Impact Factor
ISSN: 1837-9664
J Cancer 2015; 6(12):1236-1244. doi:10.7150/jca.12626 This issue Cite
Research Paper
A Fraction of CD133+ CNE2 Cells Is Made of Giant Cancer Cells with Morphological Evidence of Asymmetric Mitosis
1. Department of Pathology, Key Laboratory of Major Obstetrics Diseases of Guangdong Province, the Third Affiliated Hospital, Guangzhou Medical University, Guangzhou, China
2. Cancer Research Institute, Southern Medical University, Guangzhou 510515, China.
3. Department of Pathology, the University of Texas MD Anderson Cancer Center, Houston, TX, USA.
# These authors contributed equally to this work.
Received 2015-5-7; Accepted 2015-8-31; Published 2015-10-20
Abstract
CD133 has been suggested as a broad-spectrum marker for cancer stem cells(CSCs). The present study investigated the expression of CD133 in biopsy tissues of nasopharyngeal carcinoma (NPC), NPC cell lines and the immortalized cell line NP69 by immunohistochemistry, flow cytometry and qRT-PCR. CD133+ cancer cells were isolated using magnetic-activated cell sorting technology. The study demonstrated that CD133+ cells are rare in NPC tissues and cell lines and that their self-renewal and proliferation abilities are stronger than those of CD133- cells and suggested that CD133+ NPC cells have characteristics of cancer stem cells. We further observed CD133+ cancer cells using a light microscope and scanning electron microscope. Generally, CD133+ cells are small, regular and round with small microvilli. On the other hand, CD133- cells are more polymorphic and larger with long micromicrovilli. Additionally, in some fields, several giant cancer cells (GCCs) in the CD133+ cell group were identified under the light microscope. Most of them were polynuclear cells. Under the scanning electron microscope, we found indefinite regular small bodies on the surface of or surrounding the giant cancer cells, some of which appeared to be creeping out the parental cells. This phenomenon was not observed in the CD133- cell groups. Through comparison with descriptions of apoptotic bodies in the literature and from the results of the acridine orange test, we propose that some of the small bodies are daughter cells of the GCCs. This phenomenon is a mode of division of cancer cells called neosis, or budding, which is a form of reproduction for simple organisms. Budding is satisfied with the rapid speed of tumor development. GCCs could be isolated by CD133 beads because the daughter cells have stem-cell characteristics and express stem-cell markers.
Keywords: CD133, stem cell, giant cancer cells, budding, nasopharyngeal carcinoma
1. Introduction
Cancer stem cells (CSCs) have been identified in different hematopoietic and solid tumors, such as acute myelogenous leukemia (AML) [1], brain[2], breast[3], lung[4], colon[5] and prostate tumors[6]. CSCs possess unlimited potential for self-renewal, differentiation and resistance against [7] drugs. Additionally, CSCs can proliferate clonally into “spheres” [5, 8, 9].
Generally, self-renewal and differentiation of CSCs have been thought to occur by asymmetrical division, wherein one daughter cell is retained as the stem cell and the other daughter cell begins the process of differentiation. However, if asymmetric division is the only means of multiplying, how can CSCs maintain their numbers by the process of continuous growth and passage? Recently, it has been suggested that CSC divisions are not only asymmetrical but also symmetrical[10]. Some researchers have proposed another type of division, neosis, or budding, as a way to generate new cells[11, 12]. It means some polyploid giant cancer cells (PGCCs) in tumors produce many “Raju” cells. These “Raju” cells have some stem cell functions and divide into tumor cells.
The isolation of CSCs from non-CSCs is very important to identify tumor origin and study metastasis. CD133, a glycoprotein with a molecular weight of 117 kDa and 5 transmembrane domains, is a special biomarker for many types of tumors[13]. It has been suggested that CD133 may be a broad-spectrum marker for CSCs[14].
Nasopharyngeal carcinoma (NPC) is one of the most common types of malignancies in Southeast Asia. However, CSC studies of NPC have not been systematically performed. In the present study, CD133 protein was detected in NPC tissues and cell lines using immunohistochemistry and qRT-PCR. A CD133+ cell population was isolated from the NPC cell line CNE2 using magnetic activated cell sorting (MACS) technology. The ability of these CD133+ cells to self-renew and differentiate was compared to that of CD133- cells. Additionally, their microstructures and ultramicrostructures were compared using scanning. We also attempted to observe the budding division of GCCs.
2. Materials and methods
2.1. Paraffin biopsies
A total of 39 NPC samples were obtained from archives in the pathology department of Guangzhou Medical University, the Third Affiliated Hospital. The samples were from 19 males and 20 females who were 23-73 years old. All of the paraffin blocks were cut into 2 slides with depths of 3 µm for immunohistochemistry. Written informed consent was obtained from all of these patients to publish their case details before inclusion in the study. This research was approved by the Ethics Committee of the Third Affiliated Hospital, Guangzhou Medical University, Guangzhou, China.
