Impact Factor
ISSN: 1837-9664
J Cancer 2015; 6(12):1245-1254. doi:10.7150/jca.12825 This issue Cite
Research Paper
High Expression of FGFR4 Enhances Tumor Growth and Metastasis in Nasopharyngeal Carcinoma
1. Department of Otorhinolaryngology Head and Neck surgery, Affiliated Hospital of Nantong University, Nantong, Jiangsu Province, China
2. Department of Pathology, Medical School of Nantong University, Nantong, Jiangsu Province, China
* Si Shi and Xingyu Li contributed equally to this work.
Received 2015-5-30; Accepted 2015-8-28; Published 2015-10-20
Abstract
Background: FGF receptor (FGFR) family can be activated by FGFs and play important roles in regulating cell growth, differentiation, migration and angiogenesis. Recent studies suggested that FGFR4 could regulate several processes including tumor progression. Nasopharyngeal carcinoma (NPC) is a malignancy with a high occurrence in Southeast Asia and Southern China. However, the molecule mechanism and the potential roles of FGFR4 in NPC remain unknown
Methods: Immunohistochemistry and western blot were used to investigate the expression of FGFR4 in NPC samples. Then we used statistical analysis to evaluate the diagnostic value and the associations of FGFR4 expression with clinical parameters. In vitro studies, the effects of FGFR4 on proliferation and migration of NPC cell line CNE2 were measured by the starvation-refeeding experiment, CCK8 assay, wounding healing assay and transwell migration assay. The changes of the epithelial-mesenchymal transition (EMT) markers in CNE2 cells after knocking down the expression of FGFR4 were measured by Western blot and immunofluorescence analysis.
Results: FGFR4 was overexpressed in NPC as compared with the inflammatory tissues. High expression of FGFR4 was correlated with Ki67 expression, clinical stages and prognosis in NPC patients (P<0.05).While in vitro, the upregulation of FGFR4 was accompanied with CNE2 cells released from serum starvation. Moreover, it could increase cell proliferation and migration by regulating EMT markers in CNE2 cells.
Conclusion: Our data suggested that FGFR4 might induce NPC progression and act as a potential therapeutic target in NPC.
Keywords: nasopharyngeal carcinoma, FGFR4, prognosis, growth, metastasis
Introduction
Nasopharyngeal carcinoma (NPC), an epithelial tumor located in the nasopharynx, has distinctive ethnic and geographic distributions. It is predominantly distributed in the southern part of China and Southeast Asia [1, 2]. NPC is highly radiosensitive and chemosensitive, so the survival rate has improved markedly owing to the advances in chemoradiotherapy [3, 4]. However, different from other head and neck cancers, NPC has a tendency to metastasize, thus the prognosis of NPC patients still remain poor because of distant metastasis [5, 6].Therefore, better understanding of the molecular mechanisms of NPC is of crucial significance for effective diagnosis and treatment.
Fibroblast growth factor (FGF) signaling is induced by FGFs and FGF receptors. FGFs, which are comprised of 22 structurally related polypeptides, play important roles in embryo development, wound healing, haematopoiesis and angiogenesis [7, 8]. Most FGFs bind to and activate cell surface FGF receptor (FGFR) family. FGFRs are composed of five receptor tyrosine kinases including FGFR1-4 and a non-tyrosine kinase receptor. They consist of a cellular ligand domain, a single transmembrane helix domain and a cytoplasmic domain with tyrosine kinase activity [9, 10]. The receptor-ligand binding induces FGFR dimerization and phosphorylation, activates various down stream signal transduction cascades and plays an important part in regulating cellular functions including cell growth, differentiation, migration and angiogenesis [11-13]. As tumor progression is strongly linked to above points, dysregulation of this signaling axis might play significant roles in tumor progression.
Previous studies have reported that the alterations of FGFRs might be associated with the progression and poor prognosis in a variety of tumors including cholangiocarcinoma, ovarian cancer, gastric cancer and colorectal cancer [9, 14, 15]. Besides, depending on the tumor types, different kinds of FGFRs are highly expressed [16]. FGFR4, which is known as CD334, has been reported to interfere the signaling events and its expression level may be associated with some human tumors, such as breast cancer, colorectal cancer, human rhabdomyosarcoma, lung adenocarcinoma and melanoma [18-22]. However, the involvement of FGFR4 in the pathogenesis and progression of NPC remains to be detected.
To probe the role of FGFR4, we first took western blot analysis and immunohistochemistry and found it was remarkably highly expressed in NPC tissues and cell lines. Besides, its expression level was associated with clinical and pathologic factors of NPC patients. To further confirm the results obtained from the clinical study, we carried out the research in vitro. By anti-FGFR4 small interfering RNA (siRNA), we down-regulated FGFR4 and found that FGFR4 could promote proliferation and migration of NPC cells. Moreover, we also observed that it had impacts on NPC cells during EMT. All the data suggested that FGFR4 might induce NPC progression and act as a potential therapeutic target in NPC.
Materials and Methods
Patients and Tissue Samples
55 NPC specimens were retrieved from the Affiliated Hospital of Nantong University and chosen at random during 2009 and 2013. None of the patients enrolled had undergone the anti-tumor therapies before. The non-tumor portion was chronic inflammatory nasopharyngeal epithelium tissue, which was obtained from people who were suspected to have NPC but excluded by the pathological diagnosis. The clinical processes gained approval from the Ethics Committee of Affiliated Hospital of Nantong University and had consent from each patient. We analyzed both the tumor and non-tumor samples to determine the expression of FGFR4 in NPC. The diagnoses of the samples were made according to the 2005 WHO histological classification and the pathologic stage was subdivided by the UICC/AJCC 1997 staging system of NPC.
