 to C. Yang; 81400635 to F. Wang), Joint Projects in Major Diseases funding from Shanghai Municipal Commission of Health and Family Planning (2014ZYJB0201 to C. Yang), Joint Projects for Novel Frontier Technology in Shanghai Municipal Hospital from Shanghai Municipal Commission of Health and Family Planning (SHDC1204122 to C. Yang), Shanghai Medical Guide Project from Shanghai Science and Technology Committee (14411971500 to F. Wang), grants from Chinese Foundation for Hepatitis Prevention and Control (TQGB20140141 to F. Wang) and funds from Shanghai Innovation Program (12431901002 to C. Yang).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Jemal A, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ. Cancer statistics, 2003. CA Cancer J Clin. 2003;53:5-26
2. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97
3. Liu X, Xiao J, Zhu H, Wei X, Platt C, Damilano F. et al. miR-222 Is Necessary for Exercise-Induced Cardiac Growth and Protects against Pathological Cardiac Remodeling. Cell Metab. 2015;21:584-95
4. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D. et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-8
5. Li J, Bei Y, Liu Q, Lv D, Xu T, He Y. et al. MicroRNA-221 is required for proliferation of mouse embryonic stem cells via P57 targeting. Stem Cell Rev. 2015;11:39-49
6. Xiao J, Liang D, Zhang H, Liu Y, Zhang D, Liu Y. et al. MicroRNA-204 is required for differentiation of human-derived cardiomyocyte progenitor cells. J Mol Cell Cardiol. 2012;53:751-9
7. Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823-6
8. Xiao JJ, Shen B, Li J, Lv DC, Zhao YY, Wang F. et al. Serum microRNA-499 and microRNA-208a as biomarkers of acute myocardial infarction. Int J Clin Exp Med. 2014;7:136-41
9. Roderburg C, Luedde T. Circulating microRNAs as markers of liver inflammation, fibrosis and cancer. J Hepatol. 2014;61:1434-7
10. Xiao J, Jing ZC, Ellinor PT, Liang D, Zhang H, Liu Y. et al. MicroRNA-134 as a potential plasma biomarker for the diagnosis of acute pulmonary embolism. J Transl Med. 2011;9:159
11. Li J, Xu J, Cheng Y, Wang F, Song Y, Xiao J. Circulating microRNAs as mirrors of acute coronary syndromes: MiRacle or quagMire? J Cell Mol Med. 2013;17:1363-70
12. Giray BG, Emekdas G, Tezcan S, Ulger M, Serin MS, Sezgin O. et al. Profiles of serum microRNAs; miR-125b-5p and miR223-3p serve as novel biomarkers for HBV-positive hepatocellular carcinoma. Mol Biol Rep. 2014;41:4513-9
13. Xu J, Zhao J, Evan G, Xiao C, Cheng Y, Xiao J. Circulating microRNAs: novel biomarkers for cardiovascular diseases. J Mol Med (Berl). 2012;90:865-75
14. Chen C, Zhang Y, Zhang L, Weakley SM, Yao Q. MicroRNA-196: critical roles and clinical applications in development and cancer. J Cell Mol Med. 2011;15:14-23
15. Choi DH, Park SJ, Kim HK. miR-215 overexpression distinguishes ampullary carcinomas from pancreatic carcinomas. Hepatobiliary Pancreat Dis Int. 2015;14:325-9
16. Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L. et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009;4:e6816
17. Kwon JJ, Nabinger SC, Vega Z, Sahu SS, Alluri RK, Abdul-Sater Z. et al. Pathophysiological role of microRNA-29 in pancreatic cancer stroma. Sci Rep. 2015;5:11450
18. Moriyama T, Ohuchida K, Mizumoto K, Yu J, Sato N, Nabae T. et al. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol Cancer Ther. 2009;8:1067-74
19. Park JK, Lee EJ, Esau C, Schmittgen TD. Antisense inhibition of microRNA-21 or -221 arrests cell cycle, induces apoptosis, and sensitizes the effects of gemcitabine in pancreatic adenocarcinoma. Pancreas. 2009;38:e190-199
20. Wang P, Zhu CF, Ma MZ, Chen G, Song M, Zeng ZL. et al. Micro-RNA-155 is induced by K-Ras oncogenic signal and promotes ROS stress in pancreatic cancer. Oncotarget. 2015 [Epub ahead of print]
21. Xia X, Zhang K, Cen G, Jiang T, Cao J, Huang K. et al. MicroRNA-301a-3p promotes pancreatic cancer progression via negative regulation of SMAD4. Oncotarget. 2015 [Epub ahead of print]
22. Lee C, He H, Jiang Y, Di Y, Yang F, Li J. et al. Elevated expression of tumor miR-222 in pancreatic cancer is associated with Ki67 and poor prognosis. Med Oncol. 2013;30:700
23. Mardente S, Mari E, Massimi I, Fico F, Faggioni A, Pulcinelli F. et al. HMGB1-Induced Cross Talk between PTEN and miRs 221/222 in Thyroid Cancer. Biomed Res Int. 2015;2015:512027