2.2. Cell culture
The CNE2, HONE1 and SUNE1 cell lines were obtained from the cancer center at Sun Yat-sen University[15]. The cell culture medium used was RPMI-1640 (Guangzhou Land Bio, China) supplemented with 5% fetal calf serum (HyClone, USA). The cells were cultured in a humidified incubator with 5% CO2 at 37˚C. The immortalized cell line NP69 was also obtained from the cancer center at Sun Yat-sen University. The NP69 cells were cultured in keratinocyte-SFM (KSFM) (Gibco, USA). Additionally, for immunocytochemical staining, cells were dropped on high pressure-treated glass slides and cultured in incubators at 37˚C for 24 hours. The slides were washed with PBS after the cells were attached tightly on the slide surfaces.
2.3. Immunohistochemical and immunocytochemical staining
Immunohistochemical staining (IHC) and immunocytochemical staining were performed using the streptavidin-peroxidase method. The tissue sections were dewaxed, hydrated, and subjected to microwave antigen retrieval in 0.01 M sodium citrate buffer (pH 6.0) for 10 min. After blocking with normal sheep serum, the tissue and cell slides were incubated overnight at 4˚C in a humidity chamber with CD133/1 antibody (Miltenyi Biotec, Germany) diluted 1:50, Cytokeratin (Santa Cruz, CA) followed by IgG biotin labeling for 10 min. The signal was detected with a labeled streptavidin-peroxidase system in the presence of the chromogen 3,30-diaminobenzidine. The cellular localization of CD133 was membrane and cytoplasm, and that of cytokeratin was membrane.
Evaluation of staining. The immunohistochemically stained tissue were reviewed separately by two pathologists. Expression of CD133 and Cytokeratin was independently evaluated and scored. The scoring standards were as follows: according to cell staining, dark brown was scored 3, brown yellow scored 2, light yellow scored 1 and nonstaining scored 0; according to the proportion of positive cells in a field of vision, >75% was scored 4, 51% -75% scored 3, 11%-50% scored 2, 1%- 10% scored 1, and negative scored 0. Two scores were multiplied, in the end, the scores 0-2 were considered as negative, and ≥3 were considered as positive expression.
2.4. Western Blot Analysis
Western blot analysis was performed as previously described[16]. The following primary antibodies were used: CD133(1:500,ab16518,Abcam, Cambridge, MA, USA)and β-actin (1:30,000; Sigma, New York, USA). All horseradish peroxidase-conjugated secondary antibodies were purchased from DAKO (1:20,000). The density of the western blot signals was measured with Image J software.
2.5. Flow cytometry
Flow cytometry- Established CNE2, HONE-1and SUNE-1 stable cell lines (1 × 106) were detached by treatment with 0.25% trypsin-EDTA (Invitrogen) and washed once with phosphate-buffered saline. The cells were then resuspended in 100 μl Staining Buffer (eBioscience, San Diego, CA) containing 1% fetal bovine serum (FBS; Gibco BRL, Grand Island, NY) and place on ice for 20 min to block Fc receptors. After incubating with primary phycoerythrin (PE) anti-human CD133 for another 45 min on ice in dark, the cells were washed twice with 1 ml ice-cold Staining Buffer and centrifuged (400×g) for 5 min at 4℃. Cells resuspended in 0.5 ml 2% formaldehyde fixation buffer were analyzed using a FACS Calibur flow cytometer and CellQuest software (BD Biosciences, San Jose, CA). All flow cytometry results were obtained from three independent experiments performed in triplicate.
2.6. CD133 cell sorting using immunomagnetic beads
A single cell suspension of 1×108 CNE2 cells was used for cell sorting. Cells were incubated with 50 µl of FcR blocking agent and CD133/l (AC-133)-biotin immunomagnetic beads (Miltenyi Biotec, Germany) for 10 min at 4℃. For immunofluorescence or flow cytometric analysis, cells were subsequently incubated with 50 µl CD133/2(293C3)-PE for 5 min at 4℃ in a dark environment. A MACS cell separation column (MS column) was used to retain the CD133+ cells linked to the beads. Unlabeled cells (CD133- cells) were washed by 3×500 µl degassed buffer and collected. MS column was removed from the separator and placed on a new collection tube. 1 Ml buffer was pipetted onto the MS column, and the magically labeled CD133+ cells was flushed out by firmly applying the plunger supplied with the column. The labeled CD133+ cells were eventually obtained by centrifugation and resuspension in RPMl-1640 (with 5% inactivated fetal calf serum) or serum-free medium. CD133+ and CD133- cells were harvested, and their purity was evaluated by standard flow cytometric analysis, immunofluorescence and qRT-PCR. They were evaluated for proliferation and sphere formation activity.