Immunohistochemistry
The expression of FGFR4 in NPC was analyzed by immunohistochemistry. The specimens were firstly fixed with formalin, embedded in paraffin, and then sectioned at 4-mm thickness and transferred to microscope slides. The sections were dewaxed in xylene, followed by rehydration in graded ethanols. Then they were incubated in pH 6.0 citrate buffer and heated to 121℃ in an autoclave to retrieve the antigen. Endogenous peroxidase was inactivated after being immersed into 0.3 % hydrogen peroxide for 30 min. To eliminate any nonspecific binding, the specimens were incubated in 10 % goat serum for 1h at room temperature. The sections were subsequently incubated with primary antibody: anti-FGFR4 antibody (diluted 1:100, Santa Cruz Biotechnology) and anti-Ki-67 antibody (diluted 1:100, Santa Cruz Biotechnology) overnight. The immunoreactive staining was processed using the peroxidase-anti-peroxidase method according to the instructions (DAKO, Hamburg, Germany). 3, 39-diaminobenzidine tetrachloride (DAB) chromogen solution was used to detect the reaction. After rinsing in water for 30 minutes, the sections were counterstained with hematoxylin, and dehydrated. At last, they were mounted in mounting medium for interpretation. The specificity of the antibody was determined by using a non-specific immunoglobulin IgG (diluted 1:100, Santa Cruz Biotechnology) at the same concentration. Two pathologists were blinded to score the immunoreactivity according to the intensity of the staining and the relative abundance of positive cells. They chose five high-power (200×magnification) fields randomly, and 500 cells were counted per field. The intensity of FGFR4 staining was scored as 0 (no staining), 1 (weak), 2 (medium) and 3 (strong). Positive cells were graded as 0 (<10% positive cells), 1 (10-25%), 2 (26-75%), 3 (76-100%). FGFR4 expression level was defined as the sum of the staining-intensity and staining-abundance: "-" (negative, score of 0), "+" (weakly positive, score of 1-2), "++" (positive, score of 3-4), "+++" (strongly positive, score of 5-6). We defined "-" and "+" as low expression, "++" and "+++" as high expression. Ki-67 staining was defined as low expression group (<50%) and high expression group (≥50 %) according to the cells that had positive stained nuclei in five high-power fields.
Cell Culture and Reagents
Nasopharyngeal carcinoma cell lines CNE1, CNE2, 5-8F, 6-10B were cultured in RPMI medium 1640 (GibCo BRL, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS) (Gibco). The immortalized normal nasopharyngeal epithelial cell line NP69 was grown in Keratinocyte-SFM medium supplemented with epidermal growth factor (EGF) (Invitrogen, Carlsbad, USA). They were kindly provided by Sun Yat-sen University Cancer Center and Xiangya Hospital of Central South University. All cell lines were incubated in an atmosphere of 37°C in 5% CO2. To investigate the percentages of the cells in cell cycle, cells were harvested and rinsed with cold PBS twice, and then fixed them in 70% ethanol at 4 °C overnight. The cells were incubated with 1 mg/mL RNase A for 30 min and stained with propidiumiodide (50 μg/mL PI) (Becton Dickinson, San Jose, CA) in PBS with 0.5% Tween-20. Becton Dickinson flow cytometer BD FACSCAN (San Jose, CA), Cell Quest acquisition and analysis programs were used after that.
RNA isolation and quantitative real-time PCR
Total RNA was extracted from cells using the Trizol reagent (Sigma) according to the manufacturer's instructions. cDNA samples were produced using a Transcriptor First Strand cDNA Synthesis Kit (Roche, Germany). Gene expression was analyzed using Taqman Universal PCR Master Mix. The primers used for quantitative real-time PCR were as follows: FGFR4 forward: ACTGGAGTCTCGTGATGG, reverse: GCAGGTAGTTATAGCGGATG; GAPDH was employed as an internal control, and the primers were forward: 5'-GAAGGTGAAGGTCGGAGTC-3'; reverse: 5'-GAAGATGGTGATGGGATTTC-3', they were obtained from Biomics Biotechnologies Co, Ltd (Nantong, China).The amplification conditions consisted of 30 min at 42℃ for reversetranscription, 2 min at 94℃ for pre-denature followed by 35 cycles of 94℃ for 20 s, 58℃ for 20 s and at 72℃ for 30 s. All the reactions were performed three times. We used ΔΔCt method to calculate the Ct-value and the results were expressed as 2-ΔΔCt.
Transient Transfection with siRNAs
We obtained FGFR4 small-interfering RNA (siRNA) and silencer negative control siRNA (snc-RNA) from Biomics Biotechnologies Co. Ltd (Nantong, China). CNE2 cells were plated overnight and grew to 30-50% confluence. According to the manufacturer's instructions, the siRNAs were transfected into CNE2 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). The efficiency of siRNA was determined by the protein levels of FGFR4 in the 72-h post transfected cells.