24. Zhong C, Ding S, Xu Y, Huang H. MicroRNA-222 promotes human non-small cell lung cancer H460 growth by targeting p27. Int J Clin Exp Med. 2015;8:5534-40
25. Ludwig N, Kim YJ, Mueller SC, Backes C, Werner TV, Galata V. et al. Posttranscriptional deregulation of signaling pathways in meningioma subtypes by differential expression of miRNAs. Neuro Oncol. 2015 doi:10.1093/neuonc/nov014
26. Song C, Chen H, Wang T, Zhang W, Ru G, Lang J. Expression profile analysis of microRNAs in prostate cancer by next-generation sequencing. Prostate. 2015;75:500-16
27. He HC, Han ZD, Dai QS, Ling XH, Fu X, Lin ZY. et al. Global analysis of the differentially expressed miRNAs of prostate cancer in Chinese patients. BMC Genomics. 2013;14:757
28. Lee JC, Zhao JT, Clifton-Bligh RJ, Gill A, Gundara JS, Ip JC. et al. MicroRNA-222 and microRNA-146b are tissue and circulating biomarkers of recurrent papillary thyroid cancer. Cancer. 2013;119:4358-65
29. Zhong S, Li W, Chen Z, Xu J, Zhao J. MiR-222 and miR-29a contribute to the drug-resistance of breast cancer cells. Gene. 2013;531:8-14
30. Li Y, Liang C, Ma H, Zhao Q, Lu Y, Xiang Z. et al. miR-221/222 promotes S-phase entry and cellular migration in control of basal-like breast cancer. Molecules. 2014;19:7122-37
31. Sun Y, Zhang B, Cheng J, Wu Y, Xing F, Wang Y. et al. MicroRNA-222 promotes the proliferation and migration of cervical cancer cells. Clin Invest Med. 2014;37:E131
32. Yang X, Yang Y, Gan R, Zhao L, Li W, Zhou H. et al. Down-regulation of mir-221 and mir-222 restrain prostate cancer cell proliferation and migration that is partly mediated by activation of SIRT1. PLoS One. 2014;9:e98833
33. Yamashita R, Sato M, Kakumu T, Hase T, Yogo N, Maruyama E. et al. Growth inhibitory effects of miR-221 and miR-222 in non-small cell lung cancer cells. Cancer Med. 2015;4:551-64
34. LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS. et al. New functional activities for the p21 family of CDK inhibitors. Genes & Development. 1997;11:847-862
35. Matsuoka S, Edwards MC, Bai C, Parker S, Zhang PM, Baldini A. et al. P57(KIP2), A Structurally distinct member of the P21(CIP1) CDK inhibitor family, is a candidate tumor-suppressor gene. Genes & Development. 1995;9:650-662
36. Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA. et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651-61
37. Riccio O, van Gijn ME, Bezdek AC, Pellegrinet L, van Es JH, Zimber-Strobl U. et al. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27(Kip1) and p57(Kip2). Embo Rep. 2008;9:377-83
38. Liu X, Cheng Y, Yang J, Xu L, Zhang C. Cell-specific effects of miR-221/222 in vessels: molecular mechanism and therapeutic application. J Mol Cell Cardiol. 2012;52:245-55
39. Medina R, Zaidi SK, Liu C-G, Stein JL, vanWijnen AJ, Croce CM. et al. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 2008;68:2773-80
40. Besson A, Dowdy SF, Roberts JM. CDK inhibitors: cell cycle regulators and beyond. Dev Cell. 2008;14:159-69
41. Guo H, Tian T, Nan K, Wang W. p57: A multifunctional protein in cancer (Review). Int J Oncol. 2010;36:1321-9
Author contact
![]() Corresponding authors: Prof Changqing Yang, Division of Gastroenterology and Hepatology, Digestive Disease Institute, Shanghai Tongji Hospital, Tongji University School of Medicine, 389 Xin Cun Road, Shanghai 200065, China. Tel.: 0086-21-66111604; Fax: 0086-21-66111604; E-mail: changqingyang_tjcom. Prof Zhaoshen Li, Department of Gastroenterology, Changhai Hospital, Second Military Medical University, 168 Chang Hai Road, Shanghai 200433, China. Tel.: 0086-21-31161111; Fax: 0086-21-31161111; E-mail: zhslinet
Corresponding authors: Prof Changqing Yang, Division of Gastroenterology and Hepatology, Digestive Disease Institute, Shanghai Tongji Hospital, Tongji University School of Medicine, 389 Xin Cun Road, Shanghai 200065, China. Tel.: 0086-21-66111604; Fax: 0086-21-66111604; E-mail: changqingyang_tjcom. Prof Zhaoshen Li, Department of Gastroenterology, Changhai Hospital, Second Military Medical University, 168 Chang Hai Road, Shanghai 200433, China. Tel.: 0086-21-31161111; Fax: 0086-21-31161111; E-mail: zhslinet

 Global reach, higher impact
Global reach, higher impact