2.7. Immunofluorescence
Equal numbers of CD133+ and CD133- cells incubated with CD133/2(293C3)-PE were placed in the center of petri dishes for 20 min. When they sank to the bottom, the cells were observed under a confocal microscope.
2.8. Quantitive real-time PCR
The relative quantification of CD133 messenger RNA was performed by TaqMan technology, using the ABI PRISM 7900 DNA sequence detection system and ready-to-use primers/probe mixes (Applied Biosystems). The CD133 probes were as follows: P1: 5'-GCACTCTATACCAAAGCGTCAA-3' and P2: 5'-CTCCCATACTTCTTAGTTTCCTCA-3'. Human GAPDH was used as the reference gene for the amplifications.
2.9. Colony formation in soft agar
CD133+ cells, CD133-cells and unsorted CNE2 were disassociated and plated at a density of 200 cells/6 cm dish containing RPMI1640 (with 10% inactivated fetal calf serum). On day 14, cultured cells were stained with crystal violet and observed. The number of colonies was counted in triplicate.
2.10. Sphere formation
For the subculturing of suspended cell spheres, CD133+ and CD133- cells (lx103/ml) were cultured in 10 ml of culture butter in serum-free DMEM-F12 medium with bFGF (10 ng/ml), EGF (20 ng/ml), B27 (10 μl/ml) and insulin (5 μg/ml). The cells were cultured in 5% CO2 at 37℃ for 7 days, and the culture medium was replaced every other day.
2.11. Scanning electron microscopy (SEM)
The main steps were as follows: CD133+ and CD133- cell suspensions isolated by MACS were fixed in 2.5% glutaraldehyde for 5-6 days and washed with 0.2 mol/L PBS (pH7.4). The cells were fixed with 2.5% glutaraldehyde again for 60 min at 4˚C and washed with 0.2 mol/L PBS and distilled water. The washed cells were dehydrated using ethanol at increasing concentrations of 50%, 70%, 95%, and 100% for 5 min at each concentration. After dehydration, the cells were placed into ethanol-isoamyl acetate (1:1) and 100% isoamyl acetate for 5 min each, and then dropped into isoamyl acetate for 15 min. Finally, the specimens were dried on critical point and coated with electrically conducting material and observed using SEM.
2.12. Acridine orange/ethidium bromide (AO/EB) assay
AO and EB, 1 PPM (10-4%) respectively, were mixed in PBS. CD133+ cells were suspended at a normal counting concentration (1×106/L) and mixed with the AO/EB mixture (1:1). The mixture was placed on a hemocytometer slide and observed under a confocal microscope with 510 nm excitation light. Different states would emit different color light: live cells - green; early apoptotic cells - bright green nuclear staining with dense plaques or fragments; late apoptotic cells - orange with dense plaques or fragments; and necrotic cells - uniform red. Additionally, the cells that were undergoing apoptosis displayed characteristic membrane blebbing (bubble formation).
2.13. Statistical analysis
The statistical software SPSS13.0 was used for data processing. Data are expressed as the means ± SD. An independent sample t-test was used for performing comparisons between two groups. One-way ANOVA with Tamerlane was used for comparisons between more than two groups. A P-value of 0.05 was considered to indicate a statistically significant result.
3. Results
3.1. CD133 expression in NPC biopsies and cell lines
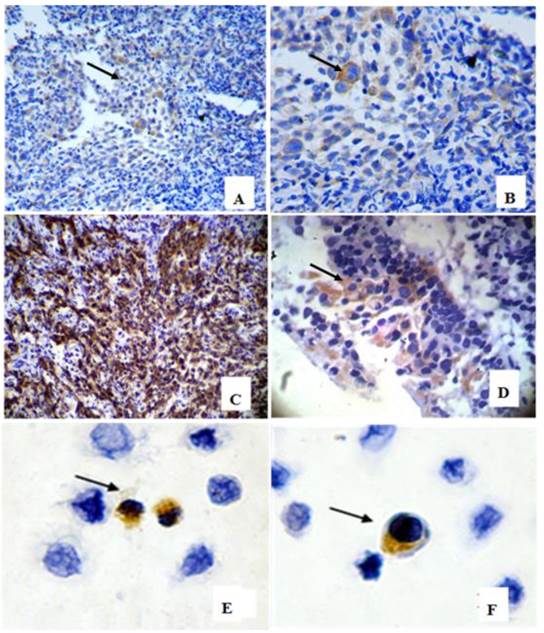
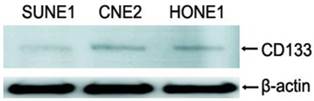
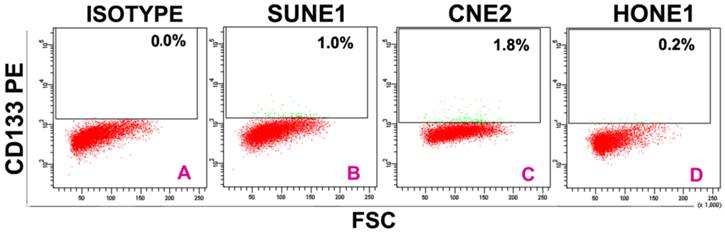
In 39 NPC cases, 8 cases showed CD133+ expression in some cells only in the marginal areas of a few cancer nests, the proportion of positive cells was about 15%(Figure 1A-B). The intensity of positive expression was brown yellow, and the total score of CD133 staining was 3. To verify that CD133+ cells were cancer cells, and not those mesenchymall cells arrounding cancer nests such as fibroblasts, macrophages, and lymphocytes which sometimes share similar morphological features with cancer cells, cytokeratin staining was done. Cytokeratin is a biomarker which extensively expressed on epithelial cells and carcinomas originated from them. The results verifed CD133+ cells located in cytokeratin positive cancer nests (Figure 1C). Meanwhile, some cells of nomal epithelial layers also expressed CD133 in 2 NPC cases (Figure 1D). In NP69, an immortalized cell line, several CD133+ cells were observed (Figure 1E-F). Analyses of CD133+ cells in SUNE1, CNE2 and HONE1 by Western Blot verified CD133 protein was expressed with different level in these three cell lines (Figure 2). Analyses of CD133+ cells in them by flow cytometry, CD133+ Cells were 0.9%, 1.8% and 0.2% (Figure 3), respectively.