Western Blot
Cells were lysed by RIPA Lysis Buffer containing protease inhibitor PMSF on ice. Protein concentration was measured with the help of the BCA Protein Assay Kit. Equal amount of total cellular protein (20 μg) was separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto poly vinyllidine difluoride filter (PVDF) membranes (Millipore, Bed ford, MA). Membrane was blocked with 5 % nonfat milk in Tris-buffered saline with Tween 20 (TBST) buffer for 1.5 h at room temperature and incubated with primary antibodies overnight at 4°C, then it was incubated with HRP-tagged secondary antibodies (1:1000, Santa Cruz Biotechnology, USA) at room temperature for 1.5 h. Immunoreactivity was detected by ECL reagent (Millipore) under the manufacturer's instructions. Quantitative data were obtained using Image J software, and β-actin was used as a loading control. The antibodies used were as follows: anti-FGFR4 primary antibody (1:500 Santa, USA), anti-E-cadherin (1:50000 Abcam, USA), anti-N-cadherin (1:10000 Abcam, USA), anti-Vimentin (1:5000 Abcam, USA). All the experiments were carried out three times.
Immunofluorescence microscopy
Cells were harvested and seeded onto 0.8 cm×0.8 cm coverslips overnight in a 24-well plate. Then the cells were fixed with 4% paraformaldehyde for 40 minutes. After three rinses in PBS, the cells were blocked with 1% normal donkey serum for 90 minutes at room temperature and then incubated with primary antibody anti-FGFR4, anti-E-cadherin, anti-N-cadherin, anti-Vimentin overnight. After being washed with PBS for three times, the cells were incubated with Alex Fluor-conjugated secondary antibodies (Invitrogen Life Technologies, 1/1000) and the nuclei were stained with Hochest. The cells were observed under a fluorescence microscope.
Cell Proliferation Assays
To determine the effect of FGFR4 on cell proliferation, CNE2 cells transfected with FGFR4-siRNA or control siRNA were seeded onto 96-well plate (Corning inc, Corning NY) at a density of 1×104 cells per well. Then at time points of 0h, 6h, 12h, 24h, 36h, 48h, the cell viability rate was assessed using cell counting kit-8 (CCK-8 Kit, beyotime institute of biotechnology). 10ul CCK-8 solution was added to each well and incubated for another 1.5 hours and a microplate reader was used to measure the absorbance of each well at 450 nm. All the experiments were independently repeated three times.
Wound Healing Assay
Wound healing assay was also used to analyze cell motility, 5×105 cells transfected with either FGFR4-siRNA or control-siRNA were seeded on 6-well plates with 10 % FBS. After the cells were grown to 80% confluence, wounds were created by scraping the cells with a 100-μl pipette tip. The 6-well plates were incubated at 37 °C and the microscope was used to observe the migrated distance of cells every 12 hours. The relative migrating distance was measured by the wound width/the distance measured at 0 h.
Transwell migration assay
Migration assays were performed by a Millipore chamber with a polycarbonate filter of 8 mm pore size (Millipore). CNE2 cells transfected with either FGFR4-siRNA or control-si RNA were harvested and resuspended in 200 ul serum-free 1640 at a density of 1×105 and added to the upper chamber while the bottom chambers were filled with 500ul complete 1640 media. After 16 h, the cells inside the upper chamber were carefully removed with a cotton swab and the reverse side of the upper chamber was washed with PBS. The migratory cells on the lower membrane surface were fixed with methanol and stained with 0.05% crystal violet. The cells were counted for 10 random 100× fields per well using an inverted microscope. All the experiments were repeated for three times.
Statistical analysis
All the data were presented after three independent experiments and reported as the means ± standard deviation (SD). Statistical analysis was performed using SPSS17.0 software (SPSS, Inc., an IBM Company, Chicago, IL). χ2 test was used to analyze the relationship between FGFR4 expression and clinicopathological features of NPC. Kaplan-Meier analysis was performed to analyze the survival data and the statistical significance was assessed using the log-rank test. Multivariate analysis was performed using Cox's proportional hazards model. Differences between groups were analyzed using Student's t-test and the P value less than 0.05 was considered to be statistically significant.
Results
The expression of FGFR4 in NPC
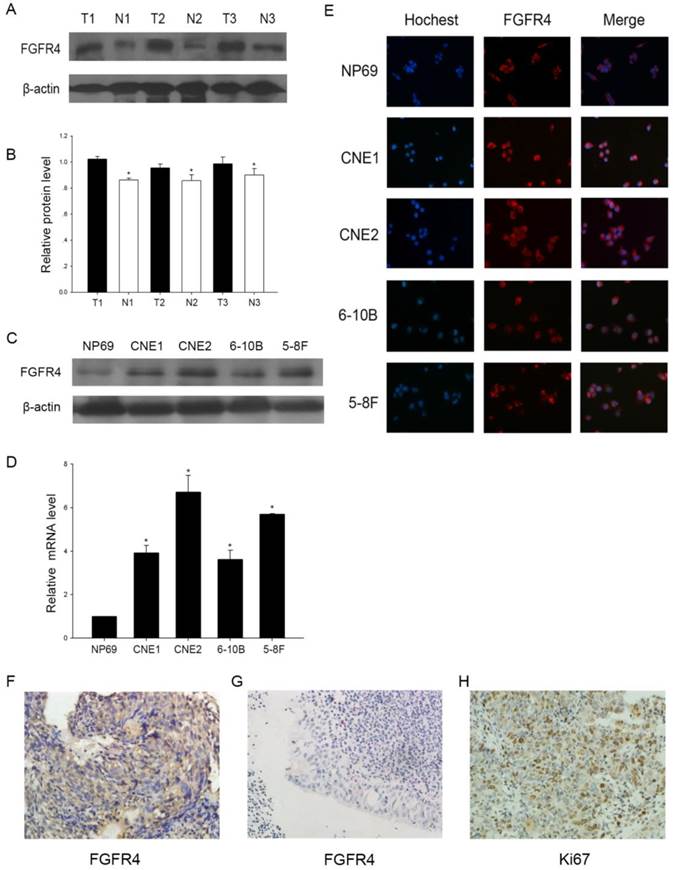
To determine the significance of FGFR4, we first detected its expression at protein levels in NPC tissues and cell lines. Western blot demonstrated that the expression of FGFR4 was totally higher in three NPC tissues and cell lines than inflammatory nasopharyngeal epithelium tissues and the normal cell line (Figure 1A, B, C). Moreover, quantitative real-time PCR analysis also showed that the upregulation occurred at the transcriptional level (Figure 1D). Then immunofluorescence analysis demonstrated FGFR4 was mainly membrane stained and highly expressed in NPC cells (Figure 1E). These results revealed that FGFR4 was elevated in NPC tissues and cell lines.