CD133 detection in nasopharyngeal epithelium, cancer nests and NP69 cell line. (arrow heads show CD133 positive cells). A-B: CD133+ cells in margin areas of cancer nests( A×200, B×400); C:Cytokeratin positive expression verified CD133+ cells located in cancer nests(×200). D: Scattered CD133+ cells in epithelial layers( ×400); E-F:CD133 positive expression in NP69 cell line( ×400).
CD133 detected in NPC cell lines by Western blot, it showed that CD133 protein was detected with low levels in SUNE1, CNE2 and HONE1.lane1:SUNE1; Lane2:CNE2; Lane3:HONE1.
CD133+ cells isolated from SUNE1, CNE2 and HONE1 by flow cytometry. A:Isotype contrast;B:SUNE1;C:CNE2;D:HONE1.Among three line cells, the rate of CD133+ cells (1.8%) in CNE2 was the highest, and that (0.2%)in HONE1 the lowest.
3.2. CD133+ cells in CNE2
The cells were analyzed after immunomagnetic sorting with CD133 beads. Three independent measurements were taken before and after sorting. The average percentage of CD133+ cells in CNE2 was 2.0±0.035%. Then qRT-PCR for CD133 were performed.
qRT-PCR detection for the same number of CD133+ and CD133- cells was performed, △Ct values of CD133+ cells and CD133- cells were 10.160 ± 0.572 and 13.100 ± 0.321, respectively. The value of 2-△△Ct was 7.656, P=0.001 (Supplemental figure 1, Supplemental table 1).
3.3. Colony formation efficiency (CFE)
A significant difference was found in CFE among three lines of unsorted CNE2, CD133+ cells and CD133- cells by three independent tests. After counting colony formation units, mean values were compared by one-way ANOVA (F=68.035, P=0.000) and CFE between three cell lines was significantly different. Through Fisher's Least Significant Difference( LSD) multiple comparison, it was found that the CFE of the CD133+ line was higher than that of unsorted CNE2 cells (P=0.000). The CFE of unsorted CNE2 cells was higher than that of the CD133- line (P=0.019). The tests verified that the CFE of CD133+ cells was highest and that of CD133- cells was the lowest (Supplemental figure 2, Supplemental table 2).
3.4. Sphere formation assay
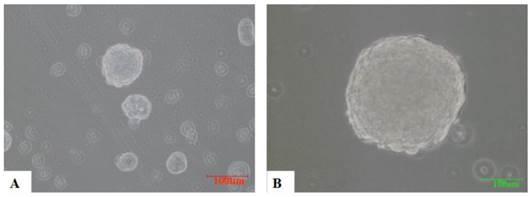
A total of 1×103/ml of CNE2, CD133+ and CD133- cells were cultured in a suspension of serum-free culture medium containing various growth factors, such as EGF. From the fourth day, CD133+ cells gradually formed spherical colonies of different sizes and irregular shapes. Most of the CD133- cells eventually died in the same serum-free medium without obvious sphere colony formation (Figure 4).
3.5. Observation under light microscope
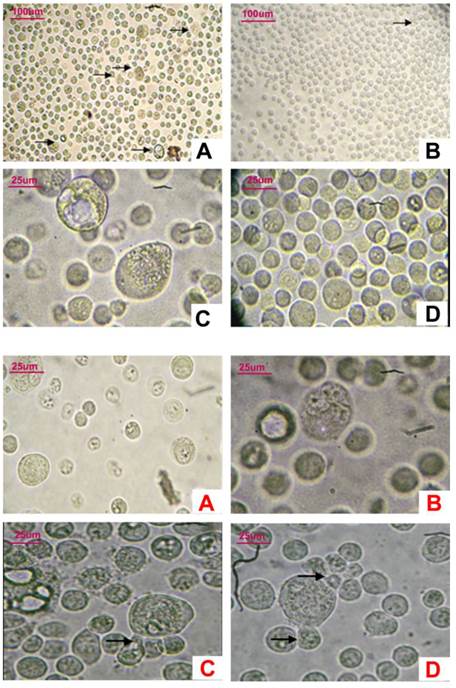
The most distinctive characteristic of the CD133+ cells was that many giant cells were dispersed among small cells. The average volume of these giant cells was 5-6 times that of the small cells. Cells in ten randomly selected high power fields were counted; approximately 1-2 large cells were discovered in every field. Polykaryon could be found in most of the giant cells. Some small nucleus-containing bodies were found surrounding some giant cells. However, most of the CD133- cells were similar and small in size (Figure 5-1,-2).
3.6. Observation under SEM
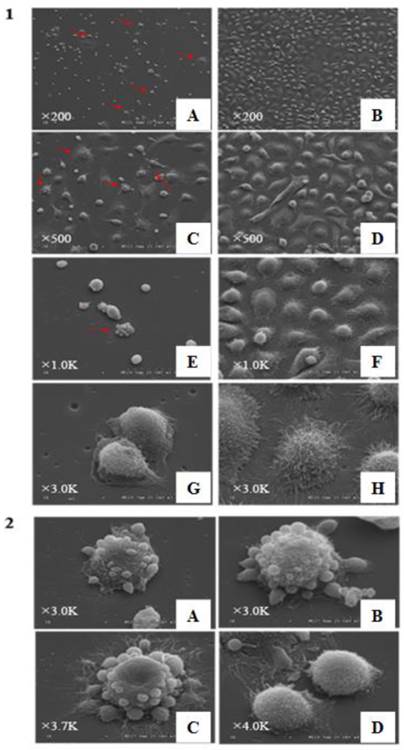
CD133+ and CD133- cells appeared different under SEM. In CD133+ cells, giant mother cells with some small spherical bodies were found, reminiscent of budding cells. The other cells were generally small and round and had short microvilli. Some bodies seemed to have originated from the parental giant cells. On the other hand, the CD133- cells were larger and polygonal with long microvilli sticking tightly to the floor. In CD133- cells, the budding phenomenon was not observed (Figure 6-1,-2).
3.7. Acridine orange/ethidium bromide (AO/EB) assay
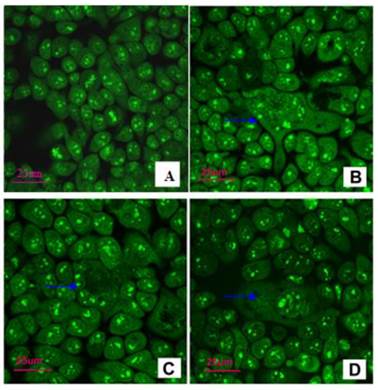
In CD133- cells, the sizes of the cells were uniform and clumpy chromatin was observed in the nucleus with bright green color. However, in CD133+ cells, GCCs were mixed with small cancer cells. GCCs possessed several nuclei. The cytoplasm of some GCCs was protuberant (Figure 7).
the sphere formation of CD133+ cells (×200). 3000/ml CD133+ cells and CD133- cells were cultured with KSFM for 7 days respectively. From the fourth day, spheres were formed with different sizes in CD133+ cells(A,B). CD133- cells have not formed obvious spheres and died eventually(not shown).
5-1, Observation of CD133+ cells and CD133- cells under light microscope. A,C: In CD133+ cells, some giant cancer cells could be seen among little cells(A:×100,C×400); B,D: In CD133-cells, cells were similar in size, giant cells hardly could be seen (B: ×100,D×400). 5-2, Observation of giant CD133+ cells under light microscope (×400). A-D: Several nuclei in CD133+ giant cells. Surrounding some giant cells, several little bodies were budding (→).
6-1, Observation of CD133+ cells and CD133- cells under scanning electroscope with different magnifications. A,C,E,G and I are images of CD133+ cells; B, D, F, and H are appearances of CD133- cells. a lot of giant cancer cells“budding” phenomena could be seen in CD133+ cells(red arrow heads), while CD133- cells almost hardly could be seen these phenomena. Besides this, sizes of most of CD133+ cells were small and round with short villi and CD133- cells were larger and heteromorphic with long villi (A-B,×200, C-D:×500, E-F:×1.0K, G-H: ×3.0K). 6-2, Giant cells with different shapes in CD133+ cells under scanning electroscope. A-C are some giant cells; D, some little bodies, “Raju cells”, were creeping off the parent giant cells(A-B:×3.0K, C:×3.7K, D:×4K).