Our results were further confirmed by the immunohistochemical analysis in 55 NPC samples obtained from the patients. Most NPC samples exhibited abnormally positive FGFR4 immunoreactivity, while inflammatory nasopharyngeal epithelium tissues had low FGFR4 expression (Figure 1F, G). At the same time, the immunoactivity of Ki67, a proliferating marker, was up-regulated in NPC tumor tissues (Figure 1H). So we hypothesized that FGFR4 might play an important role in NPC.
Correlation of FGFR4 expression with clinicopathologic variables in NPC
Then we determined the role of FGFR4 in the progression of NPC. Pearson χ2 test was used to analyze the correlation of FGFR4 expression with clinicopathologic variables in NPC. As shown in Table 1, there were positive correlations of the expression level of FGFR4 with Ki-67 and clinical stages (Table 1; P<0.05). No significant association of FGFR4 with age, gender and lymph node metastasis was observed.
The association between the expression of FGFR4 and clinicopathological parameters of NPC
| Clinicopathological parameters | Total | FGFR4 Expression | P | |
|---|---|---|---|---|
| Low | High | |||
| Gender | 55 | 0.370 | ||
| Male | 43 | 19 | 24 | |
| Female | 12 | 4 | 8 | |
| Age | 0.151 | |||
| <50 | 14 | 8 | 6 | |
| ≥50 | 41 | 15 | 26 | |
| Clinical stages | 0.038* | |||
| 1 | 2 | 2 | 0 | |
| 2 | 8 | 6 | 2 | |
| 3 | 29 | 11 | 18 | |
| 4 | 16 | 4 | 12 | |
| Lymph node metastasis | 0.125 | |||
| N0-N1 | 18 | 10 | 8 | |
| N2-N3 | 37 | 13 | 24 | |
| Ki-67 expression | 0.047* | |||
| low | 25 | 14 | 11 | |
| high | 30 | 9 | 21 | |
Statistical analyses were performed by the Pearson χ2 test. * P <0.05 was considered significant.
The expression of FGFR4 in NPC. A: Western blot of FGFR4 in 3 NPC tissues and 3 inflammatory nasopharyngeal tissues. (T) Nasopharyngeal squamous cell carcinoma tissues. (N) Inflammatory nasopharyngeal epithelium tissues.β-actin was used as a control for protein load and integrity. B: The bar demonstrated the ratio of FGFR4 protein expression to β-actin by densitometry. C: Expression level of FGFR4 in NPC cell lines or normal nasopharyngeal epithelial cells by Western blot analysis. D: qRT-PCR was used to detect the relative expression of FGFR4 in cell lines. E: Immunofluorescence analysis of FGFR4 expression in NPC cell lines and the normal cell line. DAPI was used for counterstaining of the nucleus in blue. FGFR4 staining was in red. F: High expressions of FGFR4 were observed in NPC tissues (×400). G: Low expressions of FGFR4 were observed in inflammatory nasopharyngeal epithelium tissues (×400). H: High expressions of Ki67 were found in NPC tissues (×400). The data shown were representative of at least three independent experiments. * P < 0.05.
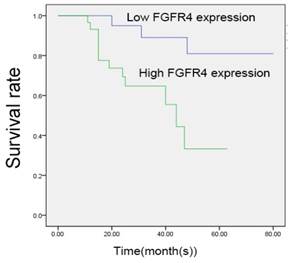
Higher expression of FGFR4 predicted poor prognosis of NPC patients
We carried out Kaplan-Meier analysis to study the association of FGFR4 expression with patients' survival time (Figure 2). The analysis was restricted to 55 patients with follow-up data and the immunohistochemistry results. The survival curves indicated that patients with high expression levels of FGFR4 achieved a lower overall survival rate. Moreover, the results of multivariate analysis using the Cox's proportional hazards model were shown in Table 2. We found that patients' overall survival rate was correlated with FGFR4 expression, clinical stages and Ki67 expression (Table 2; P<0.05).
High expression of FGFR4 predicted poor prognosis of NPC patients. Kaplan-Meier analysis of NPC patients were based on the expression of FGFR4. NPC patients with higher FGFR4 expression were significantly related to poor prognosis. * P < 0.05.
Contribution of various potential prognostic factors to survival by Cox regression analysis on 55 NPC samples
| Hazard ratio | P | 95.0% Confidence interval | |
|---|---|---|---|
| Age | 0.051 | 0.821 | 0.363-3.587 |
| Gender | 0.896 | 0.344 | 0.178-1.826 |
| Lymph node metastasis | 0.892 | 0.345 | 0.447-10.010 |
| Clinical stages | 6.522 | 0.011* | 1.292-7.020 |
| FGFR4 expression | 5.169 | 0.023* | 1.234-17.022 |
| Ki-67 | 4.473 | 0.034* | 1.105-12.935 |
Statistical analyses were performed by the Cox regression analysis. * P <0.05 was considered significant.
FGFR4 was high expressed in proliferating CNE2 cells
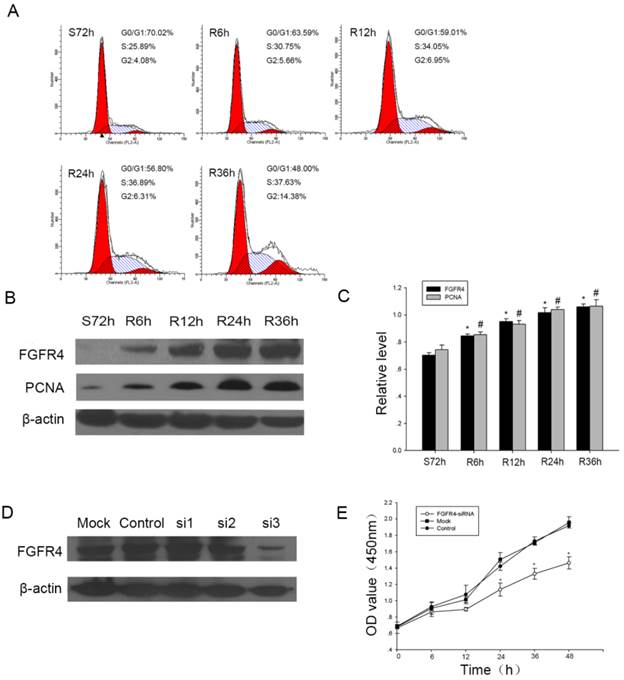
Then we carried out in vitro studies to investigate the role of FGFR4 in proliferating cells. As CNE2 was nasopharyngeal carcinoma non-keratinizing cell line, which was consistent with the main pathology subtype of NPC, we chose it for the further studies. First of all, serum starvation and refeeding assay was used during cell cycle progression in NPC cells. After growing without serum for 72 h, CNE2 cells arrested in G1 phase, then serum was added, the cells were released from the G1 phase (from 70.02% at S72h to 48% at R36h) (Figure 3A). We also found during this progression, the expression level of FGFR4 increased. PCNA, which had been used as a general marker of dividing cells, was upregulated along with FGFR4 (Figure 3B, C). So these results confirmed FGFR4 might regulate cell proliferation during cell cycle progression.
FGFR4 Knockdown reduced proliferation in NPC
To assess the role of FGFR4 in cell proliferation, we transfected CNE2 cells with three siRNAs targeting FGFR4. As illustrated in Figure 3D, FGFR4-si3 showed the highest knockdown efficiency, thus it was used for the follow-up studies. CCK8 assay showed that after downregulating the expression of FGFR4, the proliferation rate of CNE2 cells decreased significantly (Figure 3E). This suggested that FGFR4 might accelerate proliferation of CNE2 cells.
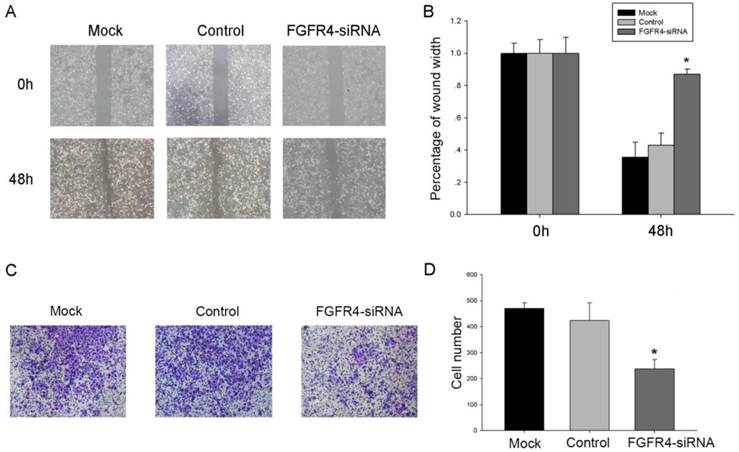
Interference of FGFR4 expression inhibited the migration of CNE2 cells
As the prognosis of NPC patients that have metastasis may be poor, we investigated the effect of FGFR4 on cell migration with wound-healing assay and transwell migration assays. After being incubated with physical-wound and cultured in serum-free medium to exclude the interference of proliferation, FGFR4-silenced cells were unable to close the wound within 48h, while the non-interfered cells had longer migrating distance (Figure 4A, B). Transwell migration assays also showed the similar results (Figure 4C, D). All the data supported that FGFR4 might stimulate cell migration.