4. Discussion
Cancer stem cells have been studied in most cancers. CD133 is universally recognized as a special biomarker for CSCs. CSC research in nasopharyngeal carcinoma has been reported in the literature. We have found label-retaining cells (LRCs) that possess stem cell characteristics. After cultivation for 14 days, the percentage of LRCs was 0.67±0.32% and there were approximately 0.55% sporadic LRCs in nude mice xenografts[7]. Likewise, another research of our group has discovered approximately 0.3% label-retaining stem cells in nude mice with human NPC xenograft tumors[17]. In 2012, the group further found ALDH1 could be a novel stem cells marker and a valuable predictor of poor survival NPC[18]. Besides of our studies, Wang et al [15] isolated side population cells (SP cells) by Hechs 33342 in NPC cell lines, and these cells show CSC functions, such as high tumorigenic ability and some chemotherapy tolerance. Recently, it was found that CNE2 cells (a NPC cell line) contain a small population of CD133+ cells with a strong potential for self-renewal, proliferation and differentiation, suggesting that these CD133+ cells are NPC stem cells[19].
Observation under confocal microscope after AO/EB staining. A: CD133- cells: the uniform sizes and clumpy chromatin in the nucleus with bright green color (×100). B-D: CD133+ cells, GCCs (blue →) were mixed with small cancer cells. GCCs possessed several nuclei, and cytoplasm of some GCCs was protuberant (×400).
It has been primarily reported that stem cells are rare and distributed in the margins of cancer nests and the basal layer of epithelium [5, 20, 21]. In the present study, CD133 expression was found in the cells of margin areas of a few cancer nests and in the basal cells of the epithelial layer in biopsies of NPC and nasopharyngitis by immunohistochemistry. Additionally, several CD133+ cells could be found in the immortalized cell line NP69. PCR showed CD133 amplification in the cancer cell lines CNE2, 6-10B and CNE2. To test if CD133+ cells possess proliferative and self-renewal capacity of CSCs, CD133+ and CD133- cells separated by MACS were used to measure colony formation efficiency and sphere formation assay. The results showed that the CFE of CD133+ cells was much higher than that of CD133- cells and that only CD133+ cells could form spheres. In accord with the literature, we verified that CD133+ cells are NPC cancer stem cells.
When CD133+ cells and CD133- cells were observed under a light microscope, a strange phenomenon was discovered. In the CD133+ cells, some GCCs were found against a background of small cells. These giant cells were approximately 1-2/HPF, and most of them were polykaryon. Some small nuclei-containing bodies were found surrounding some giant cells. On the other hand, in CD133- cells, this phenomenon was hardly observed, and all cells were almost the same size. To further validate this morphological difference, we collected these two groups of cells and observed them under SEM. Under SEM, most of the cells were uniformly small and round with very short microvilli. On the other hand, the size of CD133- cells was generally larger than small CD133+ cells, and they were polygonal and tightly sticking to the floor with long microvilli. Conclusively, appearances of CD133- cells under SEM showed that they had differentiated characteristics and verified that they were not stem cells. At the same time, the morphological manifestation of small cells in the CD133+ population could be explained as undifferentiated stem cells. Surprisingly, in CD133+ cells, we also found giant cells and several little bodies on their surface or surrounding them under SEM, similar to the observations under a light microscope. Some bodies seemed to have budded from the giant cells and had microvilli sticking to the floor. However, this phenomenon was hardly observed in the CD133- cells.
GCC presence is usually thought to indicate a state of senescence in somatic cells and cancer cells, which do not respond to growth stimulating signals[22-24]. How can the presence of GCC found using light microscopy and SEM in CD133+ stem cells be explained? Symmetrical and asymmetrical divisions are the two classical modes of cell division in eukaryotes. Usually, CSCs could divide through these two processes. Nevertheless, polyploid giant cells have been found to field small daughter cells before death[25, 26]. In 2004, this phenomenon was named neosis, a new mode of division[11]. It implies that some senescent polyploid giant cells escape the senescent checkpoint controls and bypass senescence; then distribute chromosomes to daughter cells via nuclear budding in the presence of an intact nuclear envelope, followed by asymmetric cytokinesis; and eventually become small, aneuploid, mitotically active cells termed Raju cells (Raju meaning King in the Telugu language), after which the polyploid neosis mother cell (NMC) dies. Raju cells display transient stem cell-like properties, and express stem-cell biomarkers. Before their death, they could undergo neosis three to four times. Moreover, neosis was thought be a primary cause of drug tolerance in tumor cells[27]. This process has been reported in some studies[23, 28, 29]. Recently, Zhang et al discovered that polyploid giant cancer cells (PGCCs) isolated from human ovarian cancer cell lines were highly resistant to oxygen deprivation and could form regular-sized cancer cells through budding or bursting, similar to the reproductive modes of simple organisms, such as fungi. These regular-sized cells expressed normal and cancer stem cell markers, such as CD133 and CD44. They divide asymmetrically, cycle slowly and can differentiate into adipose, cartilage and bone cells[30-31]. Microvesicles, membrane-covered fragments, can be shed from tumor stem cells. These microvesicles are very important for tumor cell survival, metastasis, escape from immune surveillance and other processes[32-34]. In some stem cells, including mesenchymal, embryonic and peripheral blood stem cells, microvesicles were found[35-38]. The present study found giant cells in CD133+ cells using light microscopy and SEM. Some small bodies were observed on and surrounding the giant cells. We speculate that some of those small bodies are microvesicles and that some of them are Raju cells with stem cell-like properties, including the expression of CD133. By MACS, giant cells were isolated along with CD133+ cells because their offspring, Raju cells bearing the CD133 marker, were still adhered to them. How can we prove that these small bodies were Raju cells and not apoptotic bodies? First, the appearance of apoptotic bodies under SEM was different from the appearance of these small bodies. According to the literature, normal cancer cells are covered with microvilli and ruffles, while there are some distinct morphological changes corresponding to typical apoptosis, including cell membrane blebbing and microvilli disappearance or reduction. These are thought to correspond to apoptotic bodies. In the present study, we found small bodies with short microvilli, and some of them had already been dropped from the parental bodies and become stuck to the floor using their structures. Additionally, for further verification, acridine orange staining was conducted. Under confocal microscopy, we found that in CD133+ cells, the nuclear chromosomes of those giant cells were asymmetrically distributed, although there was some evidence of apoptosis.
The present study suggests that the morphology of NPC cancer stem cells is different from that of other cancer cells. The study provides evidence for the existence of neosis and suggests that in addition to symmetric and asymmetric division, neosis is an important mode of division for sustaining rapid tumor growth. The drug resistance of tumor cells is a dilemma for tumor treatment. Exploration of GCCs and this special mode of division may open new areas of cancer treatment research.
Supplementary Material
Supplemental Figures 1 and 2, Supplemental tables 1 and 2.
Abbreviations
NPC, nasopharyngeal carcinoma; PGCC, polyploid giant cancer cell; SEM, scanning electron microscope; MACS, magnetic activated cell sorting; qRT-PCR, quantitative real-time polymerase chain reaction; IHC, immunohistochemistry; CFE, colony formation efficiency.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730-737
2. Venugopal C, Li N, Wang X, Manoranjan B, Hawkins C, Gunnarsson T, Hollenberg R, Klurfan P, Murty N, Kwiecien J. et al. Bmi1 marks intermediate precursors during differentiation of human brain tumor initiating cells. Stem Cell Res. 2012;8(2):141-153
3. Al-Hajj M, Wicha MS, Benito-Hernandez A. et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100(7):3983-3988
4. Eramo A, Lotti F, Sette G. et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15(3):504-514
5. Ricci-Vitiani L, Lombardi DG, Pilozzi E. et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445(7123):111-115
6. Rybak AP, He L, Kapoor A. et al. Characterization of sphere-propagating cells with stem-like properties from DU145 prostate cancer cells. Biochim Biophys Acta. 2011;1813(5):683-694
7. Jiang QP, Yao KT. Isolation and detection of label-retaining cells in a nasopharyngeal carcinoma cell line. Chin J Cancer. 2010;29(5):572-574
8. Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255(5052):1707-1710
9. Gibbs CP, Kukekov VG, Reith JD. et al. Stem-like cells in bone sarcomas: implications for tumorigenesis. Neoplasia. 2005;7(11):967-976
10. Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea--a paradigm shift. Cancer Res. 2006;66(4):1883-1890 discussion 1895-1886
11. Sundaram M, Guernsey DL, Rajaraman MM. et al. Neosis: a novel type of cell division in cancer. Cancer Biol Ther. 2004;3(2):207-218
12. Rajaraman R, Rajaraman MM, Rajaraman SR. et al. Neosis--a paradigm of self-renewal in cancer. Cell Biol Int. 2005;29(12):1084-1097
13. Huttner HB, Janich P, Kohrmann M. et al. The stem cell marker prominin-1/CD133 on membrane particles in human cerebrospinal fluid offers novel approaches for studying central nervous system disease. Stem Cells. 2008;26(3):698-705
14. Tirino V, Desiderio V, d'Aquino R. et al. Detection and characterization of CD133+ cancer stem cells in human solid tumours. PLoS One. 2008;3(10):e3469
15. Wang J, Guo LP, Chen LZ. et al. Identification of cancer stem cell-like side population cells in human nasopharyngeal carcinoma cell line. Cancer Res. 2007;67(8):3716-3724
16. Jiang QP, Liu SY, He XF. et al. Relationship between MAP3K5 and Epstein-Barr virus-encoded miR-BART22 expression in nasopharyngeal carcinoma. Nan Fang Yi Ke Da Xue Xue Bao. 2011;31(7):1146-1149
17. Zhang HB, Ren CP, Yang XY. et al. Identification of label-retaining cells in nasopharyngeal epithelia and nasopharyngeal carcinoma tissues. Histochem Cell Biol. 2007;127(3):347-354
18. Wu A, Luo W, Zhang Q. et al. Aldehyde dehydrogenase 1, a functional marker for identifying cancer stem cells in human nasopharyngeal carcinoma. Cancer Lett. 2012;330(2):181-189
19. Zhuang HW, Mo TT, Hou WJ. et al. Biological characteristics of CD133(+) cells in nasopharyngeal carcinoma. Oncol Rep. 2013;30(1):57-63
20. Michel M, Torok N, Godbout MJ. et al. Keratin 19 as a biochemical marker of skin stem cells in vivo and in vitro: keratin 19 expressing cells are differentially localized in function of anatomic sites, and their number varies with donor age and culture stage. J Cell Sci. 1996;109( Pt 5):1017-1028
21. Collins AT, Habib FK, Maitland NJ. et al. Identification and isolation of human prostate epithelial stem cells based on alpha(2)beta(1)-integrin expression. J Cell Sci. 2001;114(Pt 21):3865-3872
22. Campisi J. Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol. 2001;11(11):S27-31
23. Erenpreisa J, Ivanov A, Wheatley SP. et al. Endopolyploidy in irradiated p53-deficient tumour cell lines: persistence of cell division activity in giant cells expressing Aurora-B kinase. Cell Biol Int. 2008;32(9):1044-1056
24. Smith JR, Pereira-Smith OM.Replicative senescence. implications for in vivo aging and tumor suppression. Science. 1996;273(5271):63-67
25. Shelton DN, Chang E, Whittier PS. et al. Microarray analysis of replicative senescence. Curr Biol. 1999;9(17):939-945
26. Wright WE, Shay JW. The two-stage mechanism controlling cellular senescence and immortalization. Exp Gerontol. 1992;27(4):383-389
27. Navolanic PM, Akula SM, McCubrey JA. Neosis and its potential role in cancer development and chemoresistance. Cancer Biol Ther. 2004;3(2):219-220
28. Erenpreisa JA, Cragg MS, Fringes B. et al. Release of mitotic descendants by giant cells from irradiated Burkitt's lymphoma cell line. Cell Biol Int. 2000;24(9):635-648
29. Puig PE, Guilly MN, Bouchot A. et al. Tumor cells can escape DNA-damaging cisplatin through DNA endoreduplication and reversible polyploidy. Cell Biol Int. 2008;32(9):1031-1043
30. Zhang S, Mercado-Uribe I, Xing Z. et al. Generation of cancer stem-like cells through the formation of polyploid giant cancer cells. Oncogene. 2014;33(1):116-128
31. Zhang S, Mercado-Uribe I, Liu J. Tumor stroma and differentiated cancer cells can be originated directly from polyploid giant cancer cells induced by paclitaxel. Int J Cancer. 2014;134(3):508-518
32. Choi D, Lee HW, Hur KY. et al. Cancer stem cell markers CD133 and CD24 correlate with invasiveness and differentiation in colorectal adenocarcinoma. World J Gastroenterol. 2009;15(18):2258-2264
33. Goler-Baron V, Sladkevich I, Assaraf YG. Inhibition of the PI3K-Akt signaling pathway disrupts ABCG2-rich extracellular vesicles and overcomes multidrug resistance in breast cancer cells. Biochem Pharmacol. 2012;83(10):1340-1348
34. Liao CF, Lin SH, Chen HC. et al. CSE1L, a novel microvesicle membrane protein, mediates Ras-triggered microvesicle generation and metastasis of tumor cells. Mol Med. 2012;18:1269-1280
35. Nomura S, Ishii K, Inami N. et al. Alpha 4-integrin-positive microvesicles and SDF-1 in peripheral blood stem cell harvest. Bone Marrow Transplant. 2008;41(12):1071-1072
36. Wang XQ, Zhu XJ, Zou P. Research progress of mesenchymal stem cell-derived microvesicle. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2013;21(1):227-230
37. Bruno S, Grange C, Deregibus MC. et al. Mesenchymal stem cell-derived microvesicles protect against acute tubular injury. J Am Soc Nephrol. 2009;20(5):1053-1067
38. Yuan A, Farber EL, Rapoport AL. et al. Transfer of microRNAs by embryonic stem cell microvesicles. PLoS One. 2009;4(3):e4722
Author contact
![]() Corresponding authors: E-mail address: Jingsong liu: jliuorg, Kaitai Yao: ktyaocom
Corresponding authors: E-mail address: Jingsong liu: jliuorg, Kaitai Yao: ktyaocom

 Global reach, higher impact
Global reach, higher impact