FGFR4 knockdown inhibited the inducement of EMT in CNE2 cells
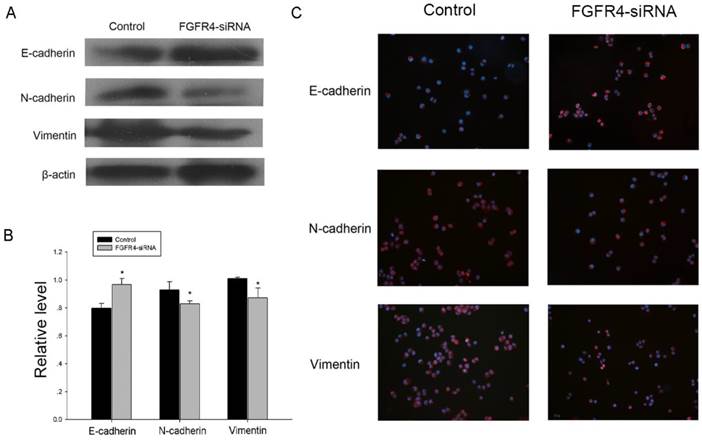
Since epithelial-mesenchymal transition (EMT) is involved in migration and invasive capacity of epithelial cells [23], we examined whether the impacts of FGFR4 on NPC cells was associated with EMT. Knocking down the expression of FGFR4 caused a significant increase of epithelial marker E-cadherin and a decrease of mesenchymal marker vimentin and N-cadherin in CNE2 cells (Figure 5A, B). Our result was also confirmed by immunofluorescence assay, and the differences of EMT markers expression were evident in the cell membrane (Figure 5C). Collectively, FGFR4 silencing could partly reverse the cells to an epithelial phenotype.
Discussion
Now, it is becoming more and more apparent that gene changes may be associated with tumor heterogeneity. Among the changes, the activation of oncogenes and the inactivation of tumor suppressor genes contribute to tumor formation. As one of the most common cancers in head and neck, the molecule mechanisms of NPC remain unknown. It is urgent for us to identify novel molecules which have important functions during NPC progression and that may help us find better prognostic markers or therapeutic targets. In the present study, we found that FGFR4 was upregulated in NPC clinical samples and cell lines. Besides, it might be associated with clinical stages and prognosis of NPC patients. In vitro studies, FGFR4 could accelerate CNE2 cells growth and migration. Taken together, these results suggested that FGFR4 might be a potential regulator in NPC progression.
Studies have reported that FGFs can bind to and activate FGFRs to induce a variety of cellular processing. The mutation and abnormal expression of FGFRs cause diverse pathologies and affect multiple solid tumors, including breast cancer, lung cancer and so on[15, 24-26]. FGFR4, belonging to the FGFR family, differs from the other FGFRs in genomic structure, ligand binding, and signal transduction [27]. Although FGFR4 was associated with some tumors, the functions and mechanisms of FGFR4 in NPC have not been fully elucidated.
A single nucleotide polymorphism (SNP) within FGFR4 transmembrane domain may be correlated with the prognosis of several human malignancies [18-22]. In these tumors, Arg388 is the most common SNP in FGFR4, which prolongs activation of the receptor. In head and neck cancer (HNSC), the FGFR4 Arg (388) allele presents an increased mortality and shorter survival, besides, it indicates a tendency to increased cisplatin sensitivity of HNSC [21, 28].
The expression of FGFR4 in proliferating CNE2 cells. A: Flow cytometry was used to quantify the cell cycle progress.Cells were subjected to serum starvation for 72h (S72h), then addition of medium containing 10% FBS for the indicated times (R6h, R12h, R24h, R36h). B: CNE2 cells were subjected to serum starvation and releasing and Western blot was used to investigate the expression of FGFR4 and PCNA in CNE2 cells during the progression.β-actin was used as a control for protein load and integrity. C: The bar demonstrated the ratio of FGFR4 and PCNA protein expression to β-actin by densitometry. The data shown were representative of at least three independent experiments. Data were analyzed by Student's t-test. *,# P < 0.05. D: Western blot showed the expression of FGFR4 after CNE2 cells transiently transfected with siRNA targeting FGFR4 (si-1, si-2, si-3) or a scrambled sequence (control siRNA). E: Proliferation was determined by CCK8 assay after CNE2 cells treated with FGFR4-siRNA or control siRNA for the indicated time. The data were means± SEM. * P <0.05.
FGFR4-silencing inhibited the migration of CNE2 cells. A: Silenced and control cells were grown until confluence and their migratory capabilities were analyzed by wound-healing assay. Representative images of the wound-healing assay are shown at 0 and 48 h with a microscope (× 200 magnifications). Migration speed of the cells was calculated by the wound width/the distance measured at 0 h. B: The histogram showed the relative migration distance of cells. C: Transwell assay was used to show the penetration of FGFR4-silencing cells through the membrane compared with the control. D: Absolute number of cells that migrated through the member. The data were means± SEM. * P <0.05.
FGFR4 knockdown inhibited the inducement of EMT in CNE2 cells. A: FGFR4-silencing and the control cells were lysed and subjected to Western blot analysis using specific antibodies against the EMT markers. B: The bar demonstrated the expression ratio of the target protein to β-actin by densitometry. The data shown were representative of at least three independent experiments. Data were analyzed by Student's t-test.*P < 0.05. C: Immunofluorescence analysis of EMT markers in CNE2 cells transfected with either FGFR4-siRNA or control siRNA. DAPI was used for counterstaining the nucleus in blue. E-Cadherin, N-cadherin and Vimentin staining was in red.
In the current study, we detected that FGFR4 expression was higher in NPC tissues and cell lines (Figure 1A-E). Then we used IHC to study the expression of FGFR4 in 55 NPC samples and found the upregulation of FGFR4 in NPC compared with inflammatory nasopharyngeal epithelium tissues. The correlation analysis also demonstrated that higher FGFR4 expression level was closely associated with Ki 67, which was often used as a proliferation marker. Besides, FGFR4 was correlated with clinical stages and patients' overall survival time of NPC (Figure 2, Table 1, 2). In our experiments, immunohistochemistry could not distinguish Gly388 and Arg388 alleles, and we would take further experiments to investigate FGFR4 gene polymorphism function in NPC. Our data were consistent with the previous studies, which had confirmed the overexpression of FGFR4 in ovarian cancer, mouth and oropharynx carcinoma and cholangiocarcinoma [9, 14, 29]. All these reminded us that FGFR4 might contribute to the progression of NPC.
To further explore the functions of FGFR4, we took studies in cell lines. We first detected the expression of FGFR4 during cell cycle and found it was increased with cell-cycle progression (Figure 3A-C). Then we used siRNA to down-regulate FGFR4 expression and the CCK8 assay showed that cells treated with siRNA exhibited a decrease of proliferation rate (Figure 3E). As FGFR4 participated in regulating cell cycle, this might partly explain the mechanism how FGFR4 accelerated cell proliferation.
NPC has a tendency to metastasize to regional lymph nodes and distant area, and metastasis is an important character which influence NPC patients' prognosis. We then used wound-healing assays and transwell migration assays to study the role of FGFR4 in NPC migration (Figure 4). The loss-of-function experiments showed that slicing FGFR4 expression could inhibit cells migrating ability.
Epithelial-mesenchymal transition (EMT) is a process that changes cells phenotype to the mesenchymal state and makes cells gain migratory and invasive properties [30]. This process is closely related with malignant phenotype of tumor. As FGFRs play important roles in carcinogenesis, previous studies had proved the correlation of FGFR with EMT. Among the family, FGFR4 was also reported to be associated with EMT to regulate cell migration. Alberto et al. found that suppression of FGFR4 expression could make a reversion of the mesenchymal to a epithelial phenotype and it could reduce the tumorigenic properties in colorectal cancer [31]. Besides, Liu et al. also found after co-cultured with tumor-associated fibroblasts (TAFs), FGFR4 was upregulated in colorectal cancer cells and played a crucial role in TAF-induced EMT. This study provided new concepts of FGFR4 in tumor-stroma interactions during tumor metastasis [32]. To explore the mechanisms of FGFR4's regulation upon NPC migration, we also explored the role of FGFR4 in regulating EMT in CNE2 cells. We knocked down the expression of FGFR4 and found the up-regulation of epithelial marker E-cadherin and the down-regulation of mesenchymal marker vimentin and N-cadherin (Figure 5). These results revealed that FGFR4 might involve in NPC migration.
In summary, we have indicated that FGFR4 had a potent oncogenic activity in NPC. It could provide prognostic information for NPC patients and was associated with growth and metastasis of NPC cells. The results represented that FGFR4 might become an important marker in NPC.
Abbreviations
NPC: nasopharyngeal carcinoma; EMT: epithelial-mesenchymal transition; FGFR: fibroblast growth factor receptor
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 81172841 and 81471603), the China Postdoctoral Science Foundation (No. 2013M541708), the Natural Science Foundation of Jiangsu Province (SBK2015022581), the project of "333 Natural Science Foundation" of Jiangsu Grant (No. BRA2013286), the "Top Six Types of Talents" Financial Assistance of Jiangsu Province Grant(No. 6), the project of Jiangsu Provincial Health Department (No. Z201005), the Outstanding Medical Academic Leader Program of Jiangsu Province (No. LJ201136), the scientific and innovative research project of Nantong (No. BK2014003, HS2014007 201HHS) and the innovative research project for Nantong University postgraduate students (No. YKS14009).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90
2. Wei WI, Sham JS. Nasopharyngeal carcinoma. Lancet. 2005;365:2041-54
3. Perri F, Dell'Oca I, Muto P, Schiavone C, Aversa C, Fulciniti F. et al. Optimal management of a patient with recurrent nasopharyngeal carcinoma. World J Clin Cases. 2014;2:297-300
4. Lai SZ, Li WF, Chen L, Luo W, Chen YY, Liu LZ. et al. How does intensity-modulated radiotherapy versus conventional two-dimensional radiotherapy influence the treatment results in nasopharyngeal carcinoma patients? Int J Radiat Oncol Biol Phys. 2011;80:661-8
5. Razak AR, Siu LL, Liu FF, Ito E, O'Sullivan B, Chan K. Nasopharyngeal carcinoma: the next challenges. Eur J Cancer. 2010;46:1967-78
6. Lo KW, To KF, Huang DP. Focus on nasopharyngeal carcinoma. Cancer Cell. 2004;5:423-8
7. Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2:REVIEWS3005
8. Cronauer MV, Schulz WA, Seifert HH, Ackermann R, Burchardt M. Fibroblast growth factors and their receptors in urological cancers: basic research and clinical implications. Eur Urol. 2003;43:309-19
9. Xu YF, Yang XQ, Lu XF, Guo S, Liu Y, Iqbal M. et al. Fibroblast growth factor receptor 4 promotes progression and correlates to poor prognosis in cholangiocarcinoma. Biochem Biophys Res Commun. 2014;446:54-60
10. Friesel R, Maciag T. Fibroblast growth factor prototype release and fibroblast growth factor receptor signaling. Thromb Haemost. 1999;82:748-54
11. Powers CJ, McLeskey SW, Wellstein A. Fibroblast growth factors, their receptors and signaling. Endocr Relat Cancer. 2000;7:165-97
12. Lin BC, Desnoyers LR. FGF19 and cancer. Adv Exp Med Biol. 2012;728:183-94
13. Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116-29
14. Zaid TM, Yeung TL, Thompson MS, Leung CS, Harding T, Co NN. et al. Identification of FGFR4 as a potential therapeutic target for advanced-stage, high-grade serous ovarian cancer. Clin Cancer Res. 2013;19:809-20
15. Jang JH, Shin KH, Park JG. Mutations in fibroblast growth factor receptor 2 and fibroblast growth factor receptor 3 genes associated with human gastric and colorectal cancers. Cancer Res. 2001;61:3541-3
16. Haugsten EM, Wiedlocha A, Olsnes S, Wesche J. Roles of fibroblast growth factor receptors in carcinogenesis. Mol Cancer Res. 2010;8:1439-52
17. Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139-49
18. Spinola M, Leoni VP, Tanuma J, Pettinicchio A, Frattini M, Signoroni S. et al. FGFR4 Gly388Arg polymorphism and prognosis of breast and colorectal cancer. Oncol Rep. 2005;14:415-9
19. Taylor JGt, Cheuk AT, Tsang PS, Chung JY, Song YK, Desai K. et al. Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J Clin Invest. 2009;119:3395-407
20. Spinola M, Leoni V, Pignatiello C, Conti B, Ravagnani F, Pastorino U. et al. Functional FGFR4 Gly388Arg polymorphism predicts prognosis in lung adenocarcinoma patients. J Clin Oncol. 2005;23:7307-11
21. da Costa Andrade VC, Parise O Jr, Hors CP, de Melo Martins PC, Silva AP, Garicochea B. The fibroblast growth factor receptor 4 (FGFR4) Arg388 allele correlates with survival in head and neck squamous cell carcinoma. Exp Mol Pathol. 2007;82:53-7
22. Streit S, Mestel DS, Schmidt M, Ullrich A, Berking C. FGFR4 Arg388 allele correlates with tumour thickness and FGFR4 protein expression with survival of melanoma patients. Br J Cancer. 2006;94:1879-86
23. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY. et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704-15
24. Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL. et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529-41
25. Behrens C, Lin HY, Lee JJ, Raso MG, Hong WK, Wistuba II. et al. Immunohistochemical expression of basic fibroblast growth factor and fibroblast growth factor receptors 1 and 2 in the pathogenesis of lung cancer. Clin Cancer Res. 2008;14:6014-22
26. Stephens P, Edkins S, Davies H, Greenman C, Cox C, Hunter C. et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590-2
27. Vainikka S, Partanen J, Bellosta P, Coulier F, Birnbaum D, Basilico C. et al. Fibroblast growth factor receptor-4 shows novel features in genomic structure, ligand binding and signal transduction. EMBO J. 1992;11:4273-80
28. Ansell A, Farnebo L, Grenman R, Roberg K, Thunell LK. Polymorphism of FGFR4 in cancer development and sensitivity to cisplatin and radiation in head and neck cancer. Oral Oncol. 2009;45:23-9
29. Dutra RL, de Carvalho MB, Dos Santos M, Mercante AM, Gazito D, de Cicco R. et al. FGFR4 profile as a prognostic marker in squamous cell carcinoma of the mouth and oropharynx. PLoS One. 2012;7:e50747
30. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871-90
31. Pelaez-Garcia A, Barderas R, Torres S, Hernandez-Varas P, Teixido J, Bonilla F. et al. FGFR4 role in epithelial-mesenchymal transition and its therapeutic value in colorectal cancer. PLoS One. 2013;8:e63695
32. Liu R, Li J, Xie K, Zhang T, Lei Y, Chen Y. et al. FGFR4 promotes stroma-induced epithelial-to-mesenchymal transition in colorectal cancer. Cancer Res. 2013;73:5926-35
Author contact
![]() Corresponding authors: Prof. Yiwen You, Department of Otorhinolaryngology Head and Neck surgery, Affiliated Hospital of Nantong University, Nantong, Jiangsu Province, China. Tel/fax: +86-0513-81168512, E-mail addresses: youyiwen_nantongcom. Prof. Xiaolei Cao, Department of Pathology, Medical School of Nantong University, Nantong, Jiangsu Province, China, E-mail addresses: xiaoleiedu.cn.
Corresponding authors: Prof. Yiwen You, Department of Otorhinolaryngology Head and Neck surgery, Affiliated Hospital of Nantong University, Nantong, Jiangsu Province, China. Tel/fax: +86-0513-81168512, E-mail addresses: youyiwen_nantongcom. Prof. Xiaolei Cao, Department of Pathology, Medical School of Nantong University, Nantong, Jiangsu Province, China, E-mail addresses: xiaoleiedu.cn.

 Global reach, higher impact
Global reach, higher